Abstract
The capacity of tumour cells to maintain continual overgrowth potential has been linked to the commandeering of normal self-renewal pathways. Using an epithelial cancer model in Drosophila melanogaster, we carried out an overexpression screen for oncogenes capable of cooperating with the loss of the epithelial apico-basal cell polarity regulator, scribbled (scrib), and identified the cell fate regulator, Abrupt, a BTB-zinc finger protein. Abrupt overexpression alone is insufficient to transform cells, but in cooperation with scrib loss of function, Abrupt promotes the formation of massive tumours in the eye/antennal disc. The steroid hormone receptor coactivator, Taiman (a homologue of SRC3/AIB1), is known to associate with Abrupt, and Taiman overexpression also drives tumour formation in cooperation with the loss of Scrib. Expression arrays and ChIP-Seq indicates that Abrupt overexpression represses a large number of genes, including steroid hormone-response genes and multiple cell fate regulators, thereby maintaining cells within an epithelial progenitor-like state. The progenitor-like state is characterised by the failure to express the conserved Eyes absent/Dachshund regulatory complex in the eye disc, and in the antennal disc by the failure to express cell fate regulators that define the temporal elaboration of the appendage along the proximo-distal axis downstream of Distalless. Loss of scrib promotes cooperation with Abrupt through impaired Hippo signalling, which is required and sufficient for cooperative overgrowth with Abrupt, and JNK (Jun kinase) signalling, which is required for tumour cell migration/invasion but not overgrowth. These results thus identify a novel cooperating oncogene, identify mammalian family members of which are also known oncogenes, and demonstrate that epithelial tumours in Drosophila can be characterised by the maintenance of a progenitor-like state.
Author Summary
Cancer is a multigenic process, involving cooperative interactions between oncogenes or tumour suppressors. In this study, in a genetic screen in the vinegar fly, Drosophila melanogaster, for genes that cooperate with a mutation in the cell polarity (shape) regulator, scribbled (scrib), we identify a novel cooperative oncogene, abrupt. Expression of abrupt in scrib mutant tissue in the developing eye/antennal epithelium results in overgrown invasive tumours. abrupt encodes a BTB-zinc finger transcription factor, which has homology to several cancer-causing proteins in humans, such as BCL6. Analysis of the Abrupt targets and misexpressed genes in abrupt expressing-tissue and abrupt-expressing scrib mutant tumours, revealed cell fate regulators as a major class of targets. Thus, our results reveal that deregulation of multiple cell fate factors by Abrupt expression in the context of polarity disruption is associated with a progenitor-like cell state and the formation of overgrown invasive tumours. Our findings suggest that defective polarity may also be a critical factor in BTB-zinc finger-driven human cancers, and warrants further investigation into this issue.
Introduction
Cancer cells with significant tumour-propagating potential are increasingly referred to as cancer stem cells. Whilst this refers to the potential of these cells to regenerate the tumour in both in vivo and in vitro assays, it also alludes to the possibility that these cells may have either hijacked self-renewal programmes involved in normal stem cell maintenance, or that they are in fact directly derived from stem or progenitor-like cells. Consistent with either of these possibilities, profiles of tumour cells show increased expression of stem cell factors and associations with progenitor-like cell states [1], [2].
In Drosophila melanogaster, tumours have long been known to be associated with the retention of stem cell states. Germ line tumours show continual overgrowth of progenitor cells that fail to initiate differentiation, and neuroblast-derived brain tumours are associated with defects in neuroblast (neural stem cell) divisions and an expansion of neuroblast numbers [reviewed in 3]. Furthermore, the overgrowth associated with l(3) malignant brain tumour mutants has been shown to depend upon the acquisition of a stem cell state associated with the germline [4]. Impaired differentiation has also been considered to be a hallmark of Drosophila epithelial tumours [5], although how differentiation is perturbed and what role this plays in maintaining tumour overgrowth is not yet known. Indeed the epithelial tissues of the imaginal discs are not thought to contain stem cells. Instead it appears that cells become progressively restricted in their developmental potential as patterning mechanisms drive greater elaboration and cell fate commitments across the epithelial field. The sequential nature of these elaborations means that epithelial progenitor-like states are generally associated with earlier developmental times and are not necessarily associated with spatially defined regions of the developing tissue. In the antennal disc, the early progenitor state is yet to be clearly characterised, although the early division between the more distally destined cells that express the homeodomain protein Distal-less (Dll) and the more proximal cells expressing the MEIS family transcription factor, Homothorax (Hth), is one of the earliest cell fate divisions to have been described within the developing appendage [reviewed in 6]. Downstream targets of these genes, including atonal (ato), dachshund (dac), distal antenna (dan), and bric-a-brac 2 (bab2), are subsequently expressed, and gradually define further cell fate divisions along the proximo-distal axis of the appendage [7]–[9]. In the eye disc, the progenitor state has been more fully defined and is thought to be characterised by the expression of Hth, which cooperates with Yorkie (Yki, or YAP in mammals), the transcriptional coactivator of the Hippo tissue growth control pathway [reviewed in 10], to maintain cells within a proliferative state [11]. The downregulation of Hth coincides with the progressive upregulation of cell fate markers such as dac, eyes absent (eya), dan, ato and embryonic lethal abnormal vision (elav), that define further differentiation [reviewed in 6]. What role, if any, these sequential cell fate restrictions play in mediating the overgrowth of eye and antennal disc tumours has not yet been investigated.
Epithelial tumours can be induced in the eye/antennal disc by using a clonal system to combine loss of the cell polarity regulator and tumour suppressor scribbled (scrib) with oncogenic Ras or Notch (N) signalling. Whilst neither genetic alteration is sufficient to transform cells, in combination they cooperate to drive the formation of invasive tumours that outcompete the surrounding untransformed tissue and massively overgrow [12]. In an overexpression screen, to identify novel cooperating oncogenes that function like oncogenic Ras or Notch, we isolated the BTB-zinc finger (BTB-ZF) domain protein Abrupt (Ab). Expression arrays and ChIP-Seq analysis of Ab binding regions and immunohistochemical analysis of the tumours indicates that Ab promotes the retention of a progenitor-like cell state in scrib mutant cells by blocking the expression of dac, eya, dan, ato and elav in the eye disc, and prevents the temporal elaboration of cell fate domains, defined by dac, cut (ct), senseless (sens), dan, bab2 and ato expression, along the proximo-distal axis in the antennal disc. The Hippo tissue growth control pathway transcriptional coactivator, Yki, is both required to promote tumour overgrowth, and sufficient to cooperate with Ab and maintain cells within the progenitor-like state.
Results
A screen for cooperating oncogenes in Drosophila
We have previously shown how loss of the epithelial cell polarity regulator and tumour suppressor scrib cooperates with oncogenic Ras (RasV12/RasACT) or Notch (Notchintra/NotchACT) signalling to promote the formation of invasive tumours [12]. To identify novel oncogenes in Drosophila we carried out an overexpression screen to identify additional genes that can cooperate with the loss of scrib to promote tumour overgrowth. This was done using a bank of Gene Search (GS) P element lines [13], which contain UAS sites to ectopically express the flanking genes. By combining this with GAL4-driven expression, we screened independent GS line insertions on the second chromosome for their ability to promote neoplastic overgrowth when combined with the loss of scrib in eye disc clones. Normally the generation of scrib mutant clones in the eye/antennal disc produces adult flies with mildly reduced and necrotic eyes due to Jun kinase (JNK)-mediated death of the mutant tissue [12]. We therefore aimed to identify genes that could either cause pupal lethality or, most importantly, act like activated alleles of either Ras or Notch to block larval pupariation and cause massive tumour overgrowth.
From screening ∼2000 GS lines, we identified over 50 that caused increased organism lethality when expressed in scrib mutant clones ( Table 1 ). As the insertion point and expressed genes have been mapped for all GS lines, it was possible to determine that this corresponded to 10 different genes. Using independent transgenes we were able to confirm that overexpression of 6 of them (abrupt (ab), dorsal (dl), escargot (esg), numb, charlatan (chn) and apontic (apt) reproduced the lethality of the GS line. For the remaining 4 genes (kismet (kis), anachronism (ana), CG3363 and CG10543), although we identified multiple independent GS lines for each, independent transgenes were not available at the time to confirm the interaction. For the confirmed interactors, we examined larval eye/antennal discs to determine the extent of clonal overgrowth induced by the transgene alone compared to the amount of overgrowth when combined with the loss of scrib ( Figure 1 and Figure S1). Some of the interactors (numb and apt) promoted very little consistent overgrowth phenotypes despite causing organism lethality at the pupal stage of development, whilst chn produced mild overgrowth, and both dl and esg were striking for producing very large antennal overgrowths before the larvae pupated at day 5/6 after egg laying (AEL). However, of the 6 confirmed genes, the strongest interactor was ab.
Table 1. Identified scrib− cooperating oncogenes.
| GS lines | Overexpressed gene, position of GS insertion | Validating transgene | Validated/Candidate gene function | Closest human homologue |
| 49 (GSV1) 81 (GSV1) | CG4807(ab)-RA, +10847 CG4807(ab)-RA, +10854 | UAS-ab | BTB-C2H2 zinc finger transcription factor | ZFP161 |
| 5075 (GSV2) 5251 (GSV2) | CG6667(dl)-RA, +505 CG6667(dl)-RA, +621 | UAS-dl | NF-kB/Rel family transcription factor | REL |
| 2077 (GSV1) 5022 (GSV2) 9490 (GSV6) 11431 (GSV6) 11506 (GSV6) 11550 (GSV6) 13437 (GSV6) 14412 (GSV6) 14394 (GSV6) | CG3758(esg)-RA, −98 CG3758(esg)-RA, −112 CG3758(esg)-RA, −265 CG3758(esg)-RA, −89 CG3758(esg)-RA, −105 CG3758(esg)-RA, −153 CG3758(esg)-RA, −93 CG3758(esg)-RA, −105 CG3758(esg)-RA, −270 | UAS-esg | Snail family C2H2 zinc finger transcription factor | SNAI2 |
| 2112 (GSV1) 11450 (GSV6) | CG11798(chn)-RA, +780 CG11798(chn)-RB, +12782 | UAS-chn | C2H2 zinc finger transcription factor | ZNF462 |
| 9032 (GSV6) 9416 (GSV6) 10126 (GSV6) 10914 (GSV6) 12693 (GSV6) 13666 (GSV6) 14392 (GSV6) | CG5393(apt)-RA, −254 CG5393(apt)-RA, −254 CG5393(apt)-RA, −394 CG5393(apt)-RA, −182 CG5393(apt)-RA, −249 CG5393(apt)-RA, −296 CG5393(apt)-RA, −315 | UAS-tdf | SANT domain transcription factor | none |
| 2149 (GSV1) 2273 (GSV1) | CG3779(numb)-RB, −37 CG3779(numb)-RB, −35 | UAS-numb | Membrane associated regulator of intracellular signalling | NUMB |
| 2198 (GSV1) 14080 (GSV6) | CG10543-RA, +2050 CG10543-RA, +1620 | Not validated | C2H2 zinc finger transcription factor | ZNF479 |
| 2067 (GSV1) 9929 (GSV6) 10455 (GSV6) 14364 (GSV6) | CG3696(kis)-RA, +28904 CG3696(kis)-RA, +1341 CG3696(kis)-RA, +28429 CG3696(kis)-RA, +28276 | Not validated | SNF2-like chromo domain protein | CHD7 |
| 9498 (GSV6) 11321 (GSV6) | CG8084(ana)-RA, −52 CG8084(ana)-RA, −51 | Not validated | Secreted glycoprotein | none |
| 7333 (GSV2) 11052 (GSV6) | No tagged gene No tagged gene | Not validated | - | - |
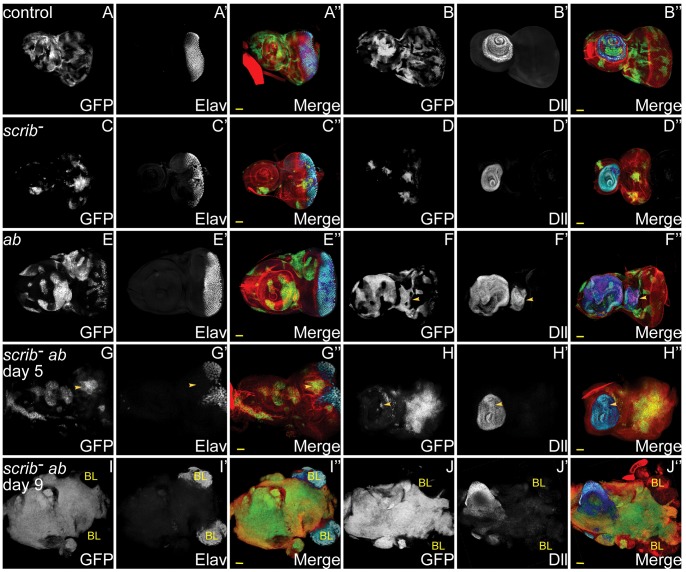
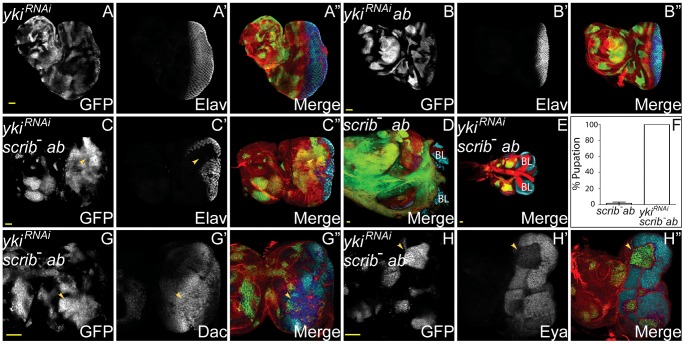
Figure 1. ab overexpression in scrib mutant clones promotes neoplastic overgrowth of eye/antennal epithelial tissue throughout an extended larval stage.
Mosaic eye/antennal discs (anterior to the left in this and all subsequent figures) generated with ey-FLP and taken from larvae 5 days (A–H) or 9 days (I,J) AEL. Clones are positively marked by GFP (white, or green in merges). Tissue morphology is shown by F-actin (red in merges), and cell fate by Elav and Dll (white, or blue in merges – dark blue when overlaid with GFP). Brain lobes in I,J are marked by BL. GFP (panels A–J), Elav (panels A′,C′,E′,G′,I′), Dll (panel B′,D′,F′H′,J′) and merges (panels A″–J″). (A,B) Control mosaic eye/antennal discs show the normal pattern of Elav expression in developing photoreceptor cells, and Dll expression within the antenna. (C,D) scrib1 cells still express Elav and Dll, although the normal pattern of Elav-expressing photoreceptor cells is disrupted by alterations in tissue morphology. (E,F) ab overexpressing clones still express Elav and Dll, but are often larger than control clones within the antennal region, and in some discs ectopic domains of Dll expression are observed (F, arrowhead). (G,H) scrib1+ab clones are larger than scrib1 clones, and do not express Elav (G, arrowhead), although Dll expression is maintained (H, arrowhead). (I,J) scrib1+ab clones at day 9 are massively overgrown and the two eye/antennal discs fuse with each other and with the Elav-expressing brain lobes (I), whilst the Dll-expressing domain in the antennal disc is maintained (J). Yellow scale bar = 50 µm.
The overexpression of ab in scrib mutant clones was unique amongst the interactors in promoting a block to pupariation and massive tumour overgrowth throughout an extended larval stage. Both GS lines were inserted within the 5′ region of ab, and orientated so as to overexpress ab, and an independent UAS-ab line reproduced the same cooperative effect as the two GS lines. Analysis of scrib−+ab larval eye disc clones at day 5 revealed that differentiation of eye disc tissue was completely abrogated, as judged by the failure to express the photoreceptor differentiation marker Elav, although expression of the antennal cell fate marker, Dll, was retained within the growing tumour ( Figure 1A–H ). By day 9, huge invasive tumours had developed and become fused with the brain lobes ( Figure 1I,J ). In contrast, the overexpression of ab in otherwise wild type eye disc clones promoted antennal disc overgrowth, and sometimes resulted in the formation of ectopic Dll-positive antennal-like structures, however, it did not block photoreceptor differentiation and the larvae pupated, although most died during pupal development ( Figure 1E,F ). Analysis of proliferation by ethynyl deoxyuridine (EdU) incorporation confirmed that whilst ab-expressing clones exhibited a relatively normal pattern of proliferation, scrib −+ab tumours ectopically proliferated and disrupted the normal pattern of cell proliferation within the eye disc (Figure S2). Furthermore, Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) stains indicated that although scrib mutant cells undergo apoptosis [12], [14], cell death in scrib−+ab discs was mainly confined to the wild type tissue surrounding the growing tumours, although interestingly, ab-expressing clones alone were also characterised by increased cell death of wild type cells bordering the clones (Figure S2). Therefore, similar to RasACT or NotchACT, the overexpression of ab can cooperate with the loss of scrib to block cell death and differentiation, and promote unrestrained and invasive tissue overgrowth, thus acting as a highly potent novel oncogene in Drosophila.
Ab acts as a global transcriptional regulator at the promoters of many genes
Ab is a transcription factor with a BTB protein interaction domain and zinc finger DNA binding domains [15]–[17]. To build a comprehensive picture of how Ab functions in its oncogenic capacity, we employed Affymetrix expression arrays to identify the transcriptional changes induced by ab overexpression both alone and in combination with the loss of scrib, and combined this with Ab Chromatin Immunoprecipitation-Sequencing (ChIP-Seq) to identify those genes that were potential direct targets of Ab-mediated regulation.
Tissue samples were prepared from mosaic eye/antennal discs overexpressing ab alone, or ab in scrib mutant clones, 5 days AEL. For the expression arrays, samples were compared back to control eye/antennal discs with wild type clones to identify differentially expressed probe sets (log base 2 fold change >1, adjusted p value [18] <0.05). This analysis indicated that Ab exerts a potent influence on gene expression, with 3028 and 3534 probe sets differentially expressed in ab and scrib −+ab discs respectively, of which 2323 probe sets were shared between the two ( Figure 2A and Dataset S1). The combined 4239 differentially expresssed probe sets encompassed 3549 annotated genes, 183 of which were represented by more than one probe set. The 183 genes with multiple probe sets were largely consistent in their pattern of expression changes in each genotype, although 59 of the 183 genes had probe sets that were both up and downregulated within the same genotype, possibly reflecting the existence of differentially expressed transcripts (Dataset S2). Quantitative real-time PCR validation of 5 representative genes confirmed the results of the expression array (Figure S3).
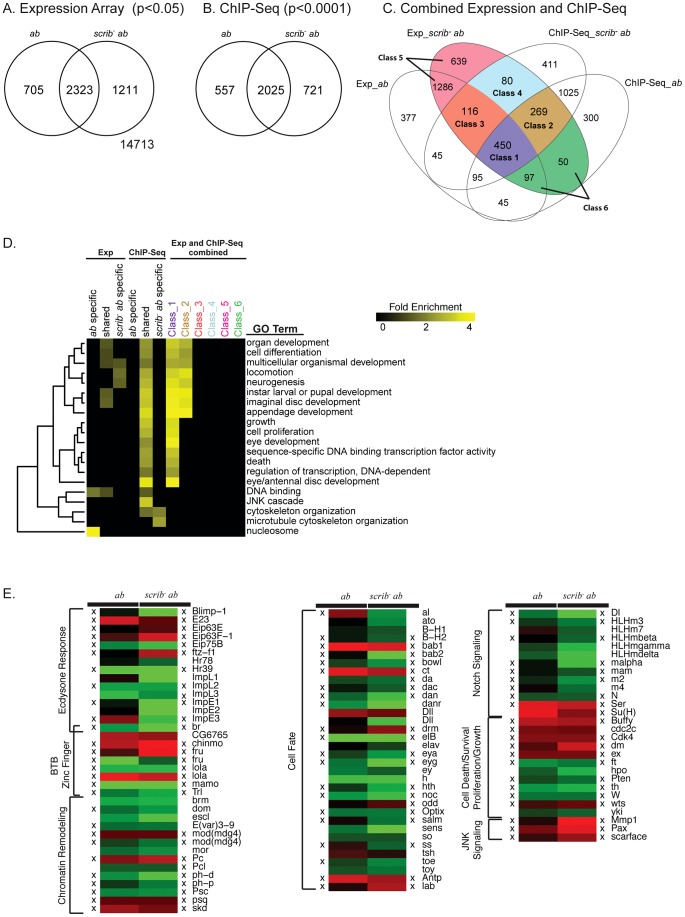
Figure 2. Potential tumourigenic targets of Ab identified from expression array and ChIP-Seq analysis.
