Abstract
Purpose
Anticancer drug development is inefficient, but genetically engineered murine models (GEMM) and orthotopic, syngeneic transplants (OST) of cancer may offer advantages to in vitro and xenograft systems.
Experimental Design
We assessed the activity of 16 treatment regimens in a RAS-driven, Ink4a/Arf-deficient melanoma GEMM. In addition, we tested a subset of treatment regimens in three breast cancer models representing distinct breast cancer subtypes: claudin-low (T11 OST), basal-like (C3-TAg GEMM), and luminal B (MMTV-Neu GEMM).
Results
Like human RAS-mutant melanoma, the melanoma GEMM was refractory to chemotherapy and single-agent small molecule therapies. Combined treatment with AZD6244 [mitogen-activated protein–extracellular signal-regulated kinase kinase (MEK) inhibitor] and BEZ235 [dual phosphoinositide-3 kinase (PI3K)/mammalian target of rapamycin (mTOR) inhibitor] was the only treatment regimen to exhibit significant antitumor activity, showed by marked tumor regression and improved survival. Given the surprising activity of the "AZD/BEZ" combination in the melanoma GEMM, we next tested this regimen in the "claudin-low" breast cancer model that shares gene expression features with melanoma. The AZD/BEZ regimen also exhibited significant activity in this model, leading us to testing in even more diverse GEMMs of basal-like and luminal breast cancer. The AZD/BEZ combination was highly active in these distinct breast cancer models, showing equal or greater efficacy compared with any other regimen tested in studies of over 700 tumor-bearing mice. This regimen even exhibited activity in lapatinib-resistant HER2+ tumors.
Conclusion
These results show the use of credentialed murine models for large-scale efficacy testing of diverse anticancer regimens and predict that combinations of PI3K/mTOR and MEK inhibitors will show antitumor activity in a wide range of human malignancies.
Introduction
The standard anticancer drug development pipeline largely relies on in vitro and xenograft assays to determine efficacy of candidate antitumor agents. This system is suboptimal as evidenced by the very high attrition rates of would-be cancer therapeutics, even in the era of rationally targeted therapies (1–4).In particular, failure at the phase II and phase III stages of human testing is common, resulting from a lack of antitumor efficacy in humans. Current drug development practices expose patients to ineffective and toxic agents, distract clinical trialists from the development of effective therapies, and force the pharmaceutical industry to subsidize the inordinate costs of late-stage failures. Thus, the preclinical assessment of efficacy is perhaps the major present challenge for the development of novel anticancer therapeutics.
Genetically engineered mouse models (GEMMs) may pose some advantages over traditional systems for this purpose (2, 5–7). In particular, a few groups have showed specific examples where GEMMs have been able to recapitulate clinical trial results of select agents or have predicted clinical outcomes before human testing has been completed.In one of the earliest comparisons, GEMMs predicted the lack of efficacy of PPAR-γ inhibitors in colon cancer (8, 9) whereas xenograft models predicted the opposite result (10). In addition, although xenograft models do not predict the influence of K-RAS mutations on response to EGFR-directed therapies and chemotherapy (11), recent analysis assessing the therapeutic response in K-Ras mutant GEMMs has found these models faithfully recapitulate the known clinical outcomes seen in patients (12). Despite these promising series, there has not been a comprehensive assessment of GEM models versus traditional preclinical efficacy testing. The GEMM approach until recently has been hampered by a variety of factors relating to experimental logistics, intellectual property, and other nonscientific concerns (covered in ref. 2). As these impediments to GEMM testing have been largely resolved, we and others have turned to the large-scale testing of novel and traditional therapeutics in credentialed and faithful murine models of human cancers.
We believe RAS-driven tumors (e.g., melanoma, carcinomas of colon, pancreas, and lung) represent a particular clinical need. As mutations of K-, N-, or H-RAS occur in 15% to 30% of all human cancers (see Compilation of Somatic Mutations in Cancer, ref. 13), RAS activation represents the foremost "undrugged" tumor-driver in cancer biology. Moreover, RAS mutation is associated with adverse outcomes in several tumor types, and targeted approaches for mutant RAS are lacking. For example, in melanoma, although mutations of B-RAF are more common (43%), mutations of N-, K-, and H-RAS are also frequent in human disease (19%, 2%, and 1%, ref. 14), and RAS-mutant tumors exhibit a worsened prognosis compared with RAF-mutant disease (15). For these reasons, we initially elected to focus on Ras-mutant tumors, particularly melanoma where easy serial assessment offers advantages for large-scale drug testing. Toward that end, we used a previously established GEMM of RAS-driven melanoma (Tyr-H-Ras (G12V) Ink4a/Arf−/− herein referred to as "TRIA"; ref. 16) that is favorable for therapeutic testing (see Materials and Methods).
In this model, we tested all cytotoxic agents reported to have single-agent activity in melanoma, as well as novel therapeutics that have been of interest in this disease. In particular, we included agents thought to be "RAS-specific" alone or in combination, as well as combined therapy with AZD6244/BEZ235, a regimen previously shown to have activity in murine models of RAS-driven lung cancer (17). Given the unexpected results in murine melanoma, we further studied the efficacy of AZD6244/BEZ235 in a related breast cancer model, and then even more diverse breast cancer GEMMs. These results in more than 700 tumor-bearing mice suggest combined inhibition of phosphoinositide-3 kinase (PI3K)/mammalian target of rapamycin (mTOR) and mitogen-activated protein–extracellular signal-regulated kinase kinase (MEK) will exhibit significant antitumor activity in a variety of human malignancies.
Materials and Methods
Animals
All animal experiments were carried out with approval of the University of North Carolina Institutional Animal Care and Use Committee. Tyr-HRas(G12V) Ink4a/Arf−/− mouse model of melanoma was studied (16). This model features an activated H-Ras codon 12 mutant transgene integrated on the Y-chromosome combined with germline Ink4a/Arf inactivation, and is faithful to the human tumor genetics: RAS activation is present in ∼20% of human melanoma, and Ink4a/Arf loss is observed in 60% to 90% of melanoma. By crossing Tyr-Ras Ink4a/Arf−/− males with Ink4a/Arf−/− females, cohort were produced where no genotyping is required (all the males get cancer) and all the progeny mice are useful (the males develop tumors and the females are used for future breeding). This model is also addicted to persistent RAS signaling (18).
In addition to the TRIA melanoma model, we studied 2 GEM models of breast cancer: the C3-TAg transgenic mouse model of basal-like breast cancer (19) and the MMTV-c-neu mouse model (20). The C3-TAg transgenic mouse model of basal-like breast cancer (19) contains a recombinant gene expressing the simian virus 40 early region transforming sequence (SV40 large T antigen), which has been shown to inactivate both p53 and RB (21–23). The MMTV-c-neu mouse model of HER2+ breast cancer (20) expresses c-neu (the mouse ortholog of human HER2) driven by the mouse mammary tumor virus (MMTV) promoter and has been shown to represent a model of the luminal breast cancer subtype (24).
Where necessary, we have included an OST models (e.g., the T11 model; ref. 25) when an adequate GEMM could not be identified for the given tumor type (e.g., Claudin-low breast cancer). When syngeneic transplant models are used, they are still assessed using the other described practices of the MP1U (e.g., for tumor regression, large cohort size, etc.). When tumors were noted to be approximately 0.2 cm2 in size, animals were treated as described and tumor response was assessed by weekly caliper measurements. Data in Fig. 1 are normalized to tumor size at the time of therapy initiation, with volumes calculated using the formula Volume=[(width)2 × length]/2. Tumor-bearing mice were euthanized at the indicated times for morbidity, tumor ulceration, or tumor size of more than 2.0 cm in diameter.