(A) Venn diagram showing the number of differentially expressed probe sets (log base 2 fold change >1, adjusted p value <0.05) within mosaic ab overexpressing eye/antennal discs compared to control mosaic discs, and scrib1+ab mosaic discs compared to the control mosaic discs. (B) Venn diagram showing the number genes identified as potential Ab targets based upon the occurrence of a significant peak (see Materials and Methods) either within 500 bp upstream of the transcription start site or within the introns of a gene, in either mosaic ab-expressing eye/antennal discs, or scrib1+ab mosaic discs, when compared to the respective input DNA controls. (C) Venn diagram that combines the results from the expression array and ChIP-Seq analysis. The six main classes of genes deregulated in scrib1+ab tumours are shown: Class 1, genes differentially expressed and associated with Ab peaks in both ab and scrib1+ab samples; Class 2, genes differentially expressed in scrib1+ab alone, but associated with Ab peaks in both samples; Class 3, genes differentially expressed in both ab and scrib1+ab samples, but associated with Ab peaks in scrib1+ab alone; Class 4, genes only differentially expressed and associated with Ab peaks in scrib1+ab alone; Class 5, genes deregulated in the scrib1+ab tumours but not associated with Ab peaks; and Class 6, genes deregulated in the scrib1+ab tumours but only associated with Ab peaks in the non-tumourigenic ab-expressing discs. Note that genes represented by multiple probe sets in the expression array are represented only once amongst the different classes, and assigned to either both genotypes if at least one probe set was significantly deregulated in both genotypes, or assigned to either ab or scrib−+ab uniques categories if at least one probe set was deregulated specifically in these genotypes. See Dataset S1 for the complete gene lists associated with the expression array, ChIP-Seq and Classes 1–6. (D) Selected GO enrichments amongst the deregulated genes and potential Ab targets. See Dataset S3 for the full listing of significantly enriched GO categories. (E) Heat map highlighting selected functional groups of deregulated genes identified from the expression array (red, upregulated; green, downregulated). For genes represented by multiple probe sets, the following probe sets are shown in the figure: br (1636931_at), chinmo (1636985_s_at), Dll (1636088_at, 1625771_at), dom (1628160_a_at), Eip75B (1635393_s_at), elB (1631207_at), fru (1641338_at, 1632859_a_at), ImpE1 (1631375_a_at), lola (1633089_a_at, 1635096_at), Mmp1 (1625761_a_at), mod(mdg4) (1627953_at, 1638041_at), toy (1633094_a_at), and Trl (1635305_s_at). See Dataset S2 for a full listing of deregulated genes with multiple probe sets, and their relative expression in ab or scrib −+ab tissue. A cross (X) denotes genes that were also identified from the ChIP-Seq analysis as potential Ab targets in either the ab alone, and/or scrib1+ab samples. See Dataset S4 for ChIP-Seq genome alignments for these genes.
To identify genes that could be direct targets of Ab regulation, we performed ChIP-Seq after pulling down Ab-associated chromatin from ab alone expressing mosaic discs and scrib −+ab mosaic tissue. The Ab antibody used for the pull-down has been widely used in the literature [17], [19], [20], and showed good specificity for Ab in eye/antennal disc tissue, as determined by reduced staining of endogenous Ab protein in ab mutant clones, and increased staining in ab over-expressing clones (Figure S4). Peak enrichments were identified by comparing each sample to input DNA controls (see Materials and Methods). Reflecting the large number of deregulated genes identified from the array, there were many peaks associated with Ab in both contexts ( Figure 2B and Dataset S1). In the ab alone sample, 8881 peaks were identified, associated with the transcriptional start site or introns of 2582 genes; whilst in the scrib −+ab tumourigenic sample, 10,892 Ab binding regions were identified, associated with 2746 genes. Validating these data, ChIP and quantitative real-time PCR for 10 candidate genes were consistent with the ChIP-Seq results (Figure S5), and there was also a high correlation between the ab and scrib−+ab samples (a correlation coefficient of ∼0.86). Of the potential target genes, 2025 were shared between the two samples, whilst 661 new binding site peaks, associated with 721 genes (some peaks overlapped more than one gene), were unique to the scrib−+ab tumourigenic sample ( Figure 2B ). Shared target genes were enriched for organismal development-related gene ontologies (GO), including “cell differentiation” (1.42E-70), “imaginal disc development” (2.39E-64), “imaginal disc morphogenesis” (1.92E-52) and “appendage development” (1.03E-42); whilst the 721 scrib−+ab unique genes, were enriched for the GO of “microtubule cytoskeleton” organization (4.58E-06) ( Figure 2D and Dataset S3).
DNA recognition sequences for Ab have previously been suggested from its isolation as a protein capable of binding to the Engrailed binding site [15], and more recently through a bacterial one-hybrid study that defined a consensus Ab binding sequence [16], however, in vivo we observed no enrichments for these motifs amongst the Ab peak sequences (data not shown). Instead, the most highly represented motif amongst the most significant Ab peaks (irrespective of genomic location) from the ab overexpression sample, exhibited significant similarity to the recognition sequence for another BTB-ZF protein and transcriptional activator, the Drosophila GAGA factor, Trithorax-like (Trl) (Figure S6). Other motifs identified within the sequences associated with the most significant Ab peaks exhibited significant similarity to recognition sequences for Scalloped (Sd), which functions with Yki to activate target genes downstream of Hippo pathway signalling [reviewed in 10], and Brinker (Brk), which is a repressor downstream of the Dpp pathway [reviewed in 21]. We also searched the Ab peak sequences within the promoter regions or introns of potential target genes, for known transcription factor recognition site motifs. This approach also identified the Trl recognition sequence as one of the most highly represented motifs in peaks common to both the ab alone and scrib −+ab samples (Table S1). Recognition sequences for the mammalian proteins MZF1 (similar to Drosophila Crooked legs (Crol), a zinc finger, ecdysone-induced gene that is also required for the expression of ecdysone response genes [22]) and VDR (similar to Drosophila hormone receptor HR96 and the ecdysone receptor EcR) were also identified, although the relevance of these sites for Drosophila proteins is not yet clear. In contrast, within peaks unique to the scrib −+ab sample (and not within the common peaks) there was significant enrichment of binding sites for AP1 (the Jun/Fos transcription factor complex that acts downstream of the JNK signalling pathway), and REL and NF-KB (transcription factors that act downstream of the Toll-like receptor inflammatory signalling pathway), suggesting that new Ab target genes could be generated through the activation of JNK and associated inflammation pathways within the tumourigenic context.
Prioritisation of Ab deregulated genes
To prioritise these data and focus upon those genes that could be transcriptional regulated by Ab and also critically required for tumour formation, we first removed from consideration all genes identified from the ChIP-Seq results that were not represented by probes on the array, and then combined the results from both analyses to identify those genes that were both associated with Ab peaks and differentially expressed from the microarray analysis ( Figure 2C ). This revealed that, for the ab alone sample, 27% of the differentially expressed genes (687 of 2511 genes), and for the scrib −+ab sample, 31% of the (915 of 2987 genes), were associated with Ab peaks, and thus potentially defined primary targets of Ab-mediated regulation.
To identify potential direct targets of Ab that could be key to promoting tumour development, we focussed upon those targets that were both deregulated and associated with Ab peaks in the scrib −+ab tumour sample. Of these, there were two classes of genes that were also associated with Ab peaks in the non-tumour samples, and either deregulated in both (Class 1), or just deregulated in the tumour sample alone (Class 2); and two further classes of genes only associated with Ab peaks in the tumour sample, but either also deregulated in both tumour and non-tumour (Class 3), or only deregulated in the tumour alone (Class 4). The largest of the four classes consisted of shared target genes deregulated in both tumour and non-tumour samples (Class 1, 450 genes), or in the scrib −+ab sample alone (Class 2, 269 genes), whilst relatively few potential new targets of Ab (which were also deregulated genes) were generated in the tumour sample (116 genes in Class 3, and 80 genes in Class 4). In contrast, 2072 genes deregulated in the tumour were likely to be either secondary downstream targets of ab or targets deregulated by the loss of scrib, since they were not associated with Ab peaks (Class 5, 1925 genes), or were only associated with Ab peaks in the ab overexpression sample alone (Class 6, 147 genes).
Classes 3 and 4 did not exhibit significant GO enrichments, however, amongst the two primary classes of potential Ab target genes (Class 1 and 2) GO categories were identified that were involved in all aspects of tumour formation, from cell fate/differentiation, cell survival/growth/proliferation, and cell migration/invasion ( Figure 2D , and Dataset S3). In contrast, genes deregulated, but not associated with Ab peaks in the tumour (Classes 5 and 6) did not show these GO enrichments. A heat map depicting the relative expression levels of selected genes from Classes 1 to 6 is shown in Figure 2E (see Dataset S4 for ChIP-Seq peak alignments to the genome for the Class 1–4 targets depicted in this figure). The functional significance of these genes will be elaborated upon below.
Ab binding is associated with the repression of multiple regulators of development and cell fate
The strongest GO enrichments amongst Ab targets included “multicellular organismal development” (1.76E-40 in Class 1 and 9.83E-22 in Class 2), “cell differentiation” (1.44E-28 in Class 1 and 7.65E-09 in Class 2) and “eye development” (3.72E-12 in Class 1) ( Figure 2D ). Amongst these targets were particular enrichments for ecdysone-response genes, other developmental genes involved with epigenetic control, Notch signalling, and the control of eye/antennal disc differentiation.
Ecdysone-response genes
Many ecdysone-response genes were identified as potential Ab targets, including Eip75B, Eip78C, broad (br), ImpL2 (all Class 1; peaks in both ab and scrib −+ab, and significantly deregulated in both), ImpE1, ftz-f1, Blimp-1 (Class 2; peaks in both ab and scrib −+ab, but significantly deregulated in scrib −+ab alone), and ImpL3 (Class 4; peaks in scrib −+ab only, and only deregulated in scrib −+ab). Confirming these data, protein levels of Br, a key ecdysone-induced gene, were substantially repressed in eye disc clones overexpressing ab (Figure S7). The involvement of Ab in the repression of ecdysone response genes is consistent with studies in the Drosophila ovary where Ab has also been shown to associate with the steroid hormone receptor coactivator, Taiman (Tai), and repress ecdysone response genes to control the timing of border cell migration [20]. Indeed, Tai is also expressed in the early 3rd instar larval eye disc ( Figure 3A ), and knockdown of tai with RNAi in scrib −+ab eye disc clones completely abrogated tumour overgrowth, restoring pupariation and resulting in the eclosion of adult flies ( Figure 3B,C ). Similar rescue of scrib −+ab tumour formation was observed by ectopically expressing a form of tai that lacks the Ab interaction domain (TaiΔB), and which has been shown to activate ecdysone response genes even in the presence of Ab [20] (Figure S8). Tai is therefore required for scrib −+ab tumour development. Furthermore, whilst the overexpression of a wild type form of tai alone in clones did not induce tumour formation ( Figure 3D ) and adult flies eclosed (data not shown), overexpression of tai in scrib mutant clones was sufficient to induce clonal overgrowth and tumour formation throughout an extended larval stage, in a similar manner, albeit with less potency, as ab overexpression ( Figure 3E–G ). Thus, whilst it is not known if Ab directly cooperates with Tai in repressing ecdysone response genes in scrib −+ab tumours, Tai is both required for ab to promote tumourigenesis, and sufficient to drive tumour formation in combination with the loss of scrib.
Figure 3. Tai is required for scrib −+ab tumour overgrowth, and sufficient to cooperate with the loss of scrib.
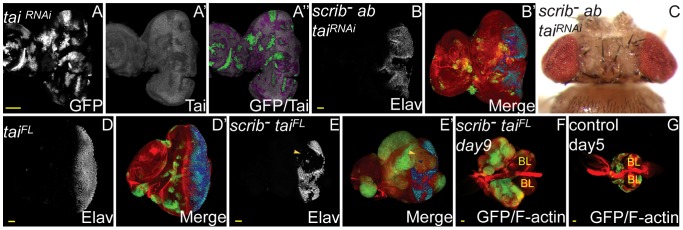
ey-FLP induced eye/antennal disc clones at 5 (A,B,D,E,G) and 9 days (F) AEL, and a dorsal view of mosaic adult eyes (C). Clones are marked by GFP (white, or green in merges). Tai (A) is white (and in merges appears magenta when overlaid with GFP), and Elav (B,D,E) is white (and blue in merges – dark blue when overlaid with GFP). F-actin is red in merges in B,D–G. Brain lobes in F,G are marked by BL. GFP (panel A), Tai (panel A′), Elav (panels B,D,E), and merges (panels A″,B′,D′,E′). (A) Expression of taiRNAi in clones reduces endogenous levels of Tai. (B–C) Expression of taiRNAi in scrib1+ab clones reduces clonal overgrowth (B) compared with scrib1+ab (see Figure 1G–J ), and results in the eclosion of adult flies (C). (D) Overexpression of taiFL in clones does not block Elav expression, nor cause clonal overgrowth throughout an extended larval stage. (E–F) Overexpression of taiFL in scrib1 clones promotes mutant tissue overgrowth and a block to Elav expression (E, arrowhead), eventually resulting in the formation of large tumours after an extended larval stage of development (F). (G) Wild type eye/antennal disc clones attached to the brain lobes at day 5, just before pupariation. Yellow scale bar = 50 µm.
Epigenetic regulators
Many epigenetic regulators were transcriptionally deregulated in both ab alone and scrib −+ab eye/antennal discs, including Pc, ph-d, Psc, mod(mdg4), psq, skd (all Class 1), ph-p (Class 2), Pcl (Class 3), and a large number of other BTB-ZF transcription factors including fru, chinmo, br, longitudinals lacking (lola) and Trl. Most of these genes were repressed by ab overexpression, with the exception of chinmo, pc, psq, skd and some isoforms of fru, lola and mod(mdg4). Other epigenetic regulators including brm, mor (both Class 5), dom and E(var)3-9 (both Class 6) were also repressed in the tumour, although they were not associated with Ab peaks in the scrib−+ab sample, and were therefore likely to be indirect targets of Ab.
Notch-regulated genes
Notch signalling is a key regulator of cell fate decisions, and many Notch-associated genes were repressed in the tumour and associated with Ab peaks, including the E(spl) region genes HLHm3, HLHmβ, m2 and mα (Class 2), the Notch transcriptional coactivator mam (Class 2) and the ligand Delta (Dl) (Class 1). Many other Notch targets, although not associated with Ab peaks, were also repressed in the tumourigenic state, including HLHm7, HLHmγ, HLHmδ (all Class 5), and m4 (Class 6). Consistent with these data, the Notch reporter E(spl)-lacZ, normally activated in the differentiating portion of the eye disc, was repressed in scrib −+ab tumours (data not shown). Furthermore, similarities between ab overexpression and Notch loss-of-function phenotypes have previously been reported [19].
Eye/antennal disc differentiation
The identification of Notch and ecdysone response genes as potential Ab targets validated our approach, however, most striking amongst the repressed Ab target genes associated with cell fate control were known regulators of eye/antennal disc differentiation, including bab2, ct, dac, dan, daughterless (da), elbow (el), eya, eyegone (eyg), hairy (h), hth, no ocelli (noc), pannier (pnr) and spineless (ss) ( Figure 2E ). This suggested that Ab could be functioning as an oncogene by maintaining cells within an undifferentiated state.
Ab promotes tumourigenesis by blocking differentiation and maintaining scrib mutant cells within a progenitor-like state
Eye disc differentiation initiates from the posterior edge of the disc in the late 2nd instar and progresses sequentially towards the anterior edge over a number of days. At the wandering third instar stage, when photoreceptor differentiation has progressed half way across the epithelium (as marked by Elav staining), the eye disc consists of a spectrum of various cellular differentiation states, from the most differentiated posterior cells to the least differentiated anterior cells. The transcription factors Hth, Eyeless (Ey) and Teashirt (Tsh) are expressed in the most anterior portion of the eye disc within the progenitor domain, whilst more posteriorly, Hth is first downregulated, followed subsequently by Ey and Tsh. The downregulation of Hth marks a transition point whereby cells begin to express Dac, Eya, Dan, Distal antenna-related (Danr), H, Da, Ato and finally Elav [reviewed in 6] ( Figure 4M ). In the antennal disc, the temporal development of the tissue is not displayed as a spatial distribution of cell fate markers at the third instar stage as it is in the eye disc, however, early to late cell fate transitions have been documented. Initial domains of Hth, Ct and Dll in the 2nd instar larvae establish the early proximo-distal axis of the antenna [23], [24], and downstream targets of these genes, including the cell fate markers Ato, Dac, Dan, Bab2, Spalt major (Salm) and Ss, are subsequently expressed throughout the 2nd and 3rd instar to elaborate the proximo-distal axis of the appendage [7]–[9] ( Figure 4M ).