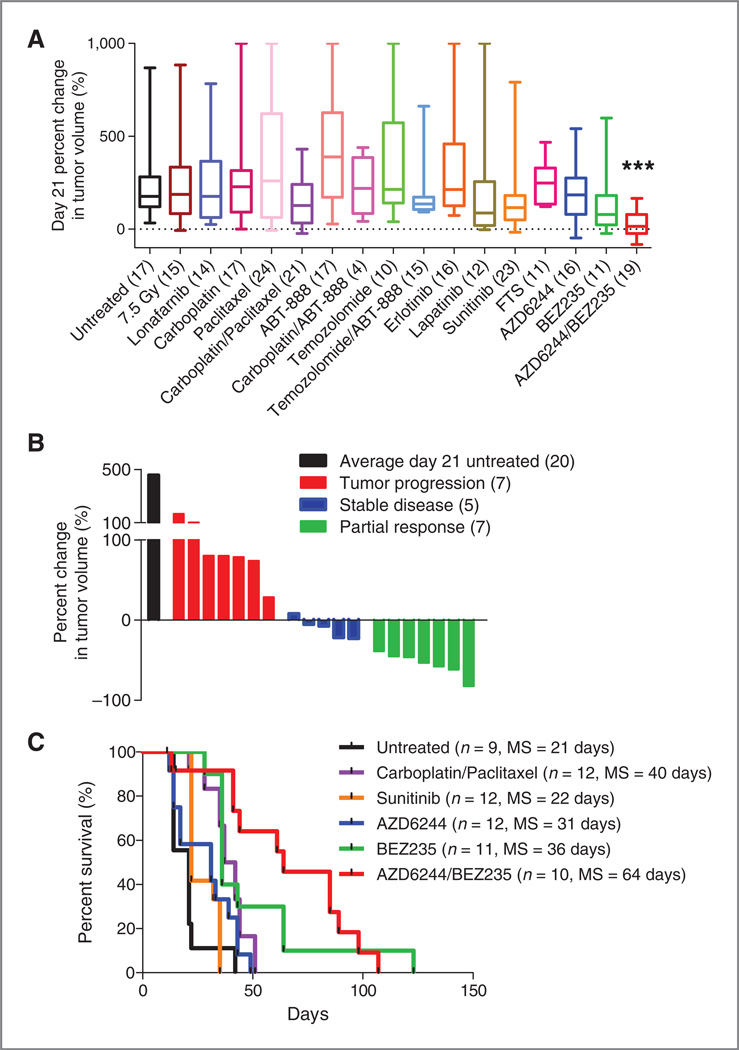
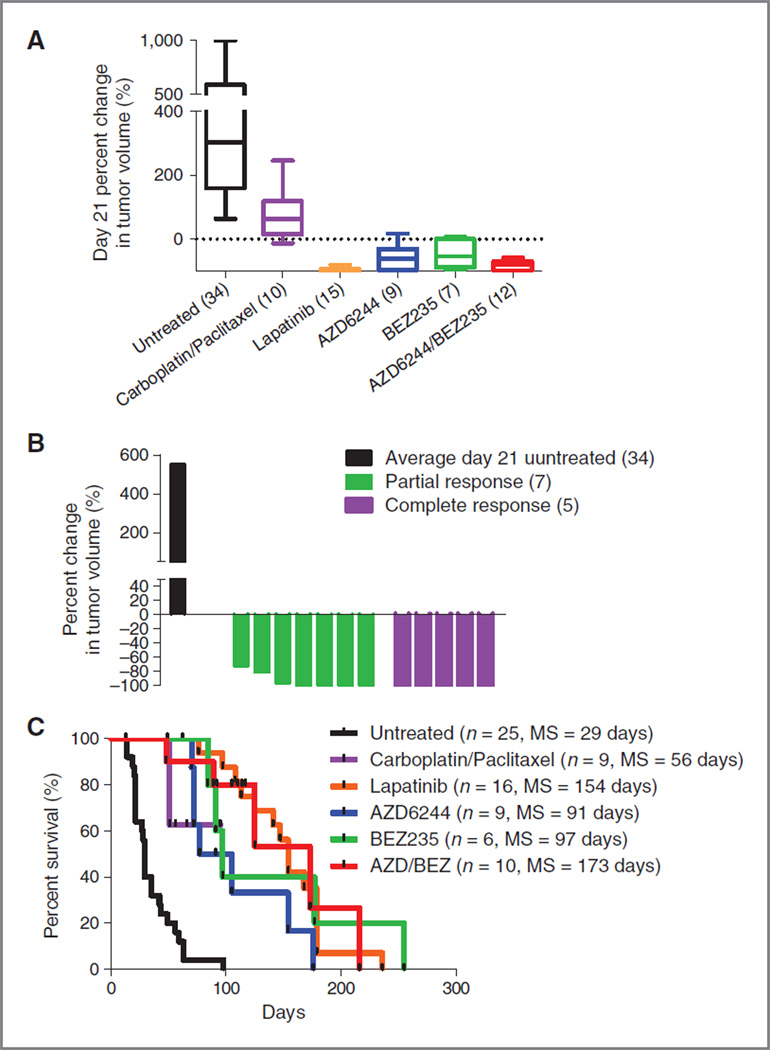
Figure 1.
RAS-driven melanoma is refractory to standard chemotherapy but is sensitive to combined PI3K/mTOR/MEK inhibition. TRIA mice were stratified by tumor size and randomly assigned to a treatment cohort. A, of the 15 treatment regimens tested, combined AZD/ BEZ treatment was the only regimen that produced tumor regression as measured by percent change in tumor volume at day 21.B,waterfall plot distribution of tumor response from combined AZD/BEZ compared with the average tumor size of untreated animals on day 21. AZD/BEZ response represents the best response seen on day 21 or beyond. Negative values indicate tumor shrinkage. C, combined AZD/BEZ treatment prolonged median survival from 21 to 61 days; MS, median survival.
Compounds
Compounds were obtained from commercial sources, by custom synthesis, or rarely under material transfer agreement from an industry partner. The MP1U has focused on conventional chemotherapeutic agents and small molecule agents rather than monoclonal antibodies because of interspecies differences in tumor epitopes ABT-888, AZD-6244, and FTS (S-trans,trans-farnesylthiosalicylic acid) were synthesized by the Center for Integrative Chemical Biology and Drug Discovery (CICBDD) at the University of North Carolina. Carboplatin (Hospira, Inc.), cyclophosphamide (Hospira, Inc.), doxorubicin (Bedford Laboratories), etopo-side (Teva Parenteral Medicines, Inc.), and paclitaxel (Ivax Pharmaceuticals, Inc.) were obtained from their respective manufacturers and handled as per standard practice. Erlotinib (Genentech, Inc.), lapatinib (GlaxoSmithKline), suni-tinib (Pfizer, Inc.), temozolomide (Merck) were obtained from clinical commercial sources. BEZ-235 and lonafarnib was obtained under material transfer agreements (MTA) with Novartis AG and Schering-Plough (now Merck/ Schering-Plough) respectively. In accord with these MTAs, pharmaceutical partners were shown these data before submission for publication, but had no role in the performance of these studies or the preparation of the MS.
Dosing and schedule
A major hurdle for the comprehensive undertaking was establishing the dose and schedule for each treatment regimen that we tested. The agent-specific approach to determine schedule and dose is extensively described in (Supplementary Table S1). In brief,we used published work to identify doses for well-studied agents (e.g., doxorubicin), and direct pharmacokinetic (PK) measurement in some instances (e.g., PD0332991, paclitaxel, carboplatin, dasatinib, data not shown).When neither of these approaches was possible, compounds were dosed at the maximally tolerated dose (MTD) as determined in the MP1U or reported by pharmaceutical partners. Most regimens required extensive dose finding in the MP1U before efficacy studies. Only results from mice treated at optimal dosing regimens are shown in the response figures. Results using dosing regimens with inadequate exposure or poor tolerability are excluded.
Carboplatin (Hospira, Inc.) was given by intraperitoneal (i.p.) injection and paclitaxel (Ivax Pharmaceuticals, Inc.) was given intravenous injection (i.v.). For temozolomide, erlotinib, and ABT-888, varying doses were used to confirm strain- and age-specific maximally tolerated dose. For oral drugs, the compound of interest was milled into chow by Research Diets, Inc. Chow was weighed daily for 1 week to calculate average daily intake.
Mice were treated with cytotoxic chemotherapy agents (e.g., carboplatin) once weekly for 21 days. A minimum of 1 week off treatment was given to all mice. Treatment only resumed when one of 2 conditions was met: (1) the primary tumor progressed by at least 2 mm in any direction by caliper measurement or (2) a secondary tumor became palpable. Orally available biologic inhibitors were dosed continuously with no dose interruption. Inhibitors were only removed in the case of complete regression of the tumor, or for weight loss. In the case of cytotoxic chemotherapy used with an oral small molecule inhibitor, the cytotoxic agent was dosed once weekly for 21 days and stopped until progression, whereas small molecule inhibitors were dosed continuously.
Response criteria
Tumor volume was calculated from 2-dimensional measurements. The percent change in volume at 14 and 21 days was used to quantify response. SD, PR, and CR were defined as per RECIST criteria. Survival was measured from first day of drug treatment.
Murine model cluster
Murine models of mammary carcinoma (GSE3165, GSE27101; refs. 24 and 26) and melanoma (GSE34866) were combined into a single dataset and compared for transcriptional similarities. Samples within each of these published datasets were analyzed on 3 different Agilent array platforms (22K, 4 × 44K, or 4 × 180K). Using normalization methods described to correct for platform effects between 22K and 4 × 44K platforms (26), we used 10 microarrays (5 MMTV-c-neu and 5 C3(1)-TAg) from each array type (30 microarrays total) to calculate a median, probe-level normalization factor using R v2.12.2. These arrays were chosen based on their high correlation to each other within each array type when clustered using a previously defined intrinsic gene list (24) to eliminate outliers from the array type correction. Principle component analysis (PCA) was used to verify proper normalization of the platforms. Unpublished MMTV-c-neu and C3(1)-TAg tumors used for platform correction were collected and microarray processed using methods previously described (24, 26). These arrays were uploaded to the Gene Expression Omnibus under series GSE35722 and to the University of North Carolina Microarray Database (27).
To identify transcriptional similarities between murine models of mammary carcinoma and melanoma, an unsupervised cluster analysis was carried out using any probe with a log2 absolute expression value greater than 2 on at least 3 microarrays (2,584 probes) using Cluster v3.0 (28). The data were viewed using Java Treeview v1.1.5r2 (29).