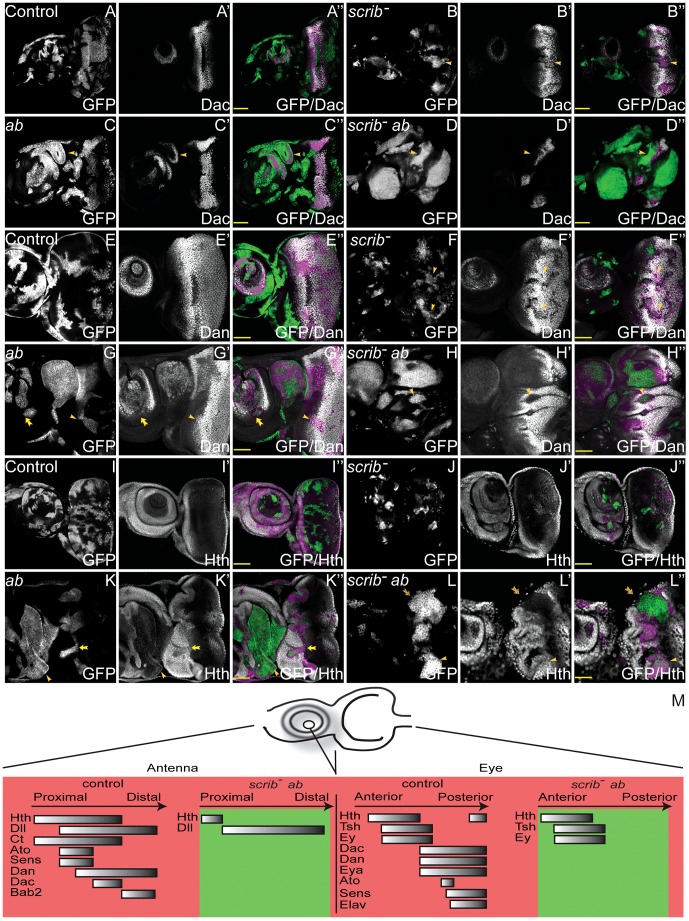
Figure 4. Overexpression of ab in scrib mutant clones promotes the retention of a progenitor-like state in the eye and antennal disc.
ey-FLP induced eye/antennal disc clones at ∼5 days AEL. Clones are marked by GFP (white, or green in merges), and cell fate is shown by the expression of Dac, Dan and Hth (white, and magenta when overlaid with GFP in the merges) in wild type control clones (A,E,I), scrib1 clones (B,F,J), ab overexpressing clones (C,G,K), and scrib1+ab clones (D,H,L). GFP (panels A–L), Dac (panels A′–D′), Dan (panels E′–H′), Hth (panels I′–L′) and merges (panels A″–L″). (A–D) Dac expression is only slightly reduced in scrib1 clones (B, yellow arrowhead), and unaffected in ab overexpressing clones in the eye disc, although ectopic Dac expressing antennal-like structures are sometimes observed in the antenna (C, arrowhead). scrib1+ab clones do not express Dac (D, arrowhead; the magenta staining observed around some clones is derived from GFP bleed-through from underlying sections. (E–H) Dan levels are reduced in scrib1 clones both in the antennal and eye disc (F, arrowheads). ab overexpressing clones do not affect Dan levels in the eye disc (G, arrowhead), although Dan is slightly repressed in the antenna (G, arrow), albeit ectopically expressed in the ectopic antennal-like structures. Dan is repressed in scrib1+ab clones (H, arrowhead). (I–L) Hth expression is generally unaffected in scrib1 clones (J). In ab overexpressing clones, levels of Hth are slightly reduced in the eye disc (K, arrow), and large clones in the antennal disc do not express Hth (K, arrowhead). In scrib1+ab clones, Hth is expressed in some clones within the eye disc (L, arrowhead), but not all clones (L, arrow), and is generally reduced in antennal disc clones (L, and data not shown). (M) Diagram summarising the expression of cell fate markers in both wild type eye/antennal discs, as well as in eye/antennal disc scrib −+ab tumours (green). In the antenna, proximal refers to the outer circular domains of the tissue, whilst distal refers to the inner, central domains. See Figures S9 and S10 for immunohistochemical images of Tsh, Ey, Eya, Ato and Sens; and Table 2 for a summary of these results. Yellow scale bar = 50 µm.
Multiple regulators of eye/antennal disc cell fate were repressed in scrib −+ab tumours. Whilst some were repressed by expression of Ab alone, most of these were substantially further repressed in combination with the absence of scrib. Potential direct targets of Ab involved in regulating cell fate in the eye/antennal disc, and repressed in the tumour state, included dan, eyg el, h and noc (Class 1), hth, dac, eya, bab2, pnr, ss (Class 2), and da (Class 4). To further validate these results we examined the expression domains of the different cell fate regulators in the tumours. We had already established that scrib −+ab tumours failed to express Elav in the eye disc ( Figure 1G ), however, examination of other cell fate markers, revealed that Dac, Dan, Eya, Sens and Ato were also repressed within the overgrowing eye disc tumour ( Figure 4A–H and Figure S9). Importantly, all of these proteins (with the exception of Ato, which was also decreased in scrib mutant and ab-overexpressing clones) were not strongly downregulated in either scrib mutant clones alone, nor in ab-expressing clones alone, but only in cooperation with both genetic lesions (summarised in Table 2 ), thus validating the results from the expression array. In contrast, the expression of the cell fate markers that define earlier states, including Hth, Tsh and Ey were relatively unaffected in scrib −+ab eye disc tumours, despite their domains of expression being enlarged and warped due to the growth of the tumour ( Figure 4I–L and Figure S10), and with the exception of Hth (see below), were not identified as Ab targets. Furthermore, Hth, Tsh and Ey were all downregulated in more posterior tumour cells indicating that they were still being subject to their normal mode of repression. scrib− clones exhibited only minor perturbations in Hth, Tsh or Ey expression, whereas ab-expressing clones showed mildly reduced Hth and Ey, and slightly upregulated Tsh, expression levels.
Table 2. Expression of cell fate regulators in eye/antennal disc clones.
| scrib− | abrupt | scrib−+abrupt | |
| Eye | |||
| Elav | Disrupted and slightly reduced | Unaffected although spacing disrupted | Repressed |
| Ato | Disrupted and slightly reduced | Reduced and delayed | Repressed |
| Sens | Disrupted and slightly reduced | Unaffected | Repressed |
| Dac | Slightly reduced | Unaffected | Repressed |
| Dan | Slightly reduced | Unaffected | Repressed |
| Eya | Unaffected | Unaffected | Repressed |
| Hth | Unaffected | Slightly reduced, especially posteriorly | Variable, but generally unaffected |
| Tsh | Generally unaffected, although some ectopic expression extends posteriorly in large disorganised clones | Increased | Unaffected |
| Ey | Slightly reduced | Slightly reduced | Unaffected |
| Antenna | |||
| Ato | - | Repressed | Repressed |
| Sens | Reduced | Repressed | Repressed |
| Dac | Slightly reduced | Ectopic in ectopic antennae-like structures | Repressed |
| Dan | Slightly reduced | Slightly reduced, but also ectopic in ectopic antennae-like structures | Repressed |
| Ct | Unaffected | Unaffected in proximal region but repressed more distally and in neck region | Repressed |
| Bab2 | Unaffected | Slightly reduced, but also ectopic in ectopic antennae-like structures | Repressed |
| Hth | Unaffected | Repressed | Variable, but often repressed |
| Dll | Unaffected | Ectopic in ectopic antennae-like structures | Unaffected and ectopic in antennal-like overgrowths |
In scrib −+ab antennal disc tumours, most of the tumour tissue expressed Dll, whilst Hth and Ct expression was repressed ( Figure 4I–L , Figure S10 and data not shown). Furthermore, the expression of subsequent cell fate markers expressed along the proximo-distal axis, including Dan (Class 1), Bab2 and Dac (Class 2), as well as Sens and Ato, were also repressed within the tumours ( Figure 4A–H , Figure S9 and Figure S10). Their expression was only slightly perturbed in scrib mutant clones, whilst ab-expressing clones alone also repressed most of these markers, with the exception of Dac (summarised in Table 2 ). The repression of Hth in the Dll-positive tumour mass within the antenna suggested that the tissue was transformed to a more leg-like state [25], and, consistent with this, the HOX genes Antennapedia (Antp) and labial (lab) were also upregulated by ab overexpression ( Figure 2E and Dataset S1).
The data therefore suggest that whilst scrib −+ab eye/antennal disc tumours are not homogeneous, and consist of a diverse population of cells, they are characterised by the maintenance of an earlier progenitor-like cell state through the continual overgrowth of tissue that, in the eye disc, fails to transition to the expression of Dac, Eya, Ato and Elav, and in the antennal disc, fails to express differentiation markers downstream of Dll that define the elaboration of the appendage along the proximo-distal axis (summarised in Figure 4M and Table 2 ).
The endogenous expression of Ab overlaps that of the progenitor state transcription factor Hth, but Hth is neither sufficient nor required for Ab-mediated tumour overgrowth
The capacity of Ab to maintain cells within a progenitor-like state suggested that its function might be linked to the eye disc progenitor state transcription factor, Hth. Indeed, the endogenous expression of Ab in the eye disc mirrored the expression of Hth ( Figure 5A ), and its downregulation in the eye disc heralds the beginning of Dac and Eya expression. Although our analysis of scrib −+ab tumours indicated that Hth expression was not maintained in the tumours, and in fact was repressed in the antennal disc, it was still possible that Hth might be sufficient for tumour formation in combination with scrib mutants or required for scrib −+ab tumour overgrowth.
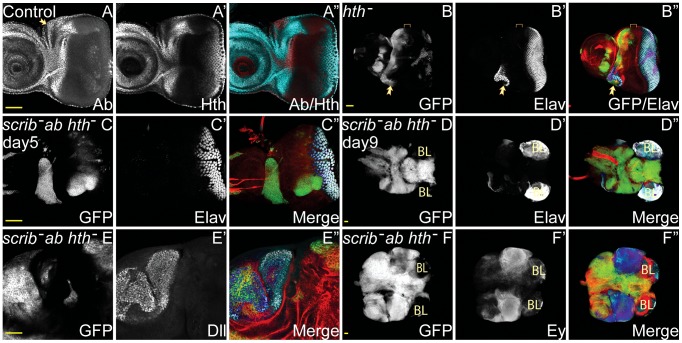
Figure 5. Hth is not essential for scrib −+ab tumour overgrowth.
ey-FLP induced eye/antennal disc clones at 5 (A–C) and 9 (D–F) days AEL. Clones are marked by GFP (white, or green in merges), and cell fate markers (Ab, Hth, Elav, Ey and Dll) are white (or blue in the merges, changing to dark blue when overlaid with GFP, except Ab, which is red in the merge in A). F-actin is shown in red in the merges of B–F. Brain lobes in D,F are marked by BL. Ab (panel A), Hth (panel A′), GFP (panels B–F), Elav (panels B′–D′), Dll (E′), Ey (F′), and merges (A″–F″). (A) Control discs show the endogenous expression of Ab and Hth in the eye/antennal disc, which overlap, except in the centre of the antennal disc and in the dorsally located ocelli (A, arrow), which express Ab but not Hth, and in the posterior part of the eye disc, which expresses Hth but not Ab. (B) hthP2 clones are absent from the progenitor domain (bracketed), although overgrowth is sometimes observed within the neck region between the eye and antennal disc, and ectopic domains of Elav expression are sometimes generated within and adjacent to these mutant clones (B, arrow). (C–F) The overexpression of ab in scrib1 hthP2 double mutant clones promotes overgrowth of clonal tissue that does not express Elav (C), and fuses with the brain lobes (BL) that are Elav positive (D). The expression of both Dll (E) and Ey (F) is maintained within the tumours. Yellow scale bar = 50 µm.
To determine if Hth was sufficient to cooperate with the loss of scrib, we ectopically expressed Hth in scrib mutant clones. However, although this promoted overgrowth of the tumour tissue and pupal lethality, it did not result in a block to pupariation and massive tumour overgrowth throughout an extended larval stage of development, indicating that Hth could not substitute for Ab in a tumour-promoting role (Figure S11). Conversely, to test for whether Hth was required for scrib −+ab tumour overgrowth, we overexpressed ab in scrib− hth− double mutant clones and assayed for tumour formation. Examination of tumour samples at day 5 revealed that overgrowth was initially confined to regions within the neck and the ventral portion of the eye disc ( Figure 5C ), regions that correspond to tissue that is least dependent upon hth for cell survival and/or proliferation, and are associated with a role for hth in repressing ventral eye formation [26], [27]. Indeed, this tissue continued to grow in scrib− hth−+ab tumours, so that whilst overgrowth was substantially delayed compared to scrib −+ab tumours, massive and invasive tumour masses eventually overtook the larvae ( Figure 5D ). These tumours consisted of characteristic Dll-positive tumour masses within the antennal region, and Ey-positive tumour tissue within the eye disc ( Figure 5E,F ). Thus, hth is neither sufficient nor absolutely required for Ab-driven tumour formation.
Yki promotes overgrowth of scrib−+ab tumours
To identify potential targets of Ab that could be important for maintaining tumour overgrowth, we analysed Class 1 and 2 genes for GO enrichments associated with cell survival and proliferation. Importantly, the GO categories of “cell death” (4.84 E-03), “growth” (1.09 E-06) and “cell proliferation” (2.03 E-05) were all enriched within Class 1 targets, which were genes associated with Ab peaks and deregulated in both ab alone and scrib −+ab tumours. Amongst potential cell death targets, the pro-survival Bcl2 homologue Buffy was upregulated, and the cell death inducer Hid (W) was downregulated by ab overexpression. Furthermore, klumpfuss (klu) and echinus (ec) that promote cell death in the pupal retina [28]–[30], were also downregulated by ab. Notable Class 1 ab targets involved in cell growth and proliferation included the cell growth and G1-S phase driver Cdk4 (upregulated), the inhibitor of the PI3K pathway Pten (downregulated) and a number of Hippo pathway components and/or targets, including expanded (ex), fat (ft), thread (th/DIAP1) and diminutive (dm), the Drosophila Myc gene. Whilst th, a survival-promoting effector of Yki activity, was repressed by ab overexpression, which was confirmed by immuno-histochemical analysis of ab-expressing larval discs (Figure S12), the Yki targets dm and ex were upregulated upon ab overexpression, and ft and hippo (Class 5), two negative tissue growth components of the Hippo pathway, were repressed. Thus, although multiple genes may contribute to ab-driven tumour overgrowth, ab-mediated impairment to the Hippo pathway could be a key factor.
A role for the Hippo pathway in scrib −+ab tumour overgrowth was tested by knocking down yki, a critical downstream transcriptional effector of impaired Hippo pathway signalling. Strikingly, and unlike loss of hth, this substantially restrained scrib −+ab tumour overgrowth and restored pupariation to the tumour-bearing larvae ( Figure 6A–F ). To determine whether the rescue in tumour overgrowth was accompanied by a restoration to differentiation we examined the expression of cell fate markers. This revealed that whilst knockdown of yki did not restore Elav and Eya expression to scrib−+ab tumours, Dac levels were substantially increased ( Figure 6G,H ). It was therefore possible that the increased levels of Dac upon yki knockdown could account for the suppression of tumour overgrowth. However, overexpressing dac within scrib −+ab tumours, using a dac transgene, failed to restrain tumour overgrowth and restore pupariation (Figure S13). Thus, the downregulation of Dac is not a key requirement for continual tumour overgrowth. Furthermore, scrib−+ab tumour cells expressing ykiRNAi, could still be observed with mesenchymal morphology between the brain lobes (Figure S14), suggesting that whilst Yki activity is required for tumour overgrowth, it is not an essential mediator of tumour cell migration and invasion.
Figure 6. Yki is required for scrib −+ab tumour overgrowth.
ey-FLP induced eye/antennal disc clones marked by GFP (white, or green in merges). Elav, Dac and Eya are in white (blue in merged images, changing to dark blue when overlaid with GFP), and F-actin for cell morphology is in red. Brain lobes are labeled BL. GFP (panels A–C,G,H), Elav (panels A′–C′), Dac (panel G′), Eya (panel H′) and merges (panels A″–C″,D,E,G″,H″). (A) ykiRNAi-expressing clones. (B) Clones overexpressing ykiRNAi+ab are similar to ab overexpressing clones. (C–E) Expressing ykiRNAi in scrib1+ab tumours does not restore Elav expression to the tumour cells (C, arrowhead), however, tumour overgrowth is substantially reduced (E, compared to scrib1+ab tumours in D), and larvae pupate instead of entering an extended larval stage. (F) Quantification of percentage of scrib1+ab and scrib1+ab+ykiRNAi tumour-bearing larvae that had pupated by 9 days AEL. (G,H) Expressing ykiRNAi in scrib1+ab tumours restores Dac expression in the clones (G, arrowhead) but Eya levels remain reduced (H, arrowhead). Yellow scale bar = 50 µm.
Impaired Hippo signalling is sufficient to cooperate with Ab and promote tumour overgrowth
The ChIP-Seq and expression array analysis had indicated that ab overexpression was capable of modulating Hippo pathway activity, however, scrib mutant cells also express Hippo pathway reporters, and ectopically proliferate in a Yki-dependent manner [31]. Thus both the overexpression of ab and the loss of scrib each had the potential to promote Yki activity, and either of these could be crucial in driving cooperative tumour overgrowth. To discern which of the two was more critical in mediating cooperation we tested for whether knockdown of wts in either ab-overexpressing clones or scrib mutant clones, was sufficient to elicit cooperative tumour overgrowth throughout an extended larval stage. Whilst knockdown of wts alone in clones did not perturb Elav expression and larvae pupated normally ( Figure 7A ), ectopically expressing ab in wtsRNAi clones was sufficient to block pupariation of larvae and promote massive overgrowth of the eye/antennal discs. Examination of Elav expression indicated that although at day 5 some wtsRNAi+ab clones were still observed to express Elav, the overgrown clonal tissue that ensued was entirely composed of Elav-negative tissue ( Figure 7B,C ). Similar cooperation was observed when ab was ectopically expressed within wtsX1 mutant clones (data not shown). In contrast, although knockdown of wts in scrib mutant clones enhanced scrib mutant tissue overgrowth causing pupal lethality, it was not sufficient to completely block Elav expression and drive cooperative tumour overgrowth throughout an extended larval stage of development ( Figure 7D and data not shown). Consistent with this interpretation, scrib-mediated impairment to Hippo signalling has been shown to be atypical Protein Kinase C (aPKC)-dependent, since it is rescued by expressing a kinase dead dominant negative (DN) version of aPKC (aPKCDN) within the mutant tissue [31], and similarly, expressing aPKCDN in scrib −+ab tumours also curtailed tumour overgrowth (Figure S15). Thus, whilst ab overexpression alone may impair Hippo pathway signalling, the deregulation of the Hippo pathway induced by the absence of scrib is likely to be a key factor in promoting susceptibility to Ab-driven tumour formation.
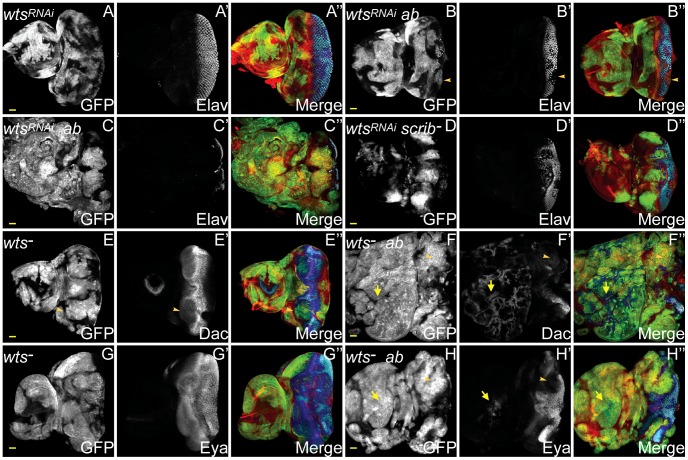
Figure 7. ab cooperates with impaired Hippo pathway signalling to drive tumour overgrowth.
ey-FLP induced eye/antennal disc clones marked by GFP (white, or green in merges). The cell fate markers Elav, Dac and Eya are shown in white (blue in merged images, changing to dark blue when overlaid with GFP). F-actin for cell morphology is in red. GFP (panels A–H), Elav (panels A′–D′), Dac (panels E′F′), Eya (panels G′H′), and merges (panels A″–H″). (A) wtsRNAi-expressing clones exhibit the normal pattern of Elav in the eye disc. (B) Coexpressing wtsRNAi+ab in clones decreases Elav expression in some (B, arrowhead), but not all clones, and some larvae enter an extended larval stage, during which massive overgrowth of Elav-negative tissue ensues (C). (D) Overexpressing wtsRNAi in scrib mutant clones increases scrib mutant clone size and reduces Elav expression, but does not result in cooperative tumour overgrowth throughout an extended larval stage. (E) wtsX1 clones exhibit mildly reduced Dac levels in anterior localised clonal tissue in the eye (E, arrowhead), and also reduced expression in the antennal disc. (F) In wtsX1+ab clones, overgrowing tissue within the eye disc does not express Dac (F, arrowhead), although extensive ectopic Dac expression is observed throughout the antennal disc (F, arrow). (G) Eya expression in wtsX1 clones is largely unperturbed. (H) wtsX1+ab clones overgrow in the eye disc, and do not express Eya (H, arrowhead), however, occasional Eya positive tissue is sometimes observed within the antennal disc region (H, arrow). Yellow scale bar = 50 µm.
To determine whether expressing ab in wts mutant clones produced tumours that were similar to scrib−+ab tumours, we examined the expression of different cell fate markers in wts−+ab clones. wts mutant clones differentiated normally, apart from a mild downregulation of Dac ( Figure 7E,G and Figure S16). However, although some of the wts−+ab clonal tissue at day 5 expressed normal, or only mildly reduced, levels of Dac, Dan and Eya (data not shown), in older larvae, the overgrown wts−+ab tumours consisted predominantly of eye disc progenitor-like tissue that did not express Dac, Dan or Eya, and antennal-like tissue that ectopically expressed Dll and Dac ( Figure 7F,H and Figure S16). Thus, the wts−+ab tumours retained a progenitor-like state that was similar to scrib −+ab tumours, with the exception that Dac expression was retained within the antennal domain of the wts−-derived tumours, but not in the scrib−-derived tumours. Furthermore, wts−+ab tumours were characterised by the generation of huge, highly-folded epithelial sheets of tissue that remained distinct and did not fuse with the brain lobes, thus indicating that cooperation between wts −+ab was unable to reproduce the invasive properties of scrib −+ab tumours.
Ab targets are involved in migration and invasion, but Ab can not promote invasion without JNK signalling
The invasive properties of RasACT and NotchACT-driven tumours are dependent upon JNK signalling, since blocking Drosophila JNK (Basket (Bsk)), within either scrib −+RasACT or scrib −+NotchACT tumours prevents tumour cell invasion [14], [32], [33]. The expression array of scrib −+ab tumours indicated that JNK signalling was also likely to be active within these tumours, as evidenced by the upregulation of known JNK-regulated genes such as Matrix metalloproteinase 1 (Mmp1) and scarface (scaf) [32], [34], which were also identified as potential Ab targets (Class 1 and 2, respectively). In addition, GO analysis of Class 1 and 2 targets of Ab indicated a significant enrichment for genes within the category of “locomotion” (6.67 E-12 in Class 1 and 2.72 E-07 in Class 2). In the scrib −+ab tumours these included wunen, wunen2 and Trapped in endoderm 1 (Tre1) that are known to promote germ cell migration, and jing and PDGF- and VEGF-related factor 1 (Pvf1) that are involved in border cell migration [reviewed in 35]. Thus, the data suggested that Ab could directly contribute to the invasive capability of scrib −+ab tumour cells by controlling the expression of migration-associated genes, including JNK targets such as Mmp1.
Using the JNK pathway reporter, misshapen (msn)-lacZ [36], we first determined whether JNK signalling was active in scrib−+ab tumours. Indeed, although ab overexpressing clones alone did not upregulate msn-lacZ expression ( Figure 8A,B ), the reporter was strongly activated within scrib−+ab tumours, most notably within basal portions of the tumour and in cells that appeared to be migrating between the brain lobes, consistent with a role for JNK in promoting invasion ( Figure 8C,D ). Immunohistochemical analysis also indicated that the JNK target, Mmp1, was ectopically expressed within scrib −+ab tumours ( Figure 8E,F ), and, in agreement with the expression array, Mmp1 levels were also slightly elevated in ab alone overexpressing clones ( Figure 8G ). To next determine whether Ab was capable of promoting invasion, independent of JNK signalling, we then examined scrib −+ab tumours in which JNK signalling was blocked, using a dominant negative JNK transgene (bskDN). Strikingly, the expression of bskDN in scrib −+ab tumours prevented the fusion of the discs to one another and to the brain lobes, thus demonstrating a critical role for JNK in mediating the invasive properties of the tumours ( Figure 8H ). To confirm the benign nature of the overgrowths we used live cell imaging to monitor the growth of the tumours over time. The scrib −+ab tumour cells were highly motile with individual cells moving rapidly into the brain (Movie S1). In contrast, the scrib −+ab+bskDN tumours remained compact, despite their massive growth throughout an extended larval stage of development (Movie S2). Thus, although ab overexpression may contribute to the invasive properties of the tumours by promoting the expression of targets such as Mmp1, it is not sufficient to promote tumour invasion in the absence of JNK signalling. In this regard, ab-driven tumours resemble RasACT and NotchACT-driven tumours, although, interestingly, expressing bskDN in RasACT and NotchACT tumours additionally restores pupariation to the tumour-bearing larvae, thus curtailing tumour overgrowth [14], [32], [33]. In contrast, the formation of massive, albeit benign, scrib−+ab+bskDN tumours during an extended larval stage, indicated that ab blocks pupariation and promotes scrib − tumour overgrowth, even in the absence of JNK signalling.
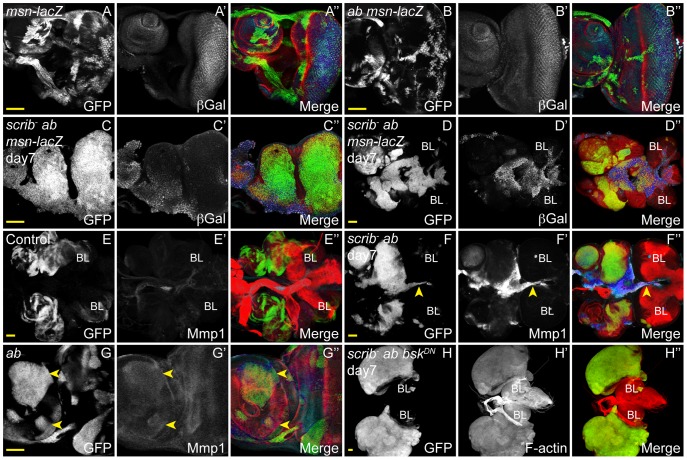
Figure 8. JNK signalling in scrib−+ab tumours is required for invasion, but not tumour overgrowth.
ey-FLP induced eye/antennal disc clones at 5 (A,B,E,G) and 7 days (C,D,F,H) AEL. Clones are marked by GFP (white, or green in merges), and JNK signalling is indicated by β-Gal expression from the msn06946-lacZ enhancer trap or Mmp1 expression (white, and blue in the merges). Tissue morphology is shown by F-actin (red in merges). Brain lobes in D,E,F,H are marked by BL. GFP (panels A–H), β-Gal (A′–D′), Mmp1 (E′–G′), F-actin (H′) and merges (A″–H″). (A) Control clones show the normal pattern of msn-lacZ expression in the eye antennal disc. (B) Clones overexpressing ab do not alter the normal pattern of msn-lacZ expression. (C,D) scrib1+ab clones show ectopic expression of msn-lacZ in some cells (C), including those that are fusing with the brain lobes (D). (E,F) Mmp1 levels are elevated in scrib 1+ab tumour cells migrating between the brain lobes (F, arrowhead), compared to control eye discs and brain lobes (E). (G) Mmp1 levels are slightly elevated in some ab-expressing clones (G, arrowheads). (H) scrib1+ab+bskDN clones massively overgrow similar to scrib 1+ab tumours, however, the eye/antennal discs do not fuse with each other or with the brain lobes, and the tumour cells show no evidence of invasive migration between the brain lobes. Yellow scale bar = 50 µm.
Overview of cooperating pathways in scrib−+ab tumours
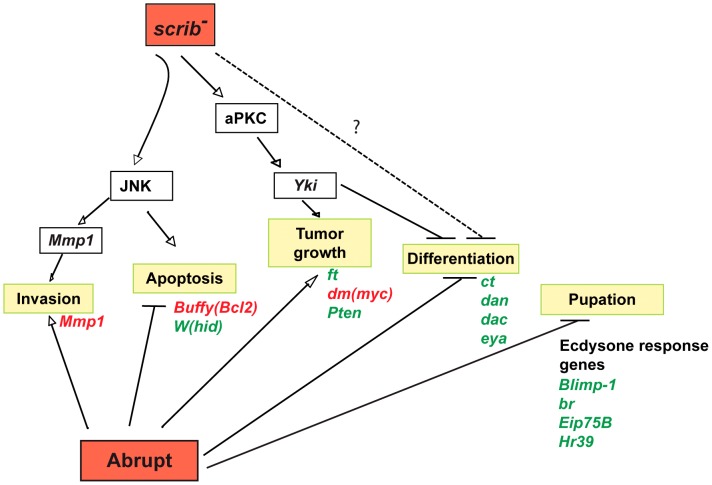
In summary, this comprehensive analysis has identified multiple modes through which the overexpression of ab and loss of scrib cooperate to promote the retention of a progenitor-like cell state and the formation of invasive tumours ( Figure 9 ). The overexpression of ab modulates the expression of a significant proportion of the genome to block differentiation, repress ecdysone signalling (potentially through direct association with the ecdysone receptor coactivator Tai), and promote cell survival and proliferation; whilst loss of scrib induces aPKC-dependent Yki activity to promote tumour overgrowth, and JNK signalling to promote invasion. Indeed, deregulation of the Hippo pathway is sufficient to cooperate with Ab and drive the formation of large, albeit benign, tumours, although the deregulation of additional pathways in scrib mutants may contribute to the complete spectrum of overgrowth and differentiation defects observed in scrib−+ab tumours.
Figure 9. Model illustrating the pathways involved in scrib −+ab cooperative tumour overgrowth.
Ab cooperates with the loss of scrib to form invasive tumours through modulating the expression of multiple genes involved in all aspects of tumour formation. Potential targets of Ab include genes involved with blocking apoptosis and promoting tumour overgrowth (eg. hid, Buffy, ft, dm, Pten), genes required for eye/antennal disc differentiation (eg. ct, dac, eya, dan), genes involved in promoting cell invasion (eg. Mmp1), and genes involved in the ecdysone-induced pupariation response (eg. Blimp-1, br, Eip75E, Hr39). Whilst not shown on the figure, the steroid hormone receptor coactivator Tai is both required for ab-driven tumour overgrowth and sufficient to cooperate with the loss of scrib, consistent with the possibility that Ab acts in concert with Tai to drive tumour formation. Loss of scrib activates JNK-mediated apoptosis, however, ab overexpression abrogates the apoptotic response, thereby unmasking a key role for JNK in promoting tumour cell migration and invasion through the expression of JNK-induced genes such as Mmp1. Loss of scrib also promotes aPKC-dependent Yki activity that is required and sufficient to cooperate with Ab by impairing differentiation and promoting tumour overgrowth. Other pathways deregulated in scrib mutants may participate in the tumour phenotype and promote the full spectrum of differentiation defects seen in scrib −+ab tumours (indicated by the dotted blocking arrow and question mark), such as Dac repression in the antenna. Green = downregulated genes, and red = upregulated genes.
Discussion
In this study we show in Drosophila that; 1) the BTB-ZF transcription factor Abrupt acts as a potent oncogene when combined with the loss of scrib in the eye/antennal disc; 2) scrib−+ab epithelial tumours are associated with an earlier developmental state; 3) impaired Hippo signalling in scrib mutants is a key factor in mediating cooperative overgrowth with ab overexpression; and 4) ab can promote tumour overgrowth, but not invasion, independently of JNK signalling. Abrupt thus joins a growing list of BTB-ZF proteins with potent oncogenic potential, including another Drosophila member of the family, Lola, which cooperates with the ectopic expression of the Notch ligand, Delta, to form metastatic tumours in Drosophila [37], as well as numerous mammalian BTB-ZF proteins that are also implicated as human oncogenes [reviewed in 38].
Underpinning the conclusions of this study is a description of the transcriptional changes and potential direct targets of Ab in scrib−+ab tumourigenesis. This has revealed a complex picture of widespread transcriptional deregulation upon Ab overexpression, and a multitude of potential Ab target genes. The in vivo binding sequence for Ab is not known, however, the most highly enriched motif from the ChIP-Seq has similarity to the binding sequence of another BTB-ZF protein, Trl. Whether this reflects a predilection for Ab to bind a similar recognition sequence as Trl, or whether Ab competes or cooperates with Trl to regulate transcription, will require further analysis. Interestingly, mammalian BTB-ZF proteins can heterodimerise, and are also known to associate with histone deacetylases (HDACs) or other corepressors to control of cell fate through transcriptional repression [reviewed in 39]. It is not known if Ab associates with HDACs, however, there was not a clear bias in Ab targets for genes that were specifically downregulated in the scrib−+ab tumour. Nevertheless, many of the most notable Ab targets were repressed in the tumourigenic state, and this included numerous Notch pathway targets and ecdysone response genes, as well as a suite of transcription factors responsible for orchestrating the differentiation of the eye and antennal disc.
Hormone signalling and developmental timing in tumourigenesis
Repressed ecdysone response genes were enriched amongst potential Ab targets, consistent with the known capacity of Ab to directly associate with the steroid hormone receptor coactivator Tai and repress the expression of ecdysone response genes in the Drosophila ovary [20]. Indeed, we show that Tai is both required for Ab to exert its oncogenic effect in the eye/antennal disc, and sufficient when overexpressed to cooperate with scrib − and promote the formation of large tumours throughout an extended larval stage. Thus, it is possible that Ab also associates with Tai in its oncogenic role, although further work will be required to determine if this is the case. The human homologue of Tai, SRC3/AIB1, is also a transcriptional coactivator of steroid hormone receptors and an oncogene [reviewed in 40], although whether it associates with BTB-ZF proteins is not yet known.
The repression of multiple ecdysone-response genes within scrib−+ab tumours is striking, yet whether this plays a cell autonomous role in promoting tumour overgrowth is unclear. However, the tumour-bearing larvae also fail to undergo an ecdysone-induced pupariation response, and this non-cell autonomous block in organismal development functions to extend the time frame available for continual tumour overgrowth. Indeed, an extended larval stage is a phenotype elicited by both neoplastic tumour overgrowth [41] and tissue damage [42], whereby it functions to give time for tissue regeneration before initiating pupariation. A key factor in mediating this delay is Drosophila insulin-like peptide 8 (Dilp8), which is secreted from tumours or damaged tissues, and acts as a diffusible signal to repress the biosynthesis of ecdysone [43], [44]. dilp8 expression can be induced by JNK signalling [43], which is consistent with previous studies indicating that JNK signalling within scrib−+RasACT and scrib−+NotchACT tumours is essential for the failure of the tumour-bearing larvae to pupate [14], [32], [33]. In contrast, we show here that ab-driven tumour overgrowth throughout an extended larval stage does not require JNK signalling. Possibly this reflects a capacity of Ab to directly or indirectly regulate dilp8 expression, independent of JNK. Indeed, the expression array indicated that dilp8 was upregulated in both ab-expressing eye/antennal discs, and in scrib −+ab tumours, although the ChIP analysis indicated that dilp8 was only associated with Ab peaks in the ab alone expressing sample (Class 6). Why ab-expressing larvae pupate (unlike scrib −+ab larvae), despite the elevated levels of dilp8 expression, remains to be determined. Interestingly, known endogenous functions of Ab are also associated with regulating the timing of hormone-induced developmental transitions, including the correct timing of border cell migration in the Drosophila ovary [20], and neuromuscular junction formation during metamorphosis [45]. In both contexts, and similar to its oncogenic role, Ab expression is associated with earlier developmental states, and its ectopic expression can inhibit temporal progression towards differentiation.
The role of Ab in mediating impaired differentiation during tumourigenesis
The cells of the adult eye are derived from progenitor cells within the 3rd instar eye disc that are characterised by the expression of a number of transcription factors including Hth, Tsh, Ey, ElB and Noc. The endogenous expression of Ab overlaps with Hth, and both Hth and Ab are downregulated prior to the downregulation of Tsh, Ey, ElB and Noc, and coincident with the upregulation of Dac, Eya, So and Dan. In ab-driven tumours, the expression of dac, eya and dan was blocked, as were other downstream posteriorly-expressed differentiation markers (ato, Elav, sens), thereby maintaining cells within an earlier progenitor-like state. The expression array also indicated that elB and noc were repressed in ab-driven tumours, and although these genes are normally expressed within the progenitor region, elB and noc mutants promote overgrowth of Hth-positive progenitor cells [46]. As the ChIP analysis indicated that Ab binding was associated with many of these genes, including eya, dac, dan, elB and noc, we suggest that Ab may promote the maintenance of a progenitor-like state by directly repressing many of these differentiation-promoting genes, although further work will be required to verify this hypothesis. Interestingly, the failure to transition to Eya, Dac and Dan expression and the maintenance of a progenitor-like state may be sufficient to promote over-proliferation of tumour cells in the eye disc, since not only is loss of elB and noc associated with over-proliferation of progenitor cells, but also loss of eya promotes tissue overgrowth in the eye, although this is eventually restrained through the induction of cell death [47], [48], and the ectopic expression of hth and tsh can block eya and dac expression and also promote eye disc overgrowth [49]. Ab, however, appears capable of maintaining eye disc tumour overgrowth independent of both hth and tsh, since Hth and Tsh levels are repressed in the posteriorly localised tumour cells, and ab overexpression can promote overgrowth of eye disc tumour tissue, even in the absence of hth. Thus, whilst the repression of multiple differentiation-promoting genes in ab-driven eye disc tumours might cooperate to elicit a default over-proliferative progenitor-like state, this state does not appear to be defined by simply maintaining the expression of the known progenitor state factors, Hth and Tsh.
Progenitor cells in the antennal disc are not as well defined as in the eye disc. However, overgrowing tumour tissue in the antennal region was characterised by the expression of Dll and Hth, whilst all other cell fate markers examined were repressed in the tumour, including Ct, Dan, Bab2, Ato and Sens. A number of other antennal cell fate markers, although not examined by immunohistochemistry, were also identified from the expression array as significantly repressed within the tumour including aristaless (al), brother of odd with entrails limited (bowl), danr, salm and ss. Although the 3rd instar antennal disc, unlike the eye disc, does not present itself as a spectrum of early to late cell fate states marked by the expression of different transcription factors, significant detail is known concerning the temporal development of the appendage from the embryonic stage onwards. The expression of hth and dll, defining the proximal and distal domains respectively, are one of the first divisions to be established in the developing appendage. Neither are required for each others expression [50], however, the expression of most other cell fate regulators that define the elaboration of the appendage along the proximodistal axis are dependent upon either or both of their activities [7], [9], [51]–[53]. Thus, in scrib −+ab tumours, the expression of cell fate markers downstream of dll and hth are repressed resulting in the overgrowth of antennal primordia tissue that has a defined proximo-distal axis that fails to transition towards a further differentiated state. In this regard, the tissue becomes indistinguishable from the developmentally related leg appendages in their early state. Indeed, within the tumours, most of the Dll-positive tissue does not express Hth, making the tumour tissue more characteristic of a leg-like, as opposed to an antennal-like, state. Consistent with this, the two HOX genes Antp, a repressor of hth [25], and lab were ectopically expressed within the tumours, and both are capable of transforming the antennae to a leg-like fate [54]. We propose that similar to the eye disc, this alteration in cell fate and block in expression of downstream cell fate regulators is directly mediated by Ab, since Ab binding was associated with most of the downstream genes. Whether this block is sufficient for tumour cells to be maintained within a proliferative state is not yet known. However, as in the eye disc, it is possible that multiple mechanisms cooperate to promote the full spectrum of tumour overgrowth. Furthermore, both ab overexpression and loss of bowl (which was repressed in the tumours) can induce the development of ectopic antennae within the eye/antennal disc [19], and whilst the cause of these phenomena is not yet clear, the eye/antennal disc is derived from the fusion of multiple segments, and it has been suggested that ectopic appendages might arise from reawakened appendage primordia that have been cryptically retained within the composite tissue [55]. Thus it is possible that within ab-driven tumours, multiple segmental appendage primordia could be contributing to tumour overgrowth.