Cell culture and western blotting
Tumor-derived C3Tag and TRIA cell lines were grown in DMEM supplemented with 10% FBS and 1% penicillin/ streptomycin. Tumor-derived T11 cells were grown in RPMI supplemented with 10% FBS and 1% penicillin/streptomycin. Cells were lysed on ice for 20 minutes in lysis buffer containing 50 mmol/L HEPES (pH 7.5), 0.5% Triton X-100, 150 mmol/L NaCl, 1 mmol/L EDTA, 1 mmol/L EGTA, 10 mmol/L sodium fluoride, 2.5 mmol/L sodium orthovanadate, 1× protease inhibitor cocktail (Roche), and 1%each of phosphatase inhibitor cocktails 2 and 3 (Sigma). Cell lysate centrifuged for 15 minutes (13,000 rpm) at 4°Cand the supernatant was collected. Proteins from cell lysates were separated by SDS-PAGE chromatography, transferred to nitrocellulose membranes, and probed with the indicated primary antibodies. Antibodies recognizing pAKT (S473), pERK1/2 (T202/Y204), and phospho-MEK1/2 antibody (S217/S221) were obtained from Cell Signaling Technology. The antibody for ERK2 was obtained from Santa Cruz Biotechnology. HRP-anti-rabbit secondary antibody was obtained from Jackson Immunoresearch Laboratories. Western blots were incubated with SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific) and exposed to film.
Statistics
Unless otherwise noted, comparisons are made with t test or 1-way ANOVA, with Bonferroni correction for multiple comparisons where appropriate. P values of less than 0.05 are considered significant. Error bars represent ±SEM.
Results
Large-scale GEMM testing
We tested 11 distinct single agents in 16 treatment regimens in the TRIA model, and a subset of these treatment regimens were also tested in models of breast cancer representing distinct clinical subtypes in the UNC Mouse Phase I Unit (MP1U). In a contemporaneous study, additional treatment regimens were tested in the 3 breast cancer models (Usary and colleagues, submitted), and the most active regimens from that study were compared with AZD/ BEZ in this work. All FDA-approved and experimental compounds used in this work are described in Supplementary Table S1. Relevant features of all MP1U testing are:
The use of credentialed GEMMs. For the breast models, credentialing was accomplished with expression profiling (see refs. 24 and 26). For melanoma, model choice was guided by RAS-dependence and other experimental features (see Materials and Methods).
The use of large (10–25) cohorts of tumor-bearing animals per therapeutic cohort, with assessment of both novel and approved agents in each disease type. Large-scale testing of combinations in the breast models will be reported elsewhere (Usary et al., submitted).
The serial assessment for tumor response as opposed to measures of nonprogression (e.g., tumor growth inhibition, TGI), as TGI does not mimic acceptable measures of efficacy in humans. The 2 primary endpoints described in this work are percent change in tumor volume at day 21 and overall survival (both calculated as described in the Materials and Methods).
Choice of murine model for efficacy testing in this work was initially guided by RAS-dependence, but given the activity of a single regimen,we subsequently turned to more diverse GEM models.
Drug testing in Ras-mutant melanomas
Current treatment approaches for advanced melanoma are largely ineffective, with no targeted agents for RASmutant disease. Temozolomide provides a meager 12% response rate (30), and even newly approved ipilimumab offers only a modest 3.5-month survival advantage (31). To test different regimens in a GEM model of human RAS-driven melanoma, we generated cohorts of 10 to 20 inbred male TRIA mice. Mice were serially assessed for tumors, and treatment regimens were initiated in tumors at a minimum size of 25 mm3. In total, 15 single-agent or combination regimens and total body irradiation (single dose TBI; 7.5 Gy) were tested (Fig. 1A). Regimens of cytotoxic therapeutics with activity in human melanoma such as temozolomide and carboplatin–paclitaxel were not effective at inducing tumor response by 21 days of therapy. Similarly, small molecule agents such as sunitinib, lapatinib, and potential anti-RAS approaches [S-trans, trans-farnesylthiosalicylic acid (FTS) and lonafarnib, a farnesyltransferase inhibitor] were ineffective in this model as single agents. Response rates of the tested agents in the TRIA model using a modified version of Response Evaluation Criteria In Solid Tumor (RECIST) correlated with the antitumor efficacy of these agents in human melanoma (Table 1). Therefore, like human RAS-mutant melanoma, this model was highly refractory to therapy and treatment studies in this GEMM correlate with an agent′s activity in human disease.
Table 1.
RECIST response rates in TRIA mice versus human melanoma
| Regimen | Mean day 0 tumor volume (mm3) |
Mean day 21 tumor volume (mm3) |
RECIST response rate (CR + PR + SD) |
Reported human response rates for melanoma |
|---|---|---|---|---|
| Untreated | 83 | 322 | 0% | |
| Carboplatin | 89 | 217 | 21% | 14%–23% |
| Paclitaxel | 62 | 182 | 21% | 14–18% |
| Carboplatin/paclitaxel | 41 | 97 | 38% | 19%–47% |
| Temozolomide | 88 | 266 | 10% | 15% (10%–17%) |
| Sunitinib | 63 | 115 | 32% | 33% |
| AZD6244/BEZ235 | 53 | 29 | 63% | Unknown |
Therapeutic response, defined as tumor regression and survival improvement, was observed with only one treatment regimen in the TRIA GEMM. The co-administration of AZD6244 (an MEK inhibitor) and BEZ235 (a dual PI3K/ mTOR inhibitor) resulted in significant tumor regression at 21 days in the majority of mice (Fig. 1A). Overall, the dual AZD/BEZ treatment cohort had a 27.5% (95% CI = −4.5–59.4) median increase in tumor volume at 21 days compared with 217.5% (95%CI= 151.6–283.4) increase in the untreated cohort (P < 0.0001). Neither AZD nor BEZ exhibited significant activity at day 21 as a single agent. A "waterfall plot" of best response (21 days or later) showed that AZD/BEZ treatment resulted in partial response (PR) or stable disease (SD) in 12 of 19 animals (63%) (Fig. 1B). This observation suggests that inhibition of single RAS downstream effector pathways, MEK or PI3K, in isolation is not sufficient to induce tumor regression, but concomitant MEK and PI3K/mTOR inhibition can cooperatively produce tumor regression in most RAS-driven melanomas.
The regimens determined to be most active in the 21-day response assay were given in long-term survival experiments (Fig. 1C). Although every agent thus far tested in the MP1U that produces a reduction in tumor growth at 21 days also enhances survival, the converse is not true. Therefore, an overall survival endpoint was assessed on therapy given that some agents showing only modest activity at 21 days still produce a significant survival benefit (see, e.g., carboplatinpaclitaxel; Fig. 1A and C). Intravenous carboplatin–paclitaxel and oral single-agents sunitinib, AZD, and BEZ all led to modest improvements in survival compared with untreated animals, with single-agent BEZ the best of this group increasing the duration of median survival by 70% (21–36 days, P < 0.001). In accord with the 21-day response data, however, AZD/BEZ treatment afforded far greater clinical benefit than any other regimen, increasing median survival by 205% compared with untreated animals (21–64 days, P < 0.001). These data show that the marked antitumor activity observed at 21 days or later (Fig. 1B) translates into a tripling of median survival in mice with established, high-grade melanomas.