In summary, parallels emerge between scrib−+ab tumour overgrowth in the eye and antennal disc, in that both are characterised by the maintenance of an earlier developmental state downstream of Hth but upstream of Dac. However, neither Hth activity, nor the downregulation of dac, appear to be essential for tumour overgrowth, thus indicating that further work is required to identify what key transcription factors define the progenitor-like state in ab-driven tumours. As many of the eye/antennal disc transcription factors targeted by Ab have human orthologues that are also implicated in cell fate regulation, organogenesis and tumourigenesis (eg. Hth (MEIS family), Dac (DACH family), Eya (EYA family), Dll (DLX family)), it is likely that this work will promote a deeper understanding of how cell fate control also influences the formation of human cancers.
The role of the Hippo pathway in maintaining the progenitor-like state
The proliferation of progenitor cells within the eye disc is yki dependent [11], and although the requirement for Yki activity in antennal disc cells has not been examined, yki is required for scrib −+ab tumour overgrowth in both the eye and antennal disc. The requirement for Yki in scrib −+ab tumours could solely reflect a basal need for Yki activity in progenitor cell proliferation, however, loss of scrib impairs Hippo pathway signalling [31], and we show here that blocking Hippo signalling is sufficient to cooperate with ab and sustain massive tumour overgrowth. Furthermore, the expression array indicated that ab overexpression may also deregulate the Hippo pathway, as can the BTB-ZF protein Trl [56], which is known to directly associate with Yki [57]. Interestingly, the mode of Hippo pathway deregulation induced by the overexpression of ab is likely to be different to that induced by the loss of scrib. The Yki targets activated in scrib mutants include CycE, DIAP1, fj-lacZ and ex-lacZ [12], [31], however, ab overexpression upregulated the Yki targets dm (Myc) and ex, but both th (DIAP1) and fj-lacZ (data not shown) were mildly repressed. As both fj and th are also targets of the JAK/STAT pathway [58], [59], Ab may additionally function to repress JAK/STAT signalling, as does another Drosophila BTB-ZF protein, Ken and barbie (Ken) [60]. Increasing complexity is being recognised in the variety of transcriptional outputs of the Hippo pathway. In the progenitor domain, Yki associates with Hth and Tsh, and instead of promoting th expression, it drives expression of the pro-survival micro-RNA bantam, which represses translation of the cell death inducer hid (W) [11]. A similar capacity could be shared by ab overexpression, and potentially it might be the bringing together of two different modes of Hippo pathway deregulation (both scrib mutant and ab overexpression dependent) that makes the combining of these two oncogenic forces so potent.
Although loss of wts was sufficient to cooperate with ab and promote massive overgrowth of undifferentiated tissue, it was not sufficient to reproduce the entire spectrum of defects in scrib −+ab tumours. The non-invasive nature of wts −+ab tumours is likely to reflect the lack of JNK pathway activity and/or the maintenance of epithelial cell polarity within the tumours. However, whether these additional defects also account for the differences in expression of cell fate regulators is not clear. Whilst overgrowth of wts −+ab tumours was characterised by the failure to express Eya and Dan, Dac was ectopically expressed within the antennal tumours. Interestingly, Dac defines the medial domain of the appendage and is one of the first markers to be expressed downstream of Hth and Dll. Thus it may be the least refractory to inhibition, relative to slightly later acting cell fate regulators. This contrasts with the eye disc in which knockdown of yki in scrib −+ab tumours restored Dac expression, but not Eya or Elav. Whilst this could indicate that, unlike the antennal disc, Dac alone is repressed by Yki activity in the eye, an alternative explanation could be that in both the antennal and eye discs Dac repression requires substantially higher levels of Yki activity than repression of the other cell fate markers. This might make Dac particularly susceptible to restoration when yki is knocked down in the tumours, and conversely, only subject to repression in scrib −+ab tumours when Yki activity is especially high.
Even though Hippo pathway mutants are not usually associated with a failure to differentiate, the potential for the Hippo pathway to elicit effects upon cell fate is not without precedence. Impaired Hippo signalling can synergise with loss of Drosophila Retinoblastoma gene (rbf) to cause dedifferentiation of photoreceptor cells in the eye disc, independent of effects on cell proliferation [61]; and in the larval brain Yki overexpression can delay differentiation of the neuroepithelia and promote overgrowth of the progenitor cells, although in this case the effects are likely to be a consequence of accelerated cell cycle progression [62]. However, most pertinent to this study, Yki overexpression throughout the eye disc is sufficient to block Eya expression and expand Hth expression [63]. Interestingly, these effects upon Hth levels were specifically linked to exceptionally high Yki activity, since it could not be reproduced by knockdown of wts alone, but only by combining wts knockdown with additional loss of ft and ex [63]. Thus, additive effects that escalate Yki activity elicit qualitatively different effects, and this is likely to be relevant to both our own analysis of ab-driven tumours, as well as more generally to understanding how cooperating pathways synergise to drive tumourigenesis.
Parallels to mammalian BTB-ZF transcription factors
There are over 40 human BTB-ZF family members, many of which are implicated in both haematopoietic and epithelial cancers, where they act as oncogenes (e.g. BCL6, ZBTB7) or tumour suppressors (e.g. PLZF, HIC1) [reviewed in 38]. They are key regulators of cell fate, most notably within the immune system [reviewed in 64], and a number of studies are also consistent with roles in regulating self-renewal and differentiation. A specific orthologue of ab is difficult to ascertain because of the low sequence conservation between Drosophila and mammalian family members, although ZFP161 exhibits the greatest amino acid sequence similarity. ZFP161 is not known to exert oncogenic activity within humans, and indeed its expression in some tumours is more consistent with a potential tumour suppressor role [65], however, Bcl6, one of the best-characterised mammalian oncogenic family members, offers striking parallels to the function of Ab. Mainly implicated in lymphomas, Bcl6 expression is also associated with epithelial cancers, and can promote self-renewal and repress differentiation of both B cells [66] and mammary epithelia [67]. A key oncogenic target of Bcl6-mediated repression in lymphomas is blimp-1, and the two proteins antagonise each other's expression to regulate lymphocyte differentiation [reviewed in 68]. Importantly, Drosophila Blimp-1 is induced by ecdysone [69], and was also identified as an Ab target, being one of the most highly repressed genes within the tumour. Bcl6 can also repress Notch signalling in Xenopus [70], similar to the repression of Notch targets by Ab in Drosophila. Although it is not yet known whether Bcl6 associates with the Tai orthologue, the activity of other BTB-ZF proteins are linked with various nuclear hormone receptors and their corepressors including NCoR and SMRT, suggesting that integration with hormone signalling pathways is a feature shared by mammalian family members. Overall, our identification of ab as an oncogene that cooperates with scrib loss of function in tumourigenesis, and analysis of cooperating pathways in Drosophila tumours, have uncovered striking parallels to mammalian tumourigenesis. It highlights the potent oncogenic potential of this class of proteins, supports prevailing views of the importance of impaired differentiation of progenitor cells as key drivers of neoplastic overgrowth, and raises the possibility that mammalian regulators of epithelial cell polarity could also act as important restraints upon the oncogenic potential of the BTB-ZF family of proteins.
Materials and Methods
Drosophila stocks
The following Drosophila stocks were used: ey-FLP1,UAS-mCD8-GFP;;Tub-GAL4,FRT82B,Tub-GAL80 [71]; y,w,hs-FLP; FRT82B,Ubi-GFP; UAS-ab55 [72]; UAS-ab79 [72]; FRT40A,ab1D [17]; UAS-bskDN [73]; UAS-chn [74]; UAS-DaPKCCAAXDN [75]; UAS-dac [76]; UAS-dl [77]; UAS-esg [78]; E(spl)m8 2.61-lacZ [79]; UAS-hth [27]; hthP2 [80]; msn06946 (msn-lacZ) [81]; UAS-numb-GFP [82]; FRT82B,scrib1 [83]; UAS-taiFL [84]; UAS-TaiDB [20]; UAS-taiRNAi (VDRC #15709); UAS-tdf (also known as apt) [85]; wtsX1 [86]; UAS-wtsRNAi (NIG #12072R-1); UAS-ykiRNAi [87].
Mosaic analysis
Clonal analysis utilised MARCM (mosaic analysis with repressible cell marker) [88] with FRT82B and ey-FLP1 to induce clones and mCD8-GFP expression to mark mutant tissue. All fly crosses were carried out at 25°C and grown on standard fly media.
F3 screen for cooperating oncogenes
A random selection of GS(mini-w+) insertions on the second chromosome (obtained from the NIG stock centre in Kyoto), carrying GAL4 sites to overexpress flanking genes, were screened for their oncogenic potential by initially crossing to w −;;TM3/TM6B flies. Male progeny of the genotype w−;+/GS(mini-w+);+/TM6B were then selected to cross to virgin w−;+/CyO;FRT82B,scrib1/TM3,Sb flies. From their progeny, w−;GS(mini-w +)/CyO;FRT82B,scrib1/TM6B male flies were then crossed to ey-FLP1,UAS-mCD8.GFP;;tub-GAL4,FRT82B,tub-GAL80/TM6B virgins to generate progeny containing scrib1 eye disc clones overexpressing the gene(s) downstream of the UAS sites and flanking the GS insertion point. The adult flies (at least 50) of the resulting progeny were examined to determine whether the expression of the GS line in scrib1 clones was lethal, or whether it enhanced the scrib1 mosaic adult eye phenotype in the non-TM6B progeny. Any crosses exhibiting lethality were then examined under fluorescent light to determine whether the non-TM6B progeny exhibited GFP-positive tumour overgrowth or a failure to pupate. The insertion point and over-expressed genes of any identified GS lines was obtained by referring to the Drosophila Gene Search Project web site (http://kyotofly.kit.jp/stocks/documents/GS_lines.html).
Immunohistochemistry
Imaginal discs were dissected in phosphate-buffered saline (PBS) from either wandering 3rd instar larvae or from staged lays for larvae of genotypes that failed to pupate and entered an extended larval stage of development. Tissues were fixed in 4% formaldehyde in PBS, and blocked in 2% goat serum in PBT (PBS 0.1% Triton X-100). For the detection of S phase cells, EdU labelling was performed for 30 min at room temperature according to the manufacturers protocol (Invitrogen). TUNEL assays were performed as described in the manufacturers protocol (Roche Applied Science). Primary antibodies were incubated with the samples in block overnight at 4°C, and were used at the following concentrations; rabbit anti-Ab (S. Crews [17], 1/200), rabbit anti-Ato ([89], 1/1000), rat anti-Bab2 ([90], 1/1000), mouse anti-β-galactosidase (Rockland, 1/400), mouse anti-Br-core (Developmental Studies Hybridoma Bank (DSHB), 1/200), mouse anti-Ct (DSHB, 1/100), mouse anti-Dac (DSHB, 1/10), rat anti-Dan ([9], 1/300), mouse anti-DIAP1 (B. Hay, 1/100), mouse anti-Dll ([8], 1/500), mouse anti-Elav (DSHB, 1/20), mouse anti-Ey ([91], 1/20), mouse anti-Eya (DSHB, 1/20), rabbit anti-GFP (Invitrogen, 1/1000), guinea pig anti-Hth ([92], 1/100), mouse anti-Mmp1 (DSHB, 1/20), guinea pig anti-Sens ([93], 1/1000), rabbit anti-Tai ([84], 1/500), rabbit anti-Tsh ([94], 1/2000). Secondary antibodies used were; anti-mouse/rat Alexa647 (Invitrogen) and anti- rabbit Alexa488 (Invitrogen) at 1/400 dilution. F-actin was detected with phalloidin–tetramethylrhodamine isothioblueate (TRITC; Sigma, 0.3 µM, 1/1000). Samples were mounted in 80% glycerol.
Microscopy and image processing
All samples were analysed by confocal microscopy on an Olympus FV1000 or Leica TCS SP5 microscope. Single optical sections were selected in FluoView software before being processed in Adobe Photoshop CS2 and assembled into figures in Adobe Illustrator CS2.
Expression array, ChIP-Seq and bioinformatic analysis
Eye/antennal discs were dissected from ∼5 day old larvae bearing ab-expressing clones, scrib1+ab-expressing clones, or FRT82B control clones. For the expression array, 20 pairs of discs for the ab and scrib−+ab samples, and 50 pairs of discs for the control FRT82B genotype, were used to prepare RNA. Samples were prepared in triplicate, and the RNA isolated using TRIZOL, before being column purified (Qiagen). Probes were hybridised to GeneChip Drosophila 2.0 Genome Arrays (Affymetrix).
For ChIP-Seq, eye/antennal discs were dissected and samples cross-linked in a 1.8% formaldehyde solution on a rotating wheel for 5 mins, prior to DNA being sheared by sonication (15 cycles of 20 seconds, 35% amplitude) to produce fragments of ∼500 bp [95]. 100 µl of extract (corresponding to ∼100 discs) was used for each immunoprecipitation. 35 µl of 50% Protein A-Sepharose CL4B was added to each sample, and cleared after 1.5 hours incubation. 2 µl of polyclonal rabbit anti-Abrupt antibody [17] was then added per sample (or, for the input DNA controls, no antibody was added), and incubated overnight with rotation. Immunocomplexes were recovered by adding 35 µl Protein A-Sepharose to each sample, incubating for 3 hours at 4°C, and then harvesting by centrifugation. Chromatin was decrosslinked by RNase and Proteinase K incubations, and the DNA column purified (Qiagen). For each sequencing sample, 3 to 6 immunoprecipitations were pooled.
High throughput sequencing was performed for the ChIP-Seq from the ab-expressing (8,643,591 reads) and scrib−+ab (8,508,640 reads) samples. ChIP-Seq reads in all cases were aligned to the Drosophila genome (dm3 genome assembly, BDGP Release 5) using Bowtie with default parameters [96]. Correlation between ChIP-Seq experiments was computed with UCSC Table browser [97]. We used PeakSeq [98] to identify the regions significantly enriched on ChIP-Seq reads from each sample in comparison to the normalised input control (READLENGTH = 40, MAXGAP = 40, MINFDR = 0.01 and PVALTHRESH = 0.0001). The resulting read maps and target lists were visualised as custom tracks in the University of California Santa Cruz (UCSC) Genome Browser [97]. Using RefSeq [99], potential Ab target genes in each ChIP-Seq experiment were identified by the presence of significant peaks either within the promoter region (from the transcriptional start sites to 500 bp upstream) or within the introns of each gene. Expression arrays, ChIP-Seq profiles and target regions were deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) repository as CEL, wiggle (WIG) and Browser Extensible Data (BED) files, respectively, under the accession numbers GSE42938 (expression arrays) and GSE42928 (ChIP-Seq profiles and target regions). Gene Ontology (GO) enrichments were identified with the “GO Term Enrichment” v1.8 from AmiGo. To annotate the list of putative transcription factor binding sites on each set of ChIP-Seq binding regions (ab and scrib −+ab), we used the MatScan program [100] with the full collection of 827 predictive matrices available in Jaspar and Transfac [101], [102]. We ranked each matrix on the basis of the number of hits in both experiments, normalizing with the total size for each set of sequences and the number of occurrences identified in the whole genome. To build the list of overrepresented matrices, we selected those models that presented a difference in each ranking of at least 200 positions and a positive normalised fold-change value. To identify a potential DNA recognition sequence for Ab binding we focussed upon peaks with a height of 40 or more reads from the ChIP-Seq profile of the ab overexpression experiment (irrespective of its genomic location), and used MEME [103] to identify enriched motifs. Potential transcription factors capable of recognizing enriched motifs were identified using the TOMTOM program (MEME suite).
Quantitative RT-PCR
Total RNA for each genotype was prepared in replicates using TRIZOL and column purification (Qiagen). After cDNA synthesis, qRT-PCR on each replicate was assayed in triplicate using StepOnePlus Real Time PCR System (Applied Biosystems). For the expression array validations, we were unable to use standard house keeping mRNAs to normalise the results since many, such as actin and GAPDH, were changed in the expression profiling. Instead, the expression levels were normalised with respect to CG6044, which is expressed in the eye/antennal disc but was not significantly deregulated in the expression arrays, and the average for the triplicates determined. The following primers were used: Blimp-1, forward TTGCGACAAGAAGTACATCAG, reverse GATGGTCTTTATCCAAACACTC; bab2, forward CAAGTTCGACATACCCATTCC, reverse GATATAGGTACCATGACCCTG; CG6044, forward ACTCAGCTTCCTCTACTTCC, reverse CGCAATACTAAAGCAATCACAC; Eip78C, forward CTAATAAAGCTGGGCTTCTTCG, reverse GTTGACAAAGTCAGAATCGTAGAG; fru, forward TCAGATACTCAGAGATGCGA, reverse TGTTGTTATCTGTGAGACCA; H15, forward GTGACTTTGATAGGGATCCCA, reverse AGGAGTCAATTGGGACATCAG. The fold changes for each sample were determined using the 2(−Delta Delta C(T)) method [104].
For the ChIP validation of a selection of representative genes, chromatin was immunoprecipitated with the Ab antibody from ab and scrib−+ab samples (as described above), and then used for quantitative real-time PCR, with rabbit IgG immunoprecipitation as a control. Primers were designed around peak regions identified from ChIP-Seq analysis by using PerlPrimer application. The following primers were used: Antp, forward AGGATCACCTATTTAACTGGAC, reverse ATGTACGTGGCATACTTTCAG; Blimp-1, forward CAAGAACCTGAGACACCTGA, reverse CAAGAACCTGAGACACCTGA; br, forward ACACATTCGCAACCAACAAT, reverse CCCTTCCAGTACCCTACTCT; Buffy, forward GGGATACATTCACCTTATATGCAC, reverse ACCGAAGTTGAAGTAAGCGA; chinmo, forward CATCTTCAACTTCCTTGCTAA, reverse TGAATACGAAATTGAGCGAA; eya, forward CACAGACAACACTCGAATCAG, reverse GCAGCAGAAGAGACAAAGAG; fru, forward GCTCTTCCATTATCGTTCTC, reverse TATACATGTGAATAGGGCAAG: ftz-f1, forward AGAGATACGAGTATCCGAGTG, reverse GACATGCACATACATATAGACGG; HLHmβ, forward CCTCCCTCCTTATGTATGTG, reverse GCACAATCAGAAGAAGTCAG; Mmp1, forward GGATAAGTGCCTATTACTAGCTG, reverse GAATAGCTTATTAGCACGGGTC.
Supporting Information
Lists of differentially expressed probe sets, identified ChIP-Seq target genes, and genes within Classes 1 to 6. Sheet 1. Probe sets deregulated in ab and scrib1+ab mosaic discs (compared to control FRT82B discs). Out of 18952 probe sets, 4239 were differentially expressed (log base 2 fold change (logFC) >1, adjusted p value <0.05) within ab and scrib1+ab mosaic discs. In ab-expressing discs, 3028 probe sets were deregulated, comprising 705 probe sets (pattern 3) unique to ab-expressing discs and 2323 probe sets (pattern 4) shared with scrib1+ab discs. In scrib1+ab mosaic discs, 3534 probe sets were deregulated, comprising 1211 unique to scrib1+ab (pattern 2), together with the 2323 shared probe sets. 14713 probe sets (pattern 1) were not significantly deregulated in either genotype. Sheet 2. Genes identified from ChIP-Seq peak enrichments from ab and scrib1+ab mosaic discs. 557 genes (pattern 2) were unique to ab; 721 genes (pattern 3) were unique to scrib1+ab; and 2025 genes (pattern 1) were shared by ab and scrib1+ab samples. Sheets 3–8. Gene lists of classes 1–6 (see Figure 2C for a Venn diagram depicting the different classes).