Testing of AZD/BEZ in a model of claudin-low breast cancer
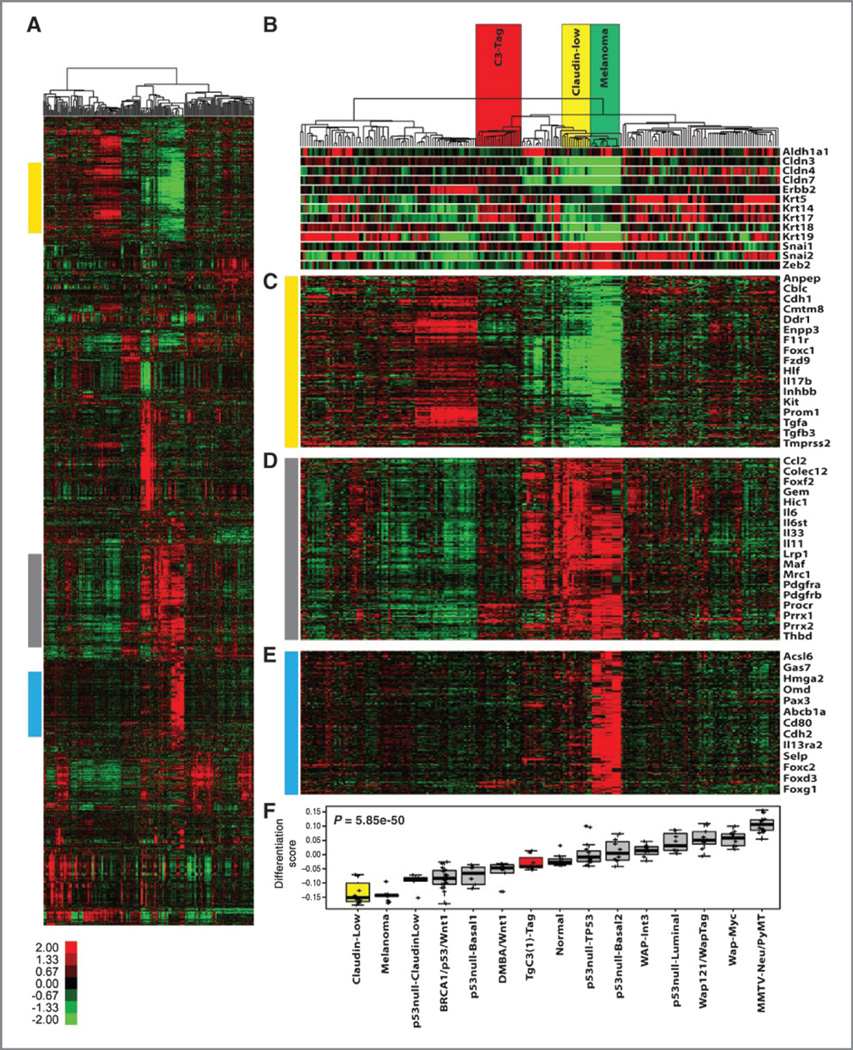
After identifying the potent activity of the AZD/BEZ drug combination in melanoma, we turned to a model of "clau-din-low" breast cancer (25). This choice was motivated by the unexpected finding that claudin-low breast models show expression features in common with melanoma. For example, unsupervised analysis of 13 murine mammary carcinoma models (24, 26) and 4 murine melanoma models showed that the melanoma samples clustered with claudin-low tumors (Fig. 2A). As seen in human claudin-low samples (25), murine melanomas exhibited low expression of claudins (e.g., Cldn3, Cldn4, Cldn7) and cytokeratins (e.g., Krt5, Krt14, Krt17) and increased expression of genes associated with epithelial-to-mesenchymal transition [e.g., Snal1/2, Zeb2, Cdh1 (E-cadherin); Fig. 2B and C). Both murine tumor types were also characterized by increased expression of inflammatory/immune genes compared with other breast models (e.g., IL6, IL33, Ccl2; Fig. 2D). Melanomas differed from claudin-low breast tumors in the expression of melanocyte lineage genes such as Pax3 as well as transcripts expressed in melanoma as opposed to breast cancer [Gas7 and Cdh2 (N-cadherin)]. Using a recently described metric to calculate a "differentiation score" in breast cancer (higher score = more differentiated; ref. 25), melanoma, and claudin-low models showed similarly low scores (Fig. 2F), suggestive of expression profiles more similar to mammary stem cells than differentiated luminal cells. Given these shared transcriptional features with the TRIA model, we elected to further test AZD/BEZ in the claudin-low breast cancer model.
Figure 2.
Transcriptional analysis of murine models of mammary carcinoma and melanoma. A, an unsupervised cluster of all probes with a log2 absolute expression value greater than 2 on at least 3 microarrays (2,584 probes). The colored bars correspond to the region of the cluster for the gene lists in parts C to E. B, highlighted is the location in the dendrogram of the C3-TAg, MMTV-c-neu, claudin-low (including T11 OST tumors), and melanoma tumors (including TRIA tumors). Below are markers of the claudin-low subtype as defined by Prat and colleagues (25). C, a cluster of genes that are downregulated in both claudin-low and melanoma tumors. D, a cluster of genes that are upregulated in tumors from 6 different claudin-low and 4 different melanoma models. E, a melanoma specific gene cluster. F, a rank of median differentiation scores for tumors from the indicated breast and melanoma models.
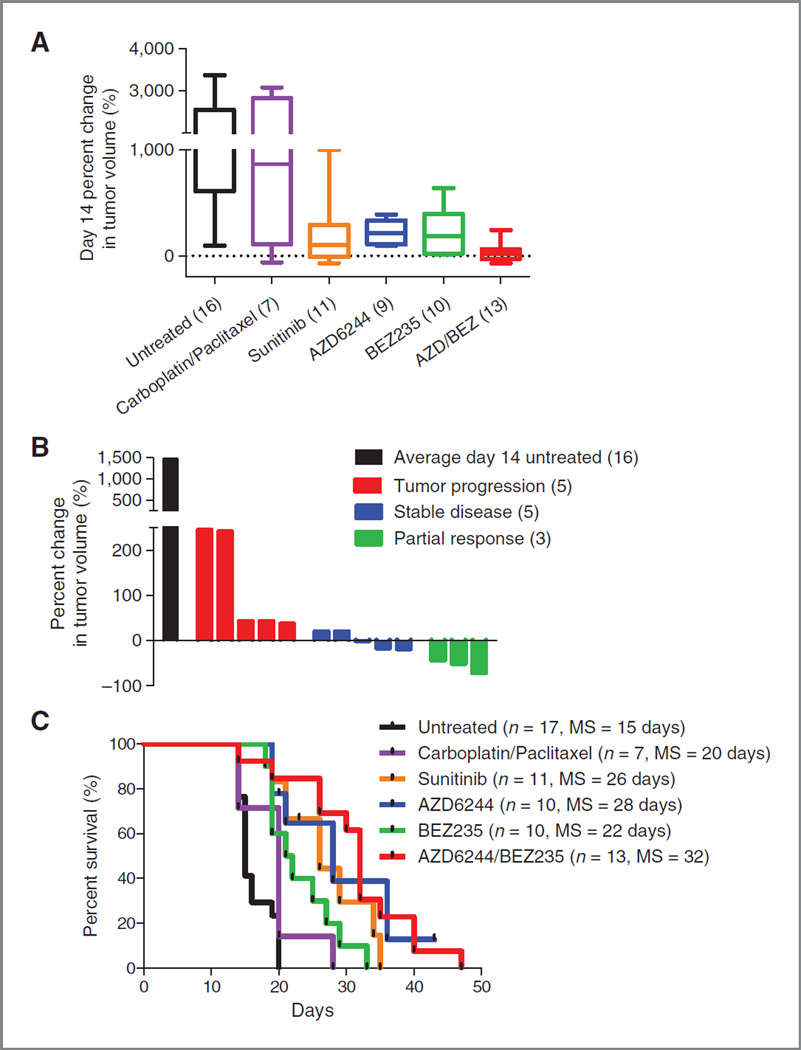
We have not identified a faithful GEM model of the claudin-low breast cancer subtype that only gives rise to just this tumor subtype, therefore have taken to the analysis of genomically selected orthotopic, syngeneic transplant (OST) models. The T11 model derives from the serial orthotopic transplantation of a murine breast tumor derived from a p53-null mouse into a syngeneic p53 competent recipient, and features sporadic, somatic K-Ras mutation (data not shown). Tumors from the T11 model display an RNA expression pattern characteristic of the human claudin-low disease (Fig. 2 and ref. 26), and are extremely aggressive, with the majority of untreated animals surviving less than 21 days from the time of enrollment in the therapy studies. Therefore, a 14-day rather than 21-day response endpoint was used to assess activity in this model. As in the TRIA melanoma model, AZD/BEZ was highly active in the T11 OST model (Fig. 3A compares AZD/BEZ with 4 of the most active other 16 tested regimens, see also Usary et al., submitted). Although no CRs were seen in the T11 model, AZD/BEZ treatment caused an SD + PRrate of 62% (Fig. 3B) and more than doubled median survival compared with untreated animals (from 15 to 36 days, P< 0.0001; Fig. 3C). Compared to other active regimens (sunitinib and carboplatin–paclitaxel) and single-agent AZD or BEZ, the combination was superior in terms of response and survival prolongation. Therefore, combined PI3K/mTOR and MEK inhibition afforded significant clinical benefit in a murine transplantation model of the claudin-low breast cancer subtype.
Figure 3.
Combined PI3K/mTOR/ MEK inhibition is the most effective treatment regimen in claudin-low breast cancer model. T11 mice were stratified by tumor size and randomly assigned to a treatment cohort. A, combined AZD/BEZ was the most effective treatment regimen as measured by percent change in tumor volume at day 14. B, waterfall plot distribution of tumor response from combined AZD/BEZ compared to the average tumor size of untreated animals on day 14. AZD/ BEZ response represents the best response seen on day 14 or beyond. Negative values indicate tumor shrinkage. C, combined AZD/BEZ treatment prolonged median survival from 15 to 32 days; MS, median survival.
Testing of AZD/BEZ in diverse GEM breast cancer models
Given the activity of this regimen in RAS-driven melanoma and claudin-low breast cancer, we determined the efficacy of this regimen in 2 other 2 breast cancer GEMMs: representing the basal-like (or BBC, C3-TAg) and luminal (MMTV-c-neu) subtypes of breast cancer. These models have been previously credentialed by unbiased gene expression analysis that indicated maximal similarity to their respective human breast cancer subtypes (BBC and luminal) from a large panel of other GEM breast models (24). The C3-TAg model (19) harbors a standard transgenic allele with a breast-specific promoter driving overexpression of the simian virus 40 Large T Antigen (TAg), which inactivates the p53 and RB tumor suppressors. This model is faithful to human BBC, which frequently features combined RB and p53 loss (32–36), and C3-TAg and human BBC show common patterns of gene expression by RNA expression profiling (24, 32). Of relevance to AZD/BEZ testing, tumors from this model frequently (∼30%), but not always, acquire K-Ras amplification with progression (37), and haploid loss of K-Ras delays progression in this model (38). Therefore, a fraction of tumors in this GEMM likely require persistent RAS activation for tumor maintenance, and we reasoned differential sensitivity to AZD/BEZ might correlate with K-Ras activation status.
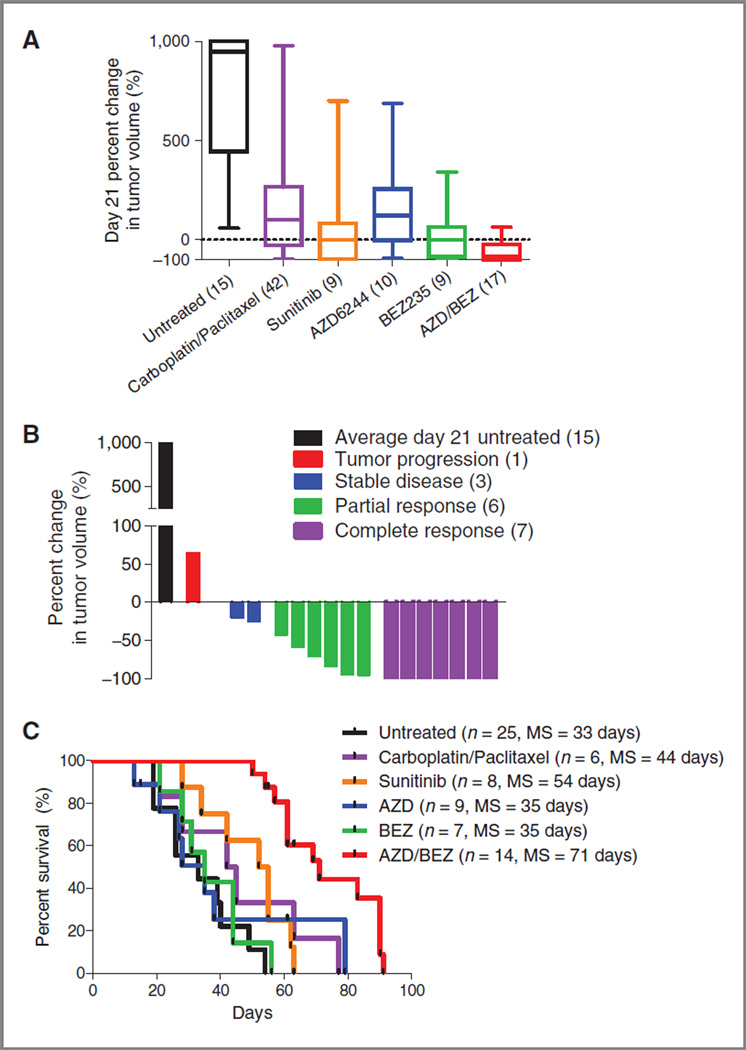
We were surprised, therefore, to note potent activity for the dual AZD/BEZ treatment in nearly all tumors from this BBC model. As in the TRIA model, AZD/BEZ treatment resulted in significant tumor regression and improved survival compared with untreated mice or mice treated with other active therapeutic regimens (Fig. 4A–C). A waterfall plot of best response (21 days or later) showed SD + PR + CR (SD, stable disease; PR, partial response; CR, complete response) in 16 of 17 treated mice, indicating a 94% response rate, with almost half of the treated mice exhibiting CR. This high response rate translated into a marked survival benefit with median survival increasing by 115% compared with untreated animals (33–71 days, P < 0.0001). As in the TRIA model, the response rate and survival advantage of AZD/BEZ was superior to any other regimen tested in this model (of N =15 regimens; Usary et al., submitted; see also Fig. 4A and C). For example, the 2 next most active regimens (sunitinib or carboplatin–paclitaxel) only modestly extended survival (64% and 33%) compared with AZD/BEZ. Similarly, neither AZD nor BEZ was nearly as active in this model as a single-agent therapy compared with combined therapy (Fig. 4A and C). These data indicate AZD/BEZ induces significant tumor response and extends survival in a credentialed GEM model of BBC, regardless of the presence or absence of K-Ras amplification.
Figure 4.
Basal-like breast cancer model is exquisitely sensitive to combined PI3K/mTOR/MEK inhibition. C3-TAg mice were stratified by tumor size and randomly assigned to a treatment cohort. A, combined AZD/BEZ was the most effective treatment regimen, resulting in a median percent change in tumor volume at day 21 of —84%. B, waterfall plot distribution of tumor response from combined AZD/BEZ compared with the average tumor size of untreated animals on day 21. AZD/BEZ response represents the best response seen on day 21 or beyond. Negative values indicate tumor shrinkage. C, combined AZD/BEZ treatment prolonged median survival from 33 to 71 days; MS, median survival.
Given this lack of general correlation between AZD/BEZ efficacy and Ras mutation in the C3-TAg model, we considered a model that does not harbor Ras mutation or amplification: the MMTV-c-neu GEMM. This mouse model of luminal breast cancer features transgenic expression of the rat ortholog of human HER2 driven by an ER-enhanced breast promoter (MMTV; ref. 20). Treatment with AZD/BEZ was also highly active in this model (Fig. 5A – C), with a PR+ CR rate of 100% (12 of 12), with 5 CRs. Several regimens (e.g., carboplatin–paclitaxel, lapatinib) induced responses and significantly prolonged survival in this model, as did single-agent treatment with AZD and BEZ (Fig. 5C, all P < 0.001 in pairwise comparison with untreated mice). As expected, this model was exquisitely sensitive to lapatinib (100% CR rate; Fig. 5A and C), a HER2 kinase inhibitor that is FDA-approved for HER2-amplified breast cancer in humans. Remarkably, however, the AZD/BEZ combination was better in terms of response than all other regimens except lapatinib (Fig. 5A), and equivalent to or better than lapatinib in terms of survival prolongation, inducing a striking 400% increase in median survival (29–173 days, P < 0.0001; Fig. 5C). Therefore, combined PI3K/mTOR and MEK inhibition offers equivalent activity in HER2-driven GEMM (expressing wild-type RAS) compared to a FDA-approved targeted anti-HER2 agent.
Figure 5.
A GEM model of luminal breast cancer is exceptionally sensitive to combined PI3K/mTOR/ MEK inhibition. MMTV-c-neu mice were stratified by tumor size and randomly assigned to a treatment cohort. A, lapatinib and combined AZD/BEZ treatment regimens provide nearly complete tumor regression as measured by percent change in tumor volume at day 21. B, waterfall plot distribution of tumor response from combined AZD/BEZ compared with the average tumor size of untreated animals on day 21. AZD/BEZ response represents the best response seen on day 21 or beyond. Negative values indicate tumor shrinkage. C, combined AZD/ BEZ treatment prolonged median survival from 29 to 173 days, whereas lapatinib prolonged median survival to only 112 days; MS, median survival.
Cross-talk between PI3K and MEK signaling
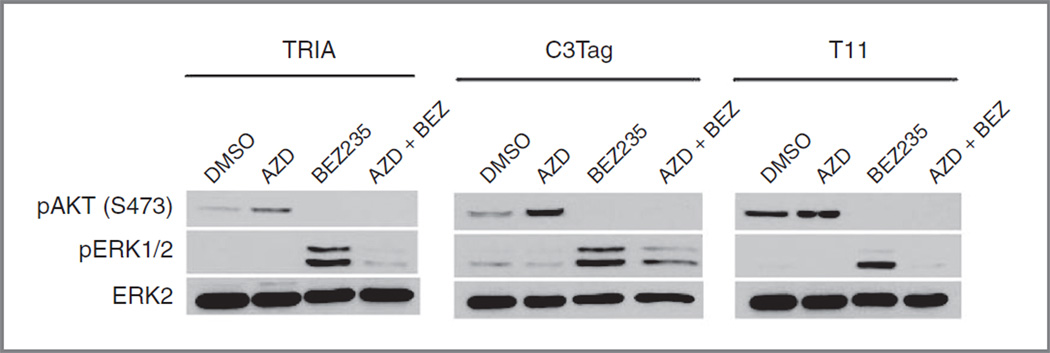
Although AZD6244 and BEZ235 both showed only modest single-agent activity in the models studied, the combined AZD/BEZ regimen was highly effective in each of these models. In an effort to understand the molecular basis for this synergy, cell lines derived from the tumor models were treated in vitro with AZD6244 (1 µmol/L), BEZ235 (250 nmol/L), or combinaiton AZD/BEZ (1 µmol/ L/250 nmol/L). Tumor-derived cell lines from TRIA, T11, and C3-TAg tumors were studied; the MMTV-c-neu model was not considered as cell lines could not be derived from this model. Cell lines were treated for 24 hours with the indicated compounds, and analyzed by Western blot for target inhibition (Fig. 6). Expression of pERK was low in all 3 cell lines, and was not affected by single-agent AZD6244 treatment. Single-agent AZD6244 treatment produced an increase in phosphorylation of AKT at serine 473, a site phosphorylated by mTOR.In contrast, single-agent BEZ235 treatment decreased S473 phosphorylation of AKT, as expected and consistent with inhibition of mTOR, but also led to a marked increase in phosphorylation of ERK1/2. Cotreatment with AZD6244 and BEZ235 inhibited phosphorylation of both AKT and ERK1/2, indicating effective target inhibition. The surprising activation of AKT by MEK inhibitors and ERK activation by PI3K inhibitors has been reported in other cell types (39–43), and reflects compensatory feedback loops between these pathways. We believe these complex inhibitory networks explain the modest activity of PI3K and MEK inhibitors as single agents in the multiple murine models thus far tested, and also explain the potent synergy of the combined AZD/BEZ regimen in vivo, which is seen in our in vivo results.