(XLS)
List of the 183 differentially expressed genes in ab or scrib −+ab mosaic eye discs compared to control discs, that are represented by more than one probe set. Of the 3549 annotated genes deregulated in ab and scrib1+ab mosaic eye discs, 183 were represented by more than one probe set. Individual probe sets showing upregulation or downregulation (log base 2 fold change >1, adjusted p value <0.05) in either ab or scrib1+ab mosaic eye discs compared to control FRT82B discs are indicated by “+1” or “−1” respectively. A “0” indicates no significant deregulation. The 59 genes that are represented by probes with conflicting expression (i.e. with probe sets showing opposite regulation in a particular genotype) are shown in red. The reasons for conflicting expression are not known, but could indicate the existence of more than one differentially regulated transcript.
(PDF)
GO enrichments (p<0.01) for differentially expressed genes, ChIP-Seq candidate genes and Classes 1, 2 and 5. Sheets 1–3. GO enrichments for differentially expressed genes unique to ab overexpression (“Abrupt Not Scrib”), unique to scrib1+ab (“Scrib Not Abrupt”), and shared between the two samples (“Abrupt and Scrib”). For the analysis, deregulated genes were identified from probe sets that could be assigned FlyBase Gene IDs. For genes with multiple deregulated probe sets, each FlyBase Gene ID was used only once per enrichment analysis. Sheets 3–6. GO enrichments for potential Ab target genes, identified by ChIP-Seq, unique to ab overexpression (“Abrupt Not Scrib”), unique to scrib1+ab (Scrib Not Abrupt”), and shared between the two samples (“Abrupt and Scrib”). Sheets 7–9. GO enrichments for genes within Classes 1, 2 and 5 (see Figure 2C for a Venn diagram depicting the different classes). Classes 3, 4 and 6 exhibited no significant GO enrichments.
(XLS)
ChIP-Seq peaks aligned to the genome for selected genes within Classes 1 to 4. Only genes depicted in Figure 2E are shown. Genes are in alphabetical order, and highlight bars beneath the peak landscape indicate significant peaks in each genotype.
(PDF)
Overexpression phenotypes of confirmed scrib − interactors in eye/antennal disc clones. Mosaic eye/antennal discs (anterior to the left in this and all subsequent figures) generated with ey-FLP and taken from larvae ∼5 days AEL. Clones are positively marked by GFP (white, or green in merges), tissue morphology is shown by F-actin (red in merges), and cell fate by Elav expression (white or pale blue, changing to magenta or dark blue when overlaid with GFP). GFP (panels A–J), GFP/Elav merges (panels A′,B″,C′,D″,E′,F″,G′,H″,I′J″), F-actin (B′,D′,F′,H′,J′) and GFP/Elav/F-actin merges (panels A″,B″′,C″,D″′,E″,F″′,G″,H″′,I″J″′). (A,B) dl-expressing clones and mosaic discs are overgrown, especially within the antennal region (A). The expression of dl in scrib1 clones also promotes overgrowth within the antennal region, and in the eye disc, clonal tissue also overgrows and does not express Elav (B). (C,D) esg-expressing clones are not overgrown (C), however, the expression of esg in scrib1 clones promotes large overgrowths especially within the antennal region (D). (E,F) chn-expressing clones are not overgrown (E), however, the expression of chn in scrib1 clones promotes antennal disc overgrowth, as well as overgrowth of eye disc tissue that does not express Elav (F, arrow). (G,H) apt-expressing clones are not overgrown (G), however, the expression of apt in scrib1 clones promotes mild clonal overgrowth although differentiation is not completely blocked (H). (I,J) Neither numb-expressing clones (I), nor scrib1 clones expressing numb, are overgrown (J). Yellow scale bar = 50 µm.
(JPG)
Proliferation and apoptosis in scrib−+ab tumours. ey-FLP induced eye/antennal disc clones marked by GFP (green). EdU (A–C) labeling is white (and magenta when overlayed with GFP in merged images), and TUNEL (D–F) is white (red in merged images, and appears yellow when overlayed with GFP in merged images). Arrowheads in A,B indicate the second mitotic wave. EdU (panel C), GFP/EdU merges (panels A,B,C′), GFP/TUNEL merges (panel D,E,F′), and TUNEL (panel F). (A,D) Wild type mosaic discs show the normal pattern of cell proliferation (A) and cell death (D). (B,E) ab overexpressing eye disc clones do not ectopically proliferate (B), but induce increased cell death in wild type cells along the clonal borders (E). (C,F) scrib1+ab clones ectopically proliferate, and disrupt the normal pattern of cell proliferation in the eye disc (C), and induce increased cell death in surrounding wild type tissue (F, arrowhead). Yellow scale bar = 50 µm.
(JPG)
Validation of the expression array by quantitative real-time PCR of selected genes. Expression levels, as determined by quantitative real-time PCR (see Materials and Methods), are shown for 5 genes (Eip78C, bab2, H15, Blimp-1, fru) in ab-expressing discs and in scrib1+ab discs, compared to the expression level in control discs containing wild type FRT82B clones (assigned an expression level of 1). The expression levels of CG6044, a gene that is expressed in the eye/antennal disc but did not significantly change in expression across the arrays, were used for normalisation. All five genes confirm the results from the expression array, with Eip78C, bab2, H15 and Blimp-1 being repressed in ab-expressing discs, and even further repressed in scrib1+ab tumours, whilst fru expression is increased upon ab overexpression, and even further increased in scrib1+ab tumours. ANOVA was performed for each primer pair; * p<0.01, ** p<0.001, *** p<0.0001 compared to the FRT82B control (n = 3). The changes in expression between ab and scrib−+ab samples were also all highly significant (p<0.0001). Error bars indicate 1 s.d.
(JPG)
The Ab antibody shows high specificity. Mosaic eye/antennal disc clones generated with ey-FLP. Clones are positively marked by GFP (white or green in merges), and Ab protein levels are shown in white (magenta when overlaid with GFP in the merges). Ab (panels A,B), GFP (panel A′B′) and merges (panels A″,B″). Tissue morphology in B is shown by F-actin (white, panel B″′). (A) Ab is endogenously expressed in the antennal disc, the anterior portion of the eye disc and the peripodial membrane, except in ab1D mutant clones, which show greatly reduced levels of Ab. (B) Clones of tissue overexpressing ab show greatly elevated levels of Ab protein. Yellow scale bar = 50 µm.
(JPG)
ChIP validation for selected representative genes. Chromatin was immunoprecipitated with the Ab antibody from ab and scrib −+ab mosaic eye/antennal discs, and then used for quantitative real-time PCR. Fold enrichment for all target genes was determined compared to rabbit IgG control immunoprecipitations, which were assigned an expression level of 1 (represented by the line on the graph). Representative genes were included from most functional categories; ecdysone response (Blimp-1, ftz-F1), BTB-ZF (br, chinmo, fru), cell fate (Antp, eya), Notch signalling (HLHmβ), cell death/survival (Buffy) and JNK signaling (Mmp1). Enrichment of all genes was observed in the Ab antibody immunoprecipitation compared to the IgG control from ab and scrib−+ab mosaic eye/antennal imaginal discs. T-test comparing each Ab immunoprecipitation to the IgG control; * p<0.01, ** p<0.001, *** p<0.0001 (n = 3 for both Ab immunoprecipitation and the IgG control). Error bars indicate 1 s.d.
(JPG)
Enriched sequence motifs amongst the top peaks from the ab alone overexpression sample. The strongest peaks were selected with a height of 40 or more reads from the ChIP-Seq profile, resulting in 1629 regions with an average length of 84.8 bp. Enriched motifs were identified using MEME, and potential transcription factors capable of recognizing the enriched motifs were identified using the TOMTOM program (MEME suite).
(JPG)
Br is repressed in ab-expressing cells. Mosaic eye/antennal disc clones generated with ey-FLP. Clones are positively marked by GFP (white, or green in merges), and tissue morphology is shown by F-actin (white). Br levels are shown in white (magenta when overlaid with GFP in the merges). GFP (panels A–C), F-actin (panels A′–C′), Br (panels A″–C″), and merges (panels A″′–C″′). (A) Br is expressed throughout the eye/antennal disc and peripodial membrane. (B,C) Br is repressed in ab-expressing clones (B, arrowheads) and in scrib1+ab clones (C, arrowheads). Yellow scale bar = 50 µm.
(JPG)
Overexpression of taiΔB in scrib−+ab tumours prevents tumour overgrowth. Mosaic eye/antennal disc clones generated with ey-FLP. Clones are positively marked by GFP (white, or green in merges), tissue morphology is shown by F-actin (red in merges), and cell fate by Elav expression (white or pale blue, changing to magenta or dark blue when overlaid with GFP). GFP (panels A–C), F-actin (panels A′–C′), Elav (panels A″–C″), GFP/Elav merges (panels A″′–C″′) and GFP/Elav/F-actin merges (panels A″″–C″″). (A) Expression of taiΔB in clones results in small clones. (B,C) Expression of taiΔB in scrib1 clones (B) or scrib1+ab clones (C) reduces clonal overgrowth and results in the eclosion of adult flies (data not shown).
(JPG)
Eya, Sens and Ato expression are reduced in scrib −+ab tumours. ey-FLP induced eye/antennal disc clones at ∼5 days AEL. Clones are marked by GFP (white, or green in merges), and cell fate is shown by the expression of Eya, Sens and Ato (all white, and magenta when overlaid with GFP in the merges) in control clones (A,E,I), scrib1 clones (B,F,J), ab-expressing clones (C,G,K), and scrib1+ab clones (D,H,L). GFP (panels A–L), Eya (panels A′–D′), Sens (panels E′–H′), Ato (panels I′–L′), and merges (panels A″–L″). (A–D) Eya expression is not altered in scrib1 clones (B, arrowhead), or ab overexpressing clones (C, arrowhead). scrib1+ab clones have greatly reduced levels of Eya (D, arrowhead). (E–H) Sens expression is disrupted and reduced in scrib1 clones, and repressed in ab overexpressing clones in the antennal but not in the eye disc (G, arrowheads). scrib1+ab clones do not express Sens (H, arrowhead). (I–L) Ato expression is disrupted and slightly reduced in scrib1 clones (J, arrowhead), and repressed in ab overexpressing clones within the antennal disc (K, arrowhead), and reduced in eye disc clones (K, arrow). scrib1+ab clones do not express Ato (L, arrowhead). Yellow scale bar = 50 µm.
(JPG)
Tsh and Ey expression is not substantially altered in scrib −+ab tumours, however, Bab2 and Ct expression is reduced. ey-FLP induced eye/antennal disc clones at ∼5 days AEL. Clones are marked by GFP (white, or green in merges), and cell fate is shown by the expression of Tsh, Ey, Bab2 and Ato (all white, and magenta when overlaid with GFP in the merges) in control clones (A,E,I,M), scrib1 clones (B,F,J,N), ab-expressing clones (C,G,K,O), and scrib1+ab clones (D,H,L,P). GFP (panels A–P), Tsh (panels A′–D′), Ey (panels E′–H′), Bab2 (panels I′–L′), Ct (panels M′–P′), and merges (panels A″–P″). (A–D) Tsh expression exhibits only slight perturbations in scrib1 clones, sometimes extending more posteriorly within large mutant clones spanning the normal expression domain (B), and is slightly increased in ab overexpressing clones (C, arrowhead). scrib1+ab clones express Tsh in the anterior portion of the eye disc, although it is repressed more posteriorly, as in control clones (D, arrowhead). (E–H) Ey expression is slightly reduced in scrib1 clones (F, arrowhead), and ab overexpressing clones (G, arrowhead). scrib1+ab clones express Ey in the anterior portion of the eye disc, although its expression is repressed, as it is in control clones, more posteriorly (H, arrow). (I–L) Bab2 expression is not altered in scrib1 clones (J), and although expanded, or ectopic, domains of Bab2 expression are sometimes associated with ab overexpressing clones in the antenna, the levels of Bab2 within the clones are slightly reduced compared to adjacent wild type tissue (K; arrowhead showing slightly enlarged Bab2 domain of expression, although levels of Bab2 in the ab-expressing clone are lower than the more highly expressing wild type tissue adjacent to the clone). scrib1+ab clones do not express Bab2 (L, arrowhead). (M–P) Ct expression is not altered in scrib1 clones (N), and is repressed in more distally located ab overexpressing clones (O, arrowhead), although expression is unaffected in the proximal region (O, arrow). scrib1+ab clones do not express Ct (P, arrowheads). Yellow scale bar = 50 µm.
(JPG)
Hth is not sufficient or required for Ab-mediated tumour overgrowth. ey-FLP induced eye/antennal disc clones at 5 days AEL. Clones are marked by GFP (white, or green in merges), and Elav is shown in white (blue in merges, and dark blue when overlaid with GFP). GFP (panels A–D), Elav (panels A′–D′), and merges (panels A″–D″). (A) hth overexpressing clones do not express Elav. (B) Overexpressing hth in scrib1 clones blocks Elav expression and promotes clonal overgrowth, however, the larvae pupate and do not undergo an extended larval stage of development. (C) scrib1 hthP2 mutant clones are similar to hthP2 mutant clones alone (see Figure 5B ), although photoreceptor differentiation is disrupted in posteriorly localised clones. (D) hthP2 mutant clones overexpressing ab are similar to hthP2 mutant clones (see Figure 5B ).
(JPG)
ab overexpression downregulates DIAP1 levels. Mosaic antennal disc clones (A) and eye disc clones (B) overexpressing ab, and positively marked by GFP (white, or green in the merges). Diap1 is shown in white (or red in merges). GFP (panels A,B), Diap1 (panels A′B′), and merges (panels A″,B″). (A,B) ab-overexpressing clones downregulate DIAP1 (red in merges), in both the antennal disc (A) and eye disc (B). Yellow scale bar = 50 µm.
(JPG)
Overexpression of dac does not restrain scrib −+ab tumour overgrowth. ey-FLP clones are positively marked by GFP (white, or green in merges) in all panels. Elav is shown in white (magenta when overlaid with GFP in the merges). Yellow lines indicate the positions of virtual cross sections (shown on the right). GFP (panels A–D), Elav (panels A′–D′), merges (panels A″–D″), and virtual cross sections of the merges (panels A″′–D”′). (A) Overexpressing dac in mosaic discs results in small cyst-like clones that disrupt the normal pattern of photoreceptor differentiation. (B) Overexpressing dac in scrib1 mutant clones produces a similar phenotype as scrib1 clones alone (the mutant clone failing to express Elav is located above the disc proper). (C) Overexpressing ab and dac together in clones generates large clones in antenna, similar to ab overexpressing clones alone, but very small clones in the eye disc. (D) Overexpressing dac in scrib1+ab clones does not restrain clone overgrowth, and large neoplastic tumours are produced, which do not express Elav. Yellow scale bar = 50 µm.
(JPG)
Knockdown of yki in scrib −+ab tumours does not prevent cell migration between the brain lobes. Eye/antennal discs still attached to the brain lobes (BL) containing ey-FLP induced scrib1 clones that express both ab and ykiRNAi (A, and a higher magnification of the region between the brain lobes in B). ey-FLP clones are positively marked by GFP (white, or green in merges) and F-actin is shown in white (red in the merges). Elav is shown in blue in the merges. GFP (panels A,B), F-actin (panels A′,B′), and GFP/F-actin/Elav merges (panels A″,B″). Mutant tissue overgrowth is restrained compared with scrib−+ab tumours, however tissue is still observed between the brain lobes (arrows), consistent with it migrating and merging with the brain lobes. Yellow scale bar = 50 µm.
(JPG)
Expression of aPKCDN in scrib −+ab tumours restrains tumour overgrowth. ey-FLP clones at day 5 AEL (A–C), and with brain lobes (BL) attached at day 7 (D). Mutant clones are positively marked by GFP (white, or green in merges). Elav is shown in white (magenta when overlaid with GFP in the merges), and cell morphology is indicated by F-actin (white). GFP (panels A–D), F-actin (panels A′–D′), Elav (panels A″–D″) and GFP/Elav merges (panels A″′–D′″). (A) Overexpressing aPKCCAAX-DN in clones does not produce a discernible phenotype. (B) Clones co-overexpressing aPKCCAAX-DN and ab are similar to ab-expressing clones alone (compare to Figure 1E ). (C,D) Overexpressing aPKCCAAX-DN in scrib1+ab clones restrains clonal tissue overgrowth and GFP specks are observed, consistent with cells undergoing cell death (C). Larvae still enter an extended larval stage of development, however, tumour overgrowth (D) is reduced compared to scrib1+ab tumours (eg. Figure 8F ). Yellow scale bar = 50 µm.
(JPG)
wtsX1+ab tumours retain Dll expression, but do not express Dan. ey-FLP induced eye/antennal disc clones marked by GFP (white, or green in merges). The cell fate markers Dll and Dan are shown in white (magenta when overlaid with GFP in merged images). F-actin (white) shows cell morphology. GFP (panels A–D), F-actin (panels A′–D′), Dll (panels A″,B″), Dan (panels C″,D″), GFP/Dll merges (panels A″′,B″′) and GFP/Dan merges (panels C″′,D′″). (A) wtsX1 clones exhibit the normal pattern of Dll in the eye/antennal disc. (B) wtsX1+ab clones retain Dll expression, often resulting in ectopic domains of Dll-expressing tissue (B, arrowhead), similar to ab expressing clones, or scrib1+ab tumours. (C) wtsX1 clones exhibit the normal pattern of Dan in the eye/antennal disc. (D) wtsX1+ab clones do not express Dan in the antenna, and the overgrowths in the eye disc are also characterised by a loss of Dan (D, arrowhead). Yellow scale bar = 50 µm.
(JPG)
Live cell imaging of scrib −+ab tumours. Movie taken at 2 minute intervals over a 20 hour period at 10× magnification. scrib1+ab tumour cells are highly motile, moving between, and over, the brain lobes. Note that, although motile hemocytes have been reported be associated with the tumours, and may be associated with tumour cell engulfment, the intensity and distribution of GFP fluorescence within the motile cells is not consistent with them being phagocytic hemocytes.
(ASF)
Live cell imaging of scrib −+ab+bskDN tumours. Movie taken at 2 minute intervals over a 20 hour period at 10× magnification. scrib1+ab+bskDN tumour cells are non invasive, and the overall tumour mass, whilst growing, remains compact.
(ASF)
Transcription factor matrices enriched amongst peak sequences associated with the promoter regions or introns of potential target genes.
(DOC)
Acknowledgments
We thank Ross Hannan, Silvia Perez-Lluch and Elaine Sanij for advice on the ChIP experiments; the Functional Genomics Core Facility of the IRB (Barcelona, Spain); Benjamin Rashleigh for developing the FileMaker Pro database that was used for compiling data from the genetic screen; Alyssa Harley and Peter Burke for help with fly crosses and maintenance for the genetic screen; H. Bellen, D. Bilder, J. Clements, S. Cohen, S. Crews, D. Duncan, B. Hay, S. Hirose, L. Jan, Y. Jan, J. Jiang, J. Knoblich, F. Laski, R. Mann, D. Montell, H. Sun, J. Treisman, the Bloomington Stock Centre, the Vienna Drosophila RNAi Centre, the National Institute of Genetics (NIG) Fly Stock Centre, the Developmental Studies Hybridoma Bank for contributing fly stocks and/or reagents, and Flybase.