Figure 6.
Inhibition of AZD6244 and BEZ235 targets in tumor-derived cell lines. To evaluate the molecular response to AZD6244, BEZ235, or combined AZD/ BEZ treatment, tumor-derived cell lines from TRIA, T11, and C3Tag tumors were treated for 24 hours with AZD6244 (1 µmol/L) and/or BEZ235 (250 nmol/L) and evaluated by Western blot analysis for target inhibition. Single-agent AZD led to PI3K activation (as evidenced by increased S473 phosphorylation of AKT), whereas single-agent BEZ led to ERK activation (as evidenced by increased phosphorylation of ERK1/2). Combined treatment with AZD/BEZ produced combined inhibition of the PI3K/mTOR and MEK/ERK pathways.
Dual PI3K/MEK inhibition in lapatinib-resistant HER2+ breast cancers
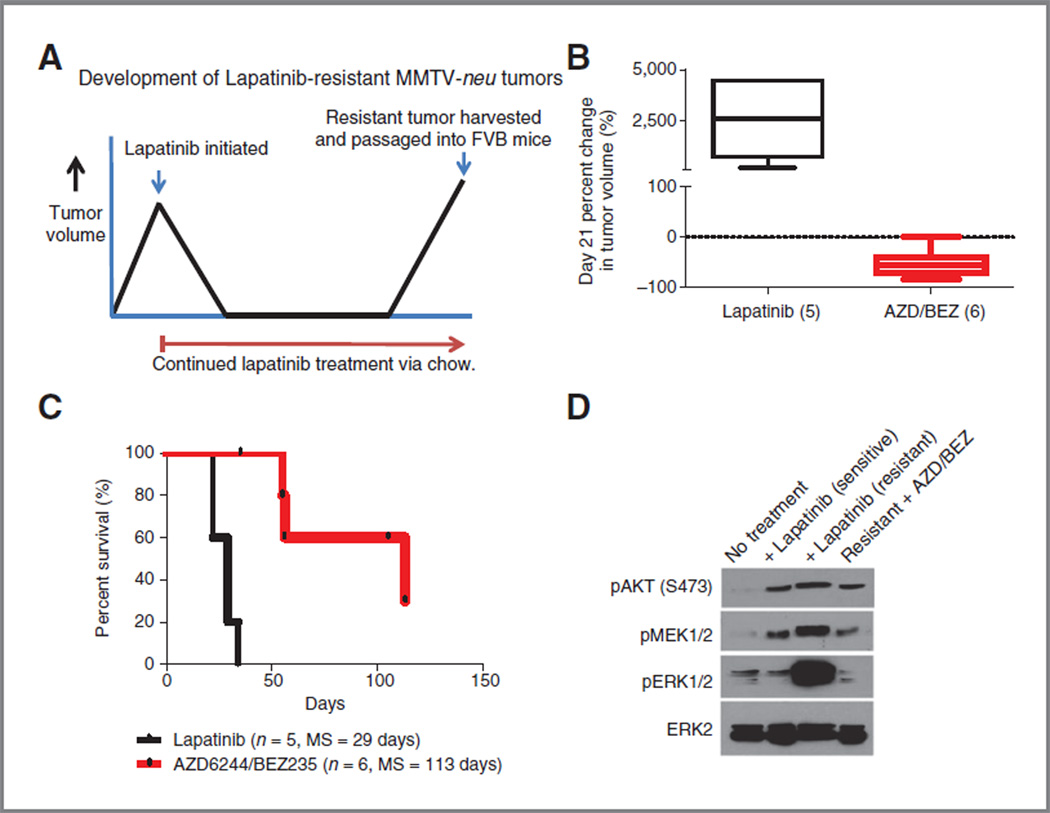
We next turned to an assessment of the AZD/BEZ regimen in the setting of resistant disease. Given the potent activity of AZD/BEZ in the MMTV-c-neu model,we decided to evaluate the efficacy of this regimen in tumors that had become completely refractory to lapatinib therapy. MMTV-c-neu tumors will always respond completely to initial lapatinib therapy (Fig. 5A), but then resistant tumors will develop on therapy causing progression and death within 150 days on treatment (Fig. 5C). Given this observation, a model to study lapatinib-resistant disease was developed as shown in Fig. 7A. In brief, at the time of progression and while on continuous lapatinib therapy, resistant tumors were harvested and orthotopically passaged into the mammary fatpad of syngeneic female mice. Tumors were allowed to establish to a size of at least 5 mm in any one direction before the initiation of lapatinib rechallenge or second-line treatment. As expected, retreatment with lapatinib in mice transplanted with lapatinib-resistant tumors was not effective (Fig. 7B and C). Remarkably, however, combined AZD/BEZ treatment was highly active in lapatinib-resistant disease, resulting in median tumor reduction of 57% (Fig. 5B, P = 0.007) and increasing survival 290% (from 29 to 113 days, P < 0.001, Fig. 7C) compared with lapatinib retreatment. These data show that AZD/BEZ therapy can be effective in tumors chosen for therapeutic refractoriness to other agents.
Figure 7.
Combined PI3K/mTOR/MEK inhibition is an effective treatment regimen in lapatinib resistant HER2+ breast cancer. A, MMTV-c-neu mice stratified by tumor size and assigned to continuous treatment with lapatinib were followed until tumors progressed through treatment. Resistant tumor were harvested and passaged into the mammary fat pad of 6-to 8-week-old female FVB mice. Following injection, tumors were allowed to reach minimum size of at least 5 mm in any one-dimension before initiating second line therapy (either rechallenge of lapatinib or dual AZD/BEZ).B, retreatment with lapatinib has no effect on tumor growth, whereas the combined AZD/BEZ treatment results in median tumor regression of −57%, as measured by percent change in tumor volume at day 21. C, combined AZD/BEZ treatment prolonged median survival from 29 to 113 days; MS, median survival. D, lapatinib-resistant MMTV-c-neu tumors display upregulated phosphorylation of AKT, MEK1/2, and ERK1/2. Treatment of mice bearing lapatinib resistant tumors with AZB/BEZ returns phosphorylation of AKT, MEK1/2, and ERK1/2 to preresistant levels.
To further evaluate the mechanism of lapatinib resistance, we isolated tumors from mice that were lapatinib naïve, responding to lapatinib treatment, lapatinib resistant, and lapatinib resistant responding to AZD/BEZ treatment. These tumors were harvested directly from mice while on therapy, and then analyzed by Western blot for target inhibition. This analysis showed that lapatinib-resistant tumors exhibited increased phosphorylation of AKT S473 (mTOR target), MEK1/2, and ERK1/2 (Fig. 7D). Treatment of lapatinib-resistant tumors with AZD/BEZ led to phosphorylation levels of these proteins equivalent to that seen in lapatinib-sensitive tumors on lapatinib (compare lanes 4 and lanes 2 of Fig. 7D). These data show that lapatinib resistance occurs in the setting of MEK/ERK and PI3K/ mTOR activation, explaining the efficacy of the AZD/BEZ regimen even in lapatinib-resistant disease.
Discussion
In this work, we show that therapeutic testing in GEM and OST models can be carried out in a medium-throughput manner at reasonable expense to identify unexpected therapeutic single-agent and combination regimens. We also show potent antitumor activity of combined therapy with PI3K/mTOR inhibitors and MEK inhibitors in a diverse array of credentialed GEM and OST models. We showed impressive activity of this regimen in the RAS-driven melanoma model. Based on a shared gene expression between melanoma and claudin-low breast tumors, we tested AZD/ BEZ in the T11 OST model, which also harbors an activating mutation of K-Ras. Given significant activity in this treatment-refractory and aggressive model, we then turned to 2 additional breast GEM models representing BBC (C3-TAg mice) and luminal breast cancer (MMTV-c-neu), which are not obligately RAS-driven. We observed broad and reproducible antitumor activity of the AZD/BEZ regimen in all of these disparate models, in accord with prior studies in GEM models of NSCLC (17, 44). In aggregate, these studies predict broad antitumor efficacy of combined MEK and PI3K/mTOR inhibitors in human cancer patients.
This large experience in animal models points to some of the strengths of GEM testing versus conventional measures to assess preclinical efficacy. For example, certain regimens (e.g., carboplatin–paclitaxel and sunitinib in TRIA mice) clearly reduced tumor growth rates, and therefore would score highly in traditional tumor-growth inhibition (TGI) assays. When assessed by more rigorous standards, however, these regimens were not capable of inducing tumor regression (Fig. 1A) and produced only modest enhancement of long-term survival (Fig. 1C). Our results using tumor regression and long-term survival correspond better to the efficacy of these agents in human patients (Table 1) than predictions based on TGI in xenograft systems. Also in contrast to xenograft models, we have generally observed significant heterogeneity among tumors within any given model in terms of rate of growth and response rates to active regimens. We believe this more faithfully models the human setting and provides an opportunity to understand primary therapeutic resistance to active agents, an area of intense ongoing study.