Funding Statement
This work was supported by grants from the Australian National Health and Medical Research Council (NHMRC) to AMB (NHMRC Grants #350396 and #509051) and HER (NHMRC Senior research Fellowship B and NHMRC Grants #400211 and #628401), NIH grant (1R21CA098997-01) to HER, Peter MacCallum Cancer Center Internal Grant to HER, a Development Travel Award to NT, and a Spanish Ministry of Education and Science grant to MC (CSD2007-00008) to support the Bioinformatics Platform. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, et al. (2008) An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet 40: 499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wong DJ, Liu H, Ridky TW, Cassarino D, Segal E, et al. (2008) Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell 2: 333–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gonzalez C (2007) Spindle orientation, asymmetric division and tumour suppression in Drosophila stem cells. Nat Rev Genet 8: 462–472. [DOI] [PubMed] [Google Scholar]
- 4. Janic A, Mendizabal L, Llamazares S, Rossell D, Gonzalez C (2010) Ectopic expression of germline genes drives malignant brain tumor growth in Drosophila. Science 330: 1824–1827. [DOI] [PubMed] [Google Scholar]
- 5. Brumby AM, Richardson HE (2005) Using Drosophila melanogaster to map human cancer pathways. Nat Rev Cancer 5: 626–639. [DOI] [PubMed] [Google Scholar]
- 6. Dominguez M, Casares F (2005) Organ specification-growth control connection: new in-sights from the Drosophila eye-antennal disc. Dev Dyn 232: 673–684. [DOI] [PubMed] [Google Scholar]
- 7. Dong PD, Dicks JS, Panganiban G (2002) Distal-less and homothorax regulate multiple targets to pattern the Drosophila antenna. Development 129: 1967–1974. [DOI] [PubMed] [Google Scholar]
- 8. Duncan DM, Burgess EA, Duncan I (1998) Control of distal antennal identity and tarsal development in Drosophila by spineless-aristapedia, a homolog of the mammalian dioxin receptor. Genes Dev 12: 1290–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Emerald BS, Curtiss J, Mlodzik M, Cohen SM (2003) Distal antenna and distal antenna related encode nuclear proteins containing pipsqueak motifs involved in antenna development in Drosophila. Development 130: 1171–1180. [DOI] [PubMed] [Google Scholar]
- 10. Zhao B, Tumaneng K, Guan KL (2011) The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat Cell Biol 13: 877–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Peng HW, Slattery M, Mann RS (2009) Transcription factor choice in the Hippo signaling pathway: homothorax and yorkie regulation of the microRNA bantam in the progenitor domain of the Drosophila eye imaginal disc. Genes Dev 23: 2307–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brumby AM, Richardson HE (2003) scribble mutants cooperate with oncogenic Ras or Notch to cause neoplastic overgrowth in Drosophila. Embo J 22: 5769–5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Toba G, Ohsako T, Miyata N, Ohtsuka T, Seong KH, et al. (1999) The gene search system. A method for efficient detection and rapid molecular identification of genes in Drosophila melanogaster. Genetics 151: 725–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Leong GR, Goulding KR, Amin N, Richardson HE, Brumby AM (2009) Scribble mutants promote aPKC and JNK-dependent epithelial neoplasia independently of Crumbs. BMC Biol 7: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kalionis B, O'Farrell PH (1993) A universal target sequence is bound in vitro by diverse homeodomains. Mech Dev 43: 57–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhu LJ, Christensen RG, Kazemian M, Hull CJ, Enuameh MS, et al. (2011) FlyFactorSurvey: a database of Drosophila transcription factor binding specificities determined using the bacterial one-hybrid system. Nucleic Acids Res 39: D111–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hu S, Fambrough D, Atashi JR, Goodman CS, Crews ST (1995) The Drosophila abrupt gene encodes a BTB-zinc finger regulatory protein that controls the specificity of neuromuscular connections. Genes Dev 9: 2936–2948. [DOI] [PubMed] [Google Scholar]
- 18. Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B 57: 289–300. [Google Scholar]
- 19. Grieder NC, Charlafti I, Kloter U, Jackle H, Schafer U, et al. (2007) Misexpression screen in Drosophila melanogaster aiming to reveal novel factors involved in formation of body parts. Genetics 175: 1707–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jang AC, Chang YC, Bai J, Montell D (2009) Border-cell migration requires integration of spatial and temporal signals by the BTB protein Abrupt. Nat Cell Biol 11: 569–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Affolter M, Marty T, Vigano MA, Jazwinska A (2001) Nuclear interpretation of Dpp signaling in Drosophila. Embo J 20: 3298–3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. D'Avino PP, Thummel CS (1998) crooked legs encodes a family of zinc finger proteins required for leg morphogenesis and ecdysone-regulated gene expression during Drosophila metamorphosis. Development 125: 1733–1745. [DOI] [PubMed] [Google Scholar]
- 23. Kenyon KL, Ranade SS, Curtiss J, Mlodzik M, Pignoni F (2003) Coordinating proliferation and tissue specification to promote regional identity in the Drosophila head. Dev Cell 5: 403–414. [DOI] [PubMed] [Google Scholar]
- 24. Wang CW, Sun YH (2012) Segregation of eye and antenna fates maintained by mutual antagonism in Drosophila. Development 139: 3413–3421. [DOI] [PubMed] [Google Scholar]
- 25. Casares F, Mann RS (1998) Control of antennal versus leg development in Drosophila. Nature 392: 723–726. [DOI] [PubMed] [Google Scholar]
- 26. Pichaud F, Casares F (2000) homothorax and iroquois-C genes are required for the establishment of territories within the developing eye disc. Mech Dev 96: 15–25. [DOI] [PubMed] [Google Scholar]
- 27. Pai CY, Kuo TS, Jaw TJ, Kurant E, Chen CT, et al. (1998) The Homothorax homeoprotein activates the nuclear localization of another homeoprotein, extradenticle, and suppresses eye development in Drosophila. Genes Dev 12: 435–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wolff T, Ready DF (1991) Cell death in normal and rough eye mutants of Drosophila. Development 113: 825–839. [DOI] [PubMed] [Google Scholar]
- 29. Copeland JM, Bosdet I, Freeman JD, Guo M, Gorski SM, et al. (2007) echinus, required for interommatidial cell sorting and cell death in the Drosophila pupal retina, encodes a protein with homology to ubiquitin-specific proteases. BMC Dev Biol 7: 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rusconi JC, Fink JL, Cagan R (2004) klumpfuss regulates cell death in the Drosophila retina. Mech Dev 121: 537–546. [DOI] [PubMed] [Google Scholar]
- 31. Doggett K, Grusche FA, Richardson HE, Brumby AM (2011) Loss of the Drosophila cell polarity regulator Scribbled promotes epithelial tissue overgrowth and cooperation with oncogenic Ras-Raf through impaired Hippo pathway signaling. BMC Dev Biol 11: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Uhlirova M, Bohmann D (2006) JNK- and Fos-regulated Mmp1 expression cooperates with Ras to induce invasive tumors in Drosophila. Embo J 25: 5294–5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Igaki T, Pagliarini RA, Xu T (2006) Loss of cell polarity drives tumor growth and invasion through JNK activation in Drosophila. Curr Biol 16: 1139–1146. [DOI] [PubMed] [Google Scholar]
- 34. Rousset R, Bono-Lauriol S, Gettings M, Suzanne M, Speder P, et al. (2010) The Drosophila serine protease homologue Scarface regulates JNK signalling in a negative-feedback loop during epithelial morphogenesis. Development 137: 2177–2186. [DOI] [PubMed] [Google Scholar]
- 35. Montell DJ (2006) The social lives of migrating cells in Drosophila. Curr Opin Genet Dev 16: 374–383. [DOI] [PubMed] [Google Scholar]
- 36. Mattila J, Omelyanchuk L, Kyttala S, Turunen H, Nokkala S (2005) Role of Jun N-terminal Kinase (JNK) signaling in the wound healing and regeneration of a Drosophila melanogaster wing imaginal disc. Int J Dev Biol 49: 391–399. [DOI] [PubMed] [Google Scholar]
- 37. Ferres-Marco D, Gutierrez-Garcia I, Vallejo DM, Bolivar J, Gutierrez-Avino FJ, et al. (2006) Epigenetic silencers and Notch collaborate to promote malignant tumours by Rb silencing. Nature 439: 430–436. [DOI] [PubMed] [Google Scholar]
- 38. Costoya JA (2007) Functional analysis of the role of POK transcriptional repressors. Brief Funct Genomic Proteomic 6: 8–18. [DOI] [PubMed] [Google Scholar]
- 39. Siggs OM, Beutler B (2012) The BTB-ZF transcription factors. Cell Cycle 11: 3358–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yan J, Tsai SY, Tsai MJ (2006) SRC-3/AIB1: transcriptional coactivator in oncogenesis. Acta Pharmacol Sin 27: 387–394. [DOI] [PubMed] [Google Scholar]
- 41. Menut L, Vaccari T, Dionne H, Hill J, Wu G, et al. (2007) A mosaic genetic screen for Drosophila neoplastic tumor suppressor genes based on defective pupation. Genetics 177: 1667–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Simpson P, Berreur P, Berreur-Bonnenfant J (1980) The initiation of pupariation in Drosophila: dependence on growth of the imaginal discs. J Embryol Exp Morphol 57: 155–165. [PubMed] [Google Scholar]
- 43. Colombani J, Andersen DS, Leopold P (2012) Secreted peptide Dilp8 coordinates Drosophila tissue growth with developmental timing. Science 336: 582–585. [DOI] [PubMed] [Google Scholar]
- 44. Garelli A, Gontijo AM, Miguela V, Caparros E, Dominguez M (2012) Imaginal discs secrete insulin-like peptide 8 to mediate plasticity of growth and maturation. Science 336: 579–582. [DOI] [PubMed] [Google Scholar]
- 45. Caygill EE, Johnston LA (2008) Temporal regulation of metamorphic processes in Drosophila by the let-7 and miR-125 heterochronic microRNAs. Curr Biol 18: 943–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Luque CM, Milan M (2007) Growth control in the proliferative region of the Drosophila eye-head primordium: the elbow-noc gene complex. Dev Biol 301: 327–339. [DOI] [PubMed] [Google Scholar]
- 47. Bonini NM, Leiserson WM, Benzer S (1993) The eyes absent gene: genetic control of cell survival and differentiation in the developing Drosophila eye. Cell 72: 379–395. [DOI] [PubMed] [Google Scholar]
- 48. Pignoni F, Hu B, Zavitz KH, Xiao J, Garrity PA, et al. (1997) The eye-specification proteins So and Eya form a complex and regulate multiple steps in Drosophila eye development. Cell 91: 881–891. [DOI] [PubMed] [Google Scholar]
- 49. Bessa J, Gebelein B, Pichaud F, Casares F, Mann RS (2002) Combinatorial control of Drosophila eye development by eyeless, homothorax, and teashirt. Genes Dev 16: 2415–2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dong PD, Chu J, Panganiban G (2000) Coexpression of the homeobox genes Distal-less and homothorax determines Drosophila antennal identity. Development 127: 209–216. [DOI] [PubMed] [Google Scholar]
- 51. Dong PD, Chu J, Panganiban G (2001) Proximodistal domain specification and interactions in developing Drosophila appendages. Development 128: 2365–2372. [DOI] [PubMed] [Google Scholar]
- 52. Godt D, Couderc JL, Cramton SE, Laski FA (1993) Pattern formation in the limbs of Drosophila: bric a brac is expressed in both a gradient and a wave-like pattern and is required for specification and proper segmentation of the tarsus. Development 119: 799–812. [DOI] [PubMed] [Google Scholar]
- 53. Chu J, Dong PD, Panganiban G (2002) Limb type-specific regulation of bric a brac contributes to morphological diversity. Development 129: 695–704. [DOI] [PubMed] [Google Scholar]
- 54. Percival-Smith A, Teft WA, Barta JL (2005) Tarsus determination in Drosophila melanogaster. Genome 48: 712–721. [DOI] [PubMed] [Google Scholar]
- 55. Bras-Pereira C, Casares F (2008) An antennal-specific role for bowl in repressing supernumerary appendage development in Drosophila. Mech Dev 125: 809–821. [DOI] [PubMed] [Google Scholar]
- 56. Bayarmagnai B, Nicolay BN, Islam AB, Lopez-Bigas N, Frolov MV (2012) Drosophila GAGA factor is required for full activation of the dE2f1-Yki/Sd transcriptional program. Cell Cycle 11: 4191–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Oh H, Slattery M, Ma L, Crofts A, White KP, et al. (2013) Genome-wide association of Yorkie with chromatin and chromatin-remodeling complexes. Cell Rep 3: 309–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Betz A, Ryoo HD, Steller H, Darnell JE Jr (2008) STAT92E is a positive regulator of Drosophila inhibitor of apoptosis 1 (DIAP/1) and protects against radiation-induced apoptosis. Proc Natl Acad Sci U S A 105: 13805–13810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zeidler MP, Perrimon N, Strutt DI (1999) The four-jointed gene is required in the Drosophila eye for ommatidial polarity specification. Curr Biol 9: 1363–1372. [DOI] [PubMed] [Google Scholar]
- 60. Arbouzova NI, Bach EA, Zeidler MP (2006) Ken & barbie selectively regulates the expression of a subset of Jak/STAT pathway target genes. Curr Biol 16: 80–88. [DOI] [PubMed] [Google Scholar]
- 61. Nicolay BN, Bayarmagnai B, Moon NS, Benevolenskaya EV, Frolov MV (2010) Combined inactivation of pRB and hippo pathways induces dedifferentiation in the Drosophila retina. PLoS Genet 6: e1000918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Reddy BV, Rauskolb C, Irvine KD (2010) Influence of fat-hippo and notch signaling on the proliferation and differentiation of Drosophila optic neuroepithelia. Development 137: 2397–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhang T, Zhou Q, Pignoni F (2011) Yki/YAP, Sd/TEAD and Hth/MEIS control tissue specification in the Drosophila eye disc epithelium. PLoS One 6: e22278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Beaulieu AM, Sant'angelo DB (2011) The BTB-ZF Family of Transcription Factors: Key Regulators of Lineage Commitment and Effector Function Development in the Immune System. J Immunol 187: 2841–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Addou-Klouche L, Adelaide J, Finetti P, Cervera N, Ferrari A, et al. (2010) Loss, mutation and deregulation of L3MBTL4 in breast cancers. Mol Cancer 9: 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Scheeren FA, Naspetti M, Diehl S, Schotte R, Nagasawa M, et al. (2005) STAT5 regulates the self-renewal capacity and differentiation of human memory B cells and controls Bcl-6 expression. Nat Immunol 6: 303–313. [DOI] [PubMed] [Google Scholar]
- 67. Logarajah S, Hunter P, Kraman M, Steele D, Lakhani S, et al. (2003) BCL-6 is expressed in breast cancer and prevents mammary epithelial differentiation. Oncogene 22: 5572–5578. [DOI] [PubMed] [Google Scholar]
- 68. Crotty S, Johnston RJ, Schoenberger SP (2010) Effectors and memories: Bcl-6 and Blimp-1 in T and B lymphocyte differentiation. Nat Immunol 11: 114–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Agawa Y, Sarhan M, Kageyama Y, Akagi K, Takai M, et al. (2007) Drosophila Blimp-1 is a transient transcriptional repressor that controls timing of the ecdysone-induced developmental pathway. Mol Cell Biol 27: 8739–8747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sakano D, Kato A, Parikh N, McKnight K, Terry D, et al. (2010) BCL6 canalizes Notch-dependent transcription, excluding Mastermind-like1 from selected target genes during left-right patterning. Dev Cell 18: 450–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lee JD, Treisman JE (2001) The role of Wingless signaling in establishing the anteroposterior and dorsoventral axes of the eye disc. Development 128: 1519–1529. [DOI] [PubMed] [Google Scholar]
- 72. Cook O, Biehs B, Bier E (2004) brinker and optomotor-blind act coordinately to initiate development of the L5 wing vein primordium in Drosophila. Development 131: 2113–2124. [DOI] [PubMed] [Google Scholar]
- 73. Adachi-Yamada T, Nakamura M, Irie K, Tomoyasu Y, Sano Y, et al. (1999) p38 mitogen-activated protein kinase can be involved in transforming growth factor beta superfamily signal transduction in Drosophila wing morphogenesis. Mol Cell Biol 19: 2322–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Escudero LM, Caminero E, Schulze KL, Bellen HJ, Modolell J (2005) Charlatan, a Zn-finger transcription factor, establishes a novel level of regulation of the proneural achaete/scute genes of Drosophila. Development 132: 1211–1222. [DOI] [PubMed] [Google Scholar]
- 75. Sotillos S, Diaz-Meco MT, Caminero E, Moscat J, Campuzano S (2004) DaPKC-dependent phosphorylation of Crumbs is required for epithelial cell polarity in Drosophila. J Cell Biol 166: 549–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Martini SR, Roman G, Meuser S, Mardon G, Davis RL (2000) The retinal determination gene, dachshund, is required for mushroom body cell differentiation. Development 127: 2663–2672. [DOI] [PubMed] [Google Scholar]
- 77. Huang L, Ohsako S, Tanda S (2005) The lesswright mutation activates Rel-related proteins, leading to overproduction of larval hemocytes in Drosophila melanogaster. Dev Biol 280: 407–420. [DOI] [PubMed] [Google Scholar]
- 78. Fuse N, Hirose S, Hayashi S (1994) Diploidy of Drosophila imaginal cells is maintained by a transcriptional repressor encoded by escargot. Genes Dev 8: 2270–2281. [DOI] [PubMed] [Google Scholar]
- 79. Kramatschek B, Campos-Ortega JA (1994) Neuroectodermal transcription of the Drosophila neurogenic genes E(spl) and HLH-m5 is regulated by proneural genes. Development 120: 815–826. [DOI] [PubMed] [Google Scholar]
- 80. Rieckhof GE, Casares F, Ryoo HD, Abu-Shaar M, Mann RS (1997) Nuclear translocation of extradenticle requires homothorax, which encodes an extradenticle-related homeodomain protein. Cell 91: 171–183. [DOI] [PubMed] [Google Scholar]
- 81. Spradling AC, Stern D, Beaton A, Rhem EJ, Laverty T, et al. (1999) The Berkeley Drosophila Genome Project gene disruption project: Single P-element insertions mutating 25% of vital Drosophila genes. Genetics 153: 135–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Roegiers F, Younger-Shepherd S, Jan LY, Jan YN (2001) Two types of asymmetric divisions in the Drosophila sensory organ precursor cell lineage. Nat Cell Biol 3: 58–67. [DOI] [PubMed] [Google Scholar]
- 83. Bilder D, Perrimon N (2000) Localization of apical epithelial determinants by the basolateral PDZ protein Scribble. Nature 403: 676–680. [DOI] [PubMed] [Google Scholar]
- 84. Bai J, Uehara Y, Montell DJ (2000) Regulation of invasive cell behavior by taiman, a Drosophila protein related to AIB1, a steroid receptor coactivator amplified in breast cancer. Cell 103: 1047–1058. [DOI] [PubMed] [Google Scholar]
- 85. Eulenberg KG, Schuh R (1997) The tracheae defective gene encodes a bZIP protein that controls tracheal cell movement during Drosophila embryogenesis. Embo J 16: 7156–7165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Xu T, Wang W, Zhang S, Stewart RA, Yu W (1995) Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development 121: 1053–1063. [DOI] [PubMed] [Google Scholar]
- 87. Zhang L, Ren F, Zhang Q, Chen Y, Wang B, et al. (2008) The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev Cell 14: 377–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Lee T, Luo L (1999) Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22: 451–461. [DOI] [PubMed] [Google Scholar]
- 89. Jarman AP, Sun Y, Jan LY, Jan YN (1995) Role of the proneural gene, atonal, in formation of Drosophila chordotonal organs and photoreceptors. Development 121: 2019–2030. [DOI] [PubMed] [Google Scholar]
- 90. Couderc JL, Godt D, Zollman S, Chen J, Li M, et al. (2002) The bric a brac locus consists of two paralogous genes encoding BTB/POZ domain proteins and acts as a homeotic and morphogenetic regulator of imaginal development in Drosophila. Development 129: 2419–2433. [DOI] [PubMed] [Google Scholar]
- 91. Clements J, Hens K, Francis C, Schellens A, Callaerts P (2008) Conserved role for the Drosophila Pax6 homolog Eyeless in differentiation and function of insulin-producing neurons. Proc Natl Acad Sci U S A 105: 16183–16188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Ryoo HD, Mann RS (1999) The control of trunk Hox specificity and activity by Extradenticle. Genes Dev 13: 1704–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Nolo R, Abbott LA, Bellen HJ (2000) Senseless, a Zn finger transcription factor, is necessary and sufficient for sensory organ development in Drosophila. Cell 102: 349–362. [DOI] [PubMed] [Google Scholar]
- 94. Ng M, Diaz-Benjumea FJ, Vincent JP, Wu J, Cohen SM (1996) Specification of the wing by localized expression of wingless protein. Nature 381: 316–318. [DOI] [PubMed] [Google Scholar]
- 95. Perez-Lluch S, Blanco E, Carbonell A, Raha D, Snyder M, et al. (2011) Genome-wide chromatin occupancy analysis reveals a role for ASH2 in transcriptional pausing. Nucleic Acids Res 39: 4628–4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, et al. (2011) The UCSC Genome Browser database: update 2011. Nucleic Acids Res 39: D876–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Rozowsky J, Euskirchen G, Auerbach RK, Zhang ZD, Gibson T, et al. (2009) PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nat Biotechnol 27: 66–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Pruitt KD, Tatusova T, Klimke W, Maglott DR (2009) NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Res 37: D32–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Blanco E, Messeguer X, Smith TF, Guigo R (2006) Transcription factor map alignment of promoter regions. PLoS Comput Biol 2: e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Portales-Casamar E, Thongjuea S, Kwon AT, Arenillas D, Zhao X, et al. (2010) JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res 38: D105–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Wingender E (2008) The TRANSFAC project as an example of framework technology that supports the analysis of genomic regulation. Brief Bioinform 9: 326–332. [DOI] [PubMed] [Google Scholar]
- 103. Bailey TL, Elkan C (1995) The value of prior knowledge in discovering motifs with MEME. Proc Int Conf Intell Syst Mol Biol 3: 21–29. [PubMed] [Google Scholar]
- 104. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Lists of differentially expressed probe sets, identified ChIP-Seq target genes, and genes within Classes 1 to 6. Sheet 1. Probe sets deregulated in ab and scrib1+ab mosaic discs (compared to control FRT82B discs). Out of 18952 probe sets, 4239 were differentially expressed (log base 2 fold change (logFC) >1, adjusted p value <0.05) within ab and scrib1+ab mosaic discs. In ab-expressing discs, 3028 probe sets were deregulated, comprising 705 probe sets (pattern 3) unique to ab-expressing discs and 2323 probe sets (pattern 4) shared with scrib1+ab discs. In scrib1+ab mosaic discs, 3534 probe sets were deregulated, comprising 1211 unique to scrib1+ab (pattern 2), together with the 2323 shared probe sets. 14713 probe sets (pattern 1) were not significantly deregulated in either genotype. Sheet 2. Genes identified from ChIP-Seq peak enrichments from ab and scrib1+ab mosaic discs. 557 genes (pattern 2) were unique to ab; 721 genes (pattern 3) were unique to scrib1+ab; and 2025 genes (pattern 1) were shared by ab and scrib1+ab samples. Sheets 3–8. Gene lists of classes 1–6 (see Figure 2C for a Venn diagram depicting the different classes).
(XLS)
List of the 183 differentially expressed genes in ab or scrib −+ab mosaic eye discs compared to control discs, that are represented by more than one probe set. Of the 3549 annotated genes deregulated in ab and scrib1+ab mosaic eye discs, 183 were represented by more than one probe set. Individual probe sets showing upregulation or downregulation (log base 2 fold change >1, adjusted p value <0.05) in either ab or scrib1+ab mosaic eye discs compared to control FRT82B discs are indicated by “+1” or “−1” respectively. A “0” indicates no significant deregulation. The 59 genes that are represented by probes with conflicting expression (i.e. with probe sets showing opposite regulation in a particular genotype) are shown in red. The reasons for conflicting expression are not known, but could indicate the existence of more than one differentially regulated transcript.
(PDF)
GO enrichments (p<0.01) for differentially expressed genes, ChIP-Seq candidate genes and Classes 1, 2 and 5. Sheets 1–3. GO enrichments for differentially expressed genes unique to ab overexpression (“Abrupt Not Scrib”), unique to scrib1+ab (“Scrib Not Abrupt”), and shared between the two samples (“Abrupt and Scrib”). For the analysis, deregulated genes were identified from probe sets that could be assigned FlyBase Gene IDs. For genes with multiple deregulated probe sets, each FlyBase Gene ID was used only once per enrichment analysis. Sheets 3–6. GO enrichments for potential Ab target genes, identified by ChIP-Seq, unique to ab overexpression (“Abrupt Not Scrib”), unique to scrib1+ab (Scrib Not Abrupt”), and shared between the two samples (“Abrupt and Scrib”). Sheets 7–9. GO enrichments for genes within Classes 1, 2 and 5 (see Figure 2C for a Venn diagram depicting the different classes). Classes 3, 4 and 6 exhibited no significant GO enrichments.
(XLS)
ChIP-Seq peaks aligned to the genome for selected genes within Classes 1 to 4. Only genes depicted in Figure 2E are shown. Genes are in alphabetical order, and highlight bars beneath the peak landscape indicate significant peaks in each genotype.
(PDF)
Overexpression phenotypes of confirmed scrib − interactors in eye/antennal disc clones. Mosaic eye/antennal discs (anterior to the left in this and all subsequent figures) generated with ey-FLP and taken from larvae ∼5 days AEL. Clones are positively marked by GFP (white, or green in merges), tissue morphology is shown by F-actin (red in merges), and cell fate by Elav expression (white or pale blue, changing to magenta or dark blue when overlaid with GFP). GFP (panels A–J), GFP/Elav merges (panels A′,B″,C′,D″,E′,F″,G′,H″,I′J″), F-actin (B′,D′,F′,H′,J′) and GFP/Elav/F-actin merges (panels A″,B″′,C″,D″′,E″,F″′,G″,H″′,I″J″′). (A,B) dl-expressing clones and mosaic discs are overgrown, especially within the antennal region (A). The expression of dl in scrib1 clones also promotes overgrowth within the antennal region, and in the eye disc, clonal tissue also overgrows and does not express Elav (B). (C,D) esg-expressing clones are not overgrown (C), however, the expression of esg in scrib1 clones promotes large overgrowths especially within the antennal region (D). (E,F) chn-expressing clones are not overgrown (E), however, the expression of chn in scrib1 clones promotes antennal disc overgrowth, as well as overgrowth of eye disc tissue that does not express Elav (F, arrow). (G,H) apt-expressing clones are not overgrown (G), however, the expression of apt in scrib1 clones promotes mild clonal overgrowth although differentiation is not completely blocked (H). (I,J) Neither numb-expressing clones (I), nor scrib1 clones expressing numb, are overgrown (J). Yellow scale bar = 50 µm.
(JPG)
Proliferation and apoptosis in scrib−+ab tumours. ey-FLP induced eye/antennal disc clones marked by GFP (green). EdU (A–C) labeling is white (and magenta when overlayed with GFP in merged images), and TUNEL (D–F) is white (red in merged images, and appears yellow when overlayed with GFP in merged images). Arrowheads in A,B indicate the second mitotic wave. EdU (panel C), GFP/EdU merges (panels A,B,C′), GFP/TUNEL merges (panel D,E,F′), and TUNEL (panel F). (A,D) Wild type mosaic discs show the normal pattern of cell proliferation (A) and cell death (D). (B,E) ab overexpressing eye disc clones do not ectopically proliferate (B), but induce increased cell death in wild type cells along the clonal borders (E). (C,F) scrib1+ab clones ectopically proliferate, and disrupt the normal pattern of cell proliferation in the eye disc (C), and induce increased cell death in surrounding wild type tissue (F, arrowhead). Yellow scale bar = 50 µm.
(JPG)
Validation of the expression array by quantitative real-time PCR of selected genes. Expression levels, as determined by quantitative real-time PCR (see Materials and Methods), are shown for 5 genes (Eip78C, bab2, H15, Blimp-1, fru) in ab-expressing discs and in scrib1+ab discs, compared to the expression level in control discs containing wild type FRT82B clones (assigned an expression level of 1). The expression levels of CG6044, a gene that is expressed in the eye/antennal disc but did not significantly change in expression across the arrays, were used for normalisation. All five genes confirm the results from the expression array, with Eip78C, bab2, H15 and Blimp-1 being repressed in ab-expressing discs, and even further repressed in scrib1+ab tumours, whilst fru expression is increased upon ab overexpression, and even further increased in scrib1+ab tumours. ANOVA was performed for each primer pair; * p<0.01, ** p<0.001, *** p<0.0001 compared to the FRT82B control (n = 3). The changes in expression between ab and scrib−+ab samples were also all highly significant (p<0.0001). Error bars indicate 1 s.d.
(JPG)
The Ab antibody shows high specificity. Mosaic eye/antennal disc clones generated with ey-FLP. Clones are positively marked by GFP (white or green in merges), and Ab protein levels are shown in white (magenta when overlaid with GFP in the merges). Ab (panels A,B), GFP (panel A′B′) and merges (panels A″,B″). Tissue morphology in B is shown by F-actin (white, panel B″′). (A) Ab is endogenously expressed in the antennal disc, the anterior portion of the eye disc and the peripodial membrane, except in ab1D mutant clones, which show greatly reduced levels of Ab. (B) Clones of tissue overexpressing ab show greatly elevated levels of Ab protein. Yellow scale bar = 50 µm.
(JPG)
ChIP validation for selected representative genes. Chromatin was immunoprecipitated with the Ab antibody from ab and scrib −+ab mosaic eye/antennal discs, and then used for quantitative real-time PCR. Fold enrichment for all target genes was determined compared to rabbit IgG control immunoprecipitations, which were assigned an expression level of 1 (represented by the line on the graph). Representative genes were included from most functional categories; ecdysone response (Blimp-1, ftz-F1), BTB-ZF (br, chinmo, fru), cell fate (Antp, eya), Notch signalling (HLHmβ), cell death/survival (Buffy) and JNK signaling (Mmp1). Enrichment of all genes was observed in the Ab antibody immunoprecipitation compared to the IgG control from ab and scrib−+ab mosaic eye/antennal imaginal discs. T-test comparing each Ab immunoprecipitation to the IgG control; * p<0.01, ** p<0.001, *** p<0.0001 (n = 3 for both Ab immunoprecipitation and the IgG control). Error bars indicate 1 s.d.
(JPG)
Enriched sequence motifs amongst the top peaks from the ab alone overexpression sample. The strongest peaks were selected with a height of 40 or more reads from the ChIP-Seq profile, resulting in 1629 regions with an average length of 84.8 bp. Enriched motifs were identified using MEME, and potential transcription factors capable of recognizing the enriched motifs were identified using the TOMTOM program (MEME suite).
(JPG)
Br is repressed in ab-expressing cells. Mosaic eye/antennal disc clones generated with ey-FLP. Clones are positively marked by GFP (white, or green in merges), and tissue morphology is shown by F-actin (white). Br levels are shown in white (magenta when overlaid with GFP in the merges). GFP (panels A–C), F-actin (panels A′–C′), Br (panels A″–C″), and merges (panels A″′–C″′). (A) Br is expressed throughout the eye/antennal disc and peripodial membrane. (B,C) Br is repressed in ab-expressing clones (B, arrowheads) and in scrib1+ab clones (C, arrowheads). Yellow scale bar = 50 µm.
(JPG)
Overexpression of taiΔB in scrib−+ab tumours prevents tumour overgrowth. Mosaic eye/antennal disc clones generated with ey-FLP. Clones are positively marked by GFP (white, or green in merges), tissue morphology is shown by F-actin (red in merges), and cell fate by Elav expression (white or pale blue, changing to magenta or dark blue when overlaid with GFP). GFP (panels A–C), F-actin (panels A′–C′), Elav (panels A″–C″), GFP/Elav merges (panels A″′–C″′) and GFP/Elav/F-actin merges (panels A″″–C″″). (A) Expression of taiΔB in clones results in small clones. (B,C) Expression of taiΔB in scrib1 clones (B) or scrib1+ab clones (C) reduces clonal overgrowth and results in the eclosion of adult flies (data not shown).
(JPG)
Eya, Sens and Ato expression are reduced in scrib −+ab tumours. ey-FLP induced eye/antennal disc clones at ∼5 days AEL. Clones are marked by GFP (white, or green in merges), and cell fate is shown by the expression of Eya, Sens and Ato (all white, and magenta when overlaid with GFP in the merges) in control clones (A,E,I), scrib1 clones (B,F,J), ab-expressing clones (C,G,K), and scrib1+ab clones (D,H,L). GFP (panels A–L), Eya (panels A′–D′), Sens (panels E′–H′), Ato (panels I′–L′), and merges (panels A″–L″). (A–D) Eya expression is not altered in scrib1 clones (B, arrowhead), or ab overexpressing clones (C, arrowhead). scrib1+ab clones have greatly reduced levels of Eya (D, arrowhead). (E–H) Sens expression is disrupted and reduced in scrib1 clones, and repressed in ab overexpressing clones in the antennal but not in the eye disc (G, arrowheads). scrib1+ab clones do not express Sens (H, arrowhead). (I–L) Ato expression is disrupted and slightly reduced in scrib1 clones (J, arrowhead), and repressed in ab overexpressing clones within the antennal disc (K, arrowhead), and reduced in eye disc clones (K, arrow). scrib1+ab clones do not express Ato (L, arrowhead). Yellow scale bar = 50 µm.
(JPG)
Tsh and Ey expression is not substantially altered in scrib −+ab tumours, however, Bab2 and Ct expression is reduced. ey-FLP induced eye/antennal disc clones at ∼5 days AEL. Clones are marked by GFP (white, or green in merges), and cell fate is shown by the expression of Tsh, Ey, Bab2 and Ato (all white, and magenta when overlaid with GFP in the merges) in control clones (A,E,I,M), scrib1 clones (B,F,J,N), ab-expressing clones (C,G,K,O), and scrib1+ab clones (D,H,L,P). GFP (panels A–P), Tsh (panels A′–D′), Ey (panels E′–H′), Bab2 (panels I′–L′), Ct (panels M′–P′), and merges (panels A″–P″). (A–D) Tsh expression exhibits only slight perturbations in scrib1 clones, sometimes extending more posteriorly within large mutant clones spanning the normal expression domain (B), and is slightly increased in ab overexpressing clones (C, arrowhead). scrib1+ab clones express Tsh in the anterior portion of the eye disc, although it is repressed more posteriorly, as in control clones (D, arrowhead). (E–H) Ey expression is slightly reduced in scrib1 clones (F, arrowhead), and ab overexpressing clones (G, arrowhead). scrib1+ab clones express Ey in the anterior portion of the eye disc, although its expression is repressed, as it is in control clones, more posteriorly (H, arrow). (I–L) Bab2 expression is not altered in scrib1 clones (J), and although expanded, or ectopic, domains of Bab2 expression are sometimes associated with ab overexpressing clones in the antenna, the levels of Bab2 within the clones are slightly reduced compared to adjacent wild type tissue (K; arrowhead showing slightly enlarged Bab2 domain of expression, although levels of Bab2 in the ab-expressing clone are lower than the more highly expressing wild type tissue adjacent to the clone). scrib1+ab clones do not express Bab2 (L, arrowhead). (M–P) Ct expression is not altered in scrib1 clones (N), and is repressed in more distally located ab overexpressing clones (O, arrowhead), although expression is unaffected in the proximal region (O, arrow). scrib1+ab clones do not express Ct (P, arrowheads). Yellow scale bar = 50 µm.
(JPG)
Hth is not sufficient or required for Ab-mediated tumour overgrowth. ey-FLP induced eye/antennal disc clones at 5 days AEL. Clones are marked by GFP (white, or green in merges), and Elav is shown in white (blue in merges, and dark blue when overlaid with GFP). GFP (panels A–D), Elav (panels A′–D′), and merges (panels A″–D″). (A) hth overexpressing clones do not express Elav. (B) Overexpressing hth in scrib1 clones blocks Elav expression and promotes clonal overgrowth, however, the larvae pupate and do not undergo an extended larval stage of development. (C) scrib1 hthP2 mutant clones are similar to hthP2 mutant clones alone (see Figure 5B ), although photoreceptor differentiation is disrupted in posteriorly localised clones. (D) hthP2 mutant clones overexpressing ab are similar to hthP2 mutant clones (see Figure 5B ).
(JPG)
ab overexpression downregulates DIAP1 levels. Mosaic antennal disc clones (A) and eye disc clones (B) overexpressing ab, and positively marked by GFP (white, or green in the merges). Diap1 is shown in white (or red in merges). GFP (panels A,B), Diap1 (panels A′B′), and merges (panels A″,B″). (A,B) ab-overexpressing clones downregulate DIAP1 (red in merges), in both the antennal disc (A) and eye disc (B). Yellow scale bar = 50 µm.
(JPG)
Overexpression of dac does not restrain scrib −+ab tumour overgrowth. ey-FLP clones are positively marked by GFP (white, or green in merges) in all panels. Elav is shown in white (magenta when overlaid with GFP in the merges). Yellow lines indicate the positions of virtual cross sections (shown on the right). GFP (panels A–D), Elav (panels A′–D′), merges (panels A″–D″), and virtual cross sections of the merges (panels A″′–D”′). (A) Overexpressing dac in mosaic discs results in small cyst-like clones that disrupt the normal pattern of photoreceptor differentiation. (B) Overexpressing dac in scrib1 mutant clones produces a similar phenotype as scrib1 clones alone (the mutant clone failing to express Elav is located above the disc proper). (C) Overexpressing ab and dac together in clones generates large clones in antenna, similar to ab overexpressing clones alone, but very small clones in the eye disc. (D) Overexpressing dac in scrib1+ab clones does not restrain clone overgrowth, and large neoplastic tumours are produced, which do not express Elav. Yellow scale bar = 50 µm.
(JPG)
Knockdown of yki in scrib −+ab tumours does not prevent cell migration between the brain lobes. Eye/antennal discs still attached to the brain lobes (BL) containing ey-FLP induced scrib1 clones that express both ab and ykiRNAi (A, and a higher magnification of the region between the brain lobes in B). ey-FLP clones are positively marked by GFP (white, or green in merges) and F-actin is shown in white (red in the merges). Elav is shown in blue in the merges. GFP (panels A,B), F-actin (panels A′,B′), and GFP/F-actin/Elav merges (panels A″,B″). Mutant tissue overgrowth is restrained compared with scrib−+ab tumours, however tissue is still observed between the brain lobes (arrows), consistent with it migrating and merging with the brain lobes. Yellow scale bar = 50 µm.
(JPG)
Expression of aPKCDN in scrib −+ab tumours restrains tumour overgrowth. ey-FLP clones at day 5 AEL (A–C), and with brain lobes (BL) attached at day 7 (D). Mutant clones are positively marked by GFP (white, or green in merges). Elav is shown in white (magenta when overlaid with GFP in the merges), and cell morphology is indicated by F-actin (white). GFP (panels A–D), F-actin (panels A′–D′), Elav (panels A″–D″) and GFP/Elav merges (panels A″′–D′″). (A) Overexpressing aPKCCAAX-DN in clones does not produce a discernible phenotype. (B) Clones co-overexpressing aPKCCAAX-DN and ab are similar to ab-expressing clones alone (compare