We believe that the implications of this work for human therapeutics are considerable, as testing of MEK and PI3K/ mTOR inhibitors has already begun in human cancer patients. Our data showing compensatory activation of ERK signaling by PI3K/mTOR inhibitors and compensatory activation of AKT by MEK inhibitors explains the observed therapeutic synergy of the AZD/BEZ combination, and suggest that single-agent trials of these agents should rapidly give way to combination trials in certain tumor types. Of note, there are approximately 17 ongoing early phase clinical trials evaluating 14 unique combinations of MEK and PI3K/mTOR inhibitors, and preliminary results for a few of these novel combinations have been promising (clinicaltrials.gov; ref. 45). In addition, our data suggest trials of combined PI3K/mTOR and MEK inhibitors should be opened in a variety of cancers, and in particular should focus on RAS-mutant cancers for which no effective targeted approaches exist. Our data also predict that resistance to combined MEK and PI3K/mTOR inhibition will develop within months of starting therapy (see Figs. 1C, 3C, and 4C), emphasizing the immediate need to study acquired resistance in human clinical specimens as well as murine models. Toward that end, these models will provide an excellent platform to study both de novo and acquired resistance to AZD/BEZ and related combinations. The finding in lapatinib-resistant tumors of ERK/AKT activation and significant sensitivity to the AZD/BEZ regimen (Fig. 7B–D) is particularly encouraging. This result suggests that mechanisms of resistance to HER2 kinase inhibitors still require activation of persistent downstream HER2 effectors such as PI3K, mTOR, and/or MEK. This observation is analogous to the finding that resistance to B-RAF inhibitors in melanoma often requires downstream activation of MEK (reviewed in ref. 46). Similarly, although lapatinib resistance in HER2-driven breast cancer is imperfectly understood, resistance is frequently associated with increased activation of the PI3K pathway (reviewed in ref. 47). Therefore, our results suggest that the AZD/BEZ combination can be active in tumors resistant to other targeted kinase inhibitors, and also suggest the possibility that early combination of these agents (e.g., with lapatinib) may limit primary refractory disease and development of secondary resistance.
A critical question based on these results relates to the toxicity of this regimen. Because of practical concerns,we do not conduct comprehensive toxicologic assessment of regimens in the MP1U. We did conduct the serial assessment of animal weights on all treated mice, as well as monitoring of blood counts in a subset of treated animals, and by these measures, toxicity of this regimen was manageable. In some mice, brief treatment breaks were given because of weight loss, and often we observed tumor regrowth during these treatment breaks, further emphasizing the activity of this regimen. Gratifyingly, the limited amount of human phase I testing thus far accomplished with combined MEK and PI3K inhibitors suggests these combinations are not too toxic for human use (48). In summary, our work predicts broad clinical activity of the combination of MEK and PI3K/mTOR inhibitors in a large variety of human malignancies with expected tolerable levels of toxicity.
Supplementary Material
Translational Relevance.
We describe a comprehensive assessment of 16 anticancer therapeutic regimens in multiple engineered murine model (GEMM) and orthotopic, syngeneic transplant (OST) cancer model. Aggregate analysis from these studies shows that, in contrast to standard preclinical efficacy testing (e.g., xenograft models), these faithful models exhibit a high predictive accuracy of the clinical efficacy of known chemotherapeutic agents used in melanoma and breast cancer. Moreover, testing of novel agents showed extraordinary anticancer activity from combined therapy with phosphoinositide-3 kinase (PI3K)/mammalian target of rapamycin (mTOR) and mitogen-activated protein–extracellular signal-regulated kinase kinase (MEK) inhibitors. These data have significant implications for the clinical development of MEK and PI3K/mTOR inhibitors and predict that this combination will have broad clinical activity across disparate human malignancies.
Grant Support
This work was supported by the University of North Carolina Lineberger Comprehensive Cancer Center Mouse Phase I Unit, and grants from the Golfers Against Cancer Foundation (to N. E. Sharpless and C. M. Perou), the Ellison Medical Foundation (to N. E. Sharpless), the Burroughs Wellcome Fund (1006673 to N. E. Sharpless), the National Institutes of Health (RO1 P01-ES014635 and UO1-CA141576 to N. E. Sharpless; P50-CA58223-09A1 and RO1-CA148761 to C. M. Perou), the University Cancer Research Fund from the University of North Carolina Lineberger Comprehensive Cancer Center (to N. E. Sharpless and W.C.Z), The Breast Cancer Research Foundation (C. M. Perou), and the American Cancer Society (PF-10-239-01-TBG to P. J. Roberts).
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Authors' Contributions
Conception and design: P. J. Roberts, D. Darr, P. M. Dillon, A. Combest, N. E. Sharpless
Development of methodology: P.J.Roberts, J. Usary, D. Darr,A.Combest, W. C. Zamboni, N. E. Sharpless
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): P. J. Roberts, J. Usary, D. Darr, P. M. Dillon, A. D. Pfefferle, M. C. Whittle, J. S. Duncan, S. M. Johnson, J. Jin, W. C. Zamboni, C. M. Perou
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): P. J. Roberts, J. Usary, D. Darr, A. D. Pfefferle, A. Combest, W. C. Zamboni, G. L. Johnson, C. M. Perou, N. E. Sharpless
Writing, review, and/or revision of the manuscript:P.J.Roberts,J.Usary, D. Darr, P. M. Dillon, A. D. Pfefferle, A. Combest, W. C. Zamboni, C. M. Perou, N. E. Sharpless
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases):P. J. Roberts, D. Darr, P. M. Dillon, J. Jin
Study supervision: P. J. Roberts, D. Darr, G. L. Johnson, N. E. Sharpless
Please enter any additional contributions here: Molecular analysis of drugs in tumors and cells via western blots (J. S. Duncan)
References
- 1.Kola I, Landis J. Can the pharmaceutical industry reduce attrition rates? Nat Rev Drug Discov. 2004;3:711–715. doi: 10.1038/nrd1470. [DOI] [PubMed] [Google Scholar]
- 2.Sharpless NE, Depinho RA. The mighty mouse: genetically engineered mouse models in cancer drug development. Nat Rev Drug Discov. 2006;5:741–754. doi: 10.1038/nrd2110. [DOI] [PubMed] [Google Scholar]
- 3.Peterson JK, Houghton PJ. Integrating pharmacology and in vivo cancer models in preclinical and clinical drug development. Eur J Cancer. 2004;40:837–844. doi: 10.1016/j.ejca.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 4.Richey EA, Lyons EA, Nebeker JR, Shankaran V, McKoy JM, Luu TH, et al. Accelerated approval of cancer drugs: improved access to therapeutic breakthroughs or early release of unsafe and ineffective drugs? J Clin Oncol. 2009;27:4398–4405. doi: 10.1200/JCO.2008.21.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olive KP, Tuveson DA. The use of targeted mouse models for preclinical testing of novel cancer therapeutics. Clin Cancer Res. 2006;12:5277–5287. doi: 10.1158/1078-0432.CCR-06-0436. [DOI] [PubMed] [Google Scholar]
- 6.Politi K, Pao W. How genetically engineered mouse tumor models provide insights into human cancers. J Clin Oncol. 2011;29:2273–2281. doi: 10.1200/JCO.2010.30.8304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh M, Johnson L. Using genetically engineered mouse models of cancer to aid drug development: an industry perspective. Clin Cancer Res. 2006;12:5312–5328. doi: 10.1158/1078-0432.CCR-06-0437. [DOI] [PubMed] [Google Scholar]
- 8.Saez E, Tontonoz P, Nelson MC, Alvarez JG, Ming UT, Baird SM, et al. Activators of the nuclear receptor PPARgamma enhance colon polyp formation. Nat Med. 1998;4:1058–1061. doi: 10.1038/2042. [DOI] [PubMed] [Google Scholar]
- 9.Kulke MH, Demetri GD, Sharpless NE, Ryan DP, Shivdasani R, Clark JS, et al. A phase II study of troglitazone, an activator of the PPAR-gamma receptor, in patients with chemotherapy-resistant metastatic colorectal cancer. Cancer J. 2002;8:395–399. doi: 10.1097/00130404-200209000-00010. [DOI] [PubMed] [Google Scholar]
- 10.Sarraf P, Mueller E, Jones D, King FJ, DeAngelo DJ, Partridge JB, et al. Differentiation and reversal of malignant changes in colon cancer through PPARgamma. Nat Med. 1998;4:1046–1052. doi: 10.1038/2030. [DOI] [PubMed] [Google Scholar]
- 11.Troiani T, Schettino C, Martinelli E, Morgillo F, Tortora G, Ciardiello F. The use of xenograft models for the selection of cancer treatments with the EGFR as an example. Crit Rev Oncol Hematol. 2008;65:200–211. doi: 10.1016/j.critrevonc.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 12.Singh M, Lima A, Molina R, Hamilton P, Clermont AC, Devasthali V, et al. Assessing therapeutic responses in Kras mutant cancers using genetically engineered mouse models. Nat Biotechnol. 2010;28:585–593. doi: 10.1038/nbt.1640. [DOI] [PubMed] [Google Scholar]
- 13.Forbes SA, Bhamra G, Bamford S, Dawson E, Kok C, Clements J, et al. The catalogue of somatic mutations in cancer (COSMIC) Curr Protoc Hum Genet. 2008 Apr; doi: 10.1002/0471142905.hg1011s57. Chapter 10:Unit 10.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forbes S, Clements J, Dawson E, Bamford S, Webb T, Dogan A, et al. Cosmic 2005. Br J Cancer. 2006;94:318–322. doi: 10.1038/sj.bjc.6602928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Devitt B, Liu W, Salemi R, Wolfe R, Kelly J, Tzen CY, et al. Clinical outcome and pathological features associated with NRAS mutation in cutaneous melanoma. Pigment Cell Melanoma Res. 2011;24:666–672. doi: 10.1111/j.1755-148X.2011.00873.x. [DOI] [PubMed] [Google Scholar]
- 16.Chin L, Pomerantz J, Polsky D, Jacobson M, Cohen C, Cordon-Cardo C, et al. Cooperative effects of INK4a and ras in melanoma susceptibility in vivo . Genes Dev. 1997;11:2822–2834. doi: 10.1101/gad.11.21.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engelman JA, Chen L, Tan X, Crosby K, Guimaraes AR, Upadhyay R, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008;14:1351–1356. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chin L, Tam A, Pomerantz J, Wong M, Holash J, Bardeesy N, et al. Essential role for oncogenic Ras in tumour maintenance. Nature. 1999;400:468–472. doi: 10.1038/22788. [DOI] [PubMed] [Google Scholar]
- 19.Maroulakou IG, Anver M, Garrett L, Green JE. Prostate and mammary adenocarcinoma in transgenic mice carrying a rat C3(1) simian virus 40 large tumor antigen fusion gene. Proc Natl Acad Sci U S A. 1994;91:11236–11240. doi: 10.1073/pnas.91.23.11236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muller WJ, Sinn E, Pattengale PK, Wallace R, Leder P. Single-step induction of mammary adenocarcinoma in transgenic mice bearing the activated c-neu oncogene. Cell. 1988;54:105–115. doi: 10.1016/0092-8674(88)90184-5. [DOI] [PubMed] [Google Scholar]
- 21.Ahuja D, Saenz-Robles MT, Pipas JM. SV40 large T antigen targets multiple cellular pathways to elicit cellular transformation. Oncogene. 2005;24:7729–7745. doi: 10.1038/sj.onc.1209046. [DOI] [PubMed] [Google Scholar]
- 22.Ali SH, DeCaprio JA. Cellular transformation by SV40 large T antigen: interaction with host proteins. Semin Cancer Biol. 2001;11:15–23. doi: 10.1006/scbi.2000.0342. [DOI] [PubMed] [Google Scholar]
- 23.Deeb KK, Michalowska AM, Yoon CY, Krummey SM, Hoenerhoff MJ, Kavanaugh C, et al. Identification of an integrated SV40 T/tantigen cancer signature in aggressive human breast, prostate, and lung carcinomas with poor prognosis. Cancer Res. 2007;67:8065–8080. doi: 10.1158/0008-5472.CAN-07-1515. [DOI] [PubMed] [Google Scholar]
- 24.Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8:R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herschkowitz JI, Zhao W, Zhang M, Usary J, Murrow G, Edwards D, et al. Breast cancer special feature: comparative oncogenomics identifies breast tumors enriched in functional tumor-initiating cells. Proc Natl Acad Sci U S A. 2011;109:2778–2783. doi: 10.1073/pnas.1018862108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.University of North Carolina Microarray Database. [cited 2012 Aug 28]; Available from: https://genome.unc.edu/pubsup/breastGEO/clinical-Data.shtml.
- 28.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics (Oxford, England) 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 29.Saldanha AJ. Java Treeview–extensible visualization of microarray data. Bioinformatics (Oxford, England) 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 30.Middleton MR, Grob JJ, Aaronson N, Fierlbeck G, Tilgen W, Seiter S, et al. Randomized phase III study of temozolomide versus dacarbazine in the treatment of patients with advanced metastatic malignant melanoma. J Clin Oncol. 2000;18:158–166. doi: 10.1200/JCO.2000.18.1.158. [DOI] [PubMed] [Google Scholar]
- 31.Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363:711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herschkowitz JI, He X, Fan C, Perou CM. The functional loss of the retinoblastoma tumour suppressor is a common event in basal-like and luminal B breast carcinomas. Breast Cancer Res. 2008;10:R75. doi: 10.1186/bcr2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gauthier ML, Berman HK, Miller C, Kozakeiwicz K, Chew K, Moore D, et al. Abrogated response to cellular stress identifies DCIS associated with subsequent tumor events and defines basal-like breast tumors. Cancer Cell. 2007;12:479–491. doi: 10.1016/j.ccr.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang Z, Deng T, Jones R, Li H, Herschkowitz JI, Liu JC, et al. Rb deletion in mouse mammary progenitors induces luminal-B or basal-like/EMT tumor subtypes depending on p53 status. J Clin Invest. 2010;120:3296–3309. doi: 10.1172/JCI41490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hollestelle A, Nagel JH, Smid M, Lam S, Elstrodt F, Wasielewski M, et al. Distinct gene mutation profiles among luminal-type and basal-type breast cancer cell lines. Breast Cancer Res Treat. 2010;121:53–64. doi: 10.1007/s10549-009-0460-8. [DOI] [PubMed] [Google Scholar]
- 36.Subhawong AP, Subhawong T, Nassar H, Kouprina N, Begum S, Vang R, et al. Most basal-like breast carcinomas demonstrate the same Rb-/ p16+ immunophenotype as the HPV-related poorly differentiated squamous cell carcinomas which they resemble morphologically. Am J Surg Pathol. 2009;33:163–175. doi: 10.1097/PAS.0b013e31817f9790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu ML, Von Lintig FC, Liyanage M, Shibata MA, Jorcyk CL, Ried T, et al. Amplification of Ki-ras and elevation of MAP kinase activity during mammary tumor progression in C3(1)/SV40 Tag transgenic mice. Oncogene. 1998;17:2403–2411. doi: 10.1038/sj.onc.1202456. [DOI] [PubMed] [Google Scholar]
- 38.Liu ML, Shibata MA, Von Lintig FC, Wang W, Cassenaer S, Boss GR, et al. Haploid loss of Ki-ras delays mammary tumor progression in C3 (1)/SV40 Tag transgenic mice. Oncogene. 2001;20:2044–2049. doi: 10.1038/sj.onc.1204280. [DOI] [PubMed] [Google Scholar]
- 39.Carracedo A, Ma L, Teruya-Feldstein J, Rojo F, Salmena L, Alimonti A, et al. Inhibition of mTORC1 leads to MAPK pathway activation through a PI3K–dependent feedback loop in human cancer. J Clin Invest. 2008;118:3065–3074. doi: 10.1172/JCI34739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ebi H, Corcoran RB, Singh A, Chen Z, Song Y, Lifshits E, et al. Receptor tyrosine kinases exert dominant control over PI3K signaling in human KRAS mutant colorectal cancers. J Clin Invest. 2011;121:4311–4321. doi: 10.1172/JCI57909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Serra V, Scaltriti M, Prudkin L, Eichhorn PJ, Ibrahim YH, Chandarlapaty S, et al. PI3K inhibition results in enhanced HER signaling and acquired ERK dependency in HER2-overexpressing breast cancer. Oncogene. 2011;30:2547–2557. doi: 10.1038/onc.2010.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Turke AB, Song Y, Costa C, Cook R, Arteaga CL, Asara JM, et al. MEK inhibition leads to PI3K/AKT activation by relieving a negative feedback on ERBB receptors. Cancer Res. 2012;72:3228–3237. doi: 10.1158/0008-5472.CAN-11-3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Faber AC, Li D, Song Y, Liang MC, Yeap BY, Bronson RT, et al. Differential induction of apoptosis in HER2 and EGFR addicted cancers following PI3K inhibition. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0905056106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carretero J, Shimamura T, Rikova K, Jackson AL, Wilkerson MD, Borgman CL, et al. Integrative genomic and proteomic analyses identify targets for Lkb1-deficient metastatic lung tumors. Cancer Cell. 2010;17:547–559. doi: 10.1016/j.ccr.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roberts PJ, Stinchcombe TE. KRAS mutations: Should we test for it? Does it matter? What are the new possible treatments? J Clin Oncol. 2012 In press. [Google Scholar]
- 46.Solit DB, Rosen N. Resistance to BRAF inhibition in melanomas. N Engl J Med. 2011;364:772–774. doi: 10.1056/NEJMcibr1013704. [DOI] [PubMed] [Google Scholar]
- 47.Chen FL, Xia W, Spector NL. Acquired resistance to small molecule ErbB2 tyrosine kinase inhibitors. Clin Cancer Res. 2008;14:6730–6734. doi: 10.1158/1078-0432.CCR-08-0581. [DOI] [PubMed] [Google Scholar]
- 48.Bendell J, LoRusso P, Kwak E, Pandya S, Musib L, Jones C, et al. Clinical combination of the MEK inhibitor GDC-0973 and the PI3K inhibitor GDC-0941: a first-in-human phase Ib study in patients with advanced solid tumors [abstract]; Proceedings of the 102nd Annual Meeting of the American Association for Cancer Research; 2011 Apr 2–6; Orlando, FL. Philadelphia (PA): AACR; 2011. Abstract nr. LB-89. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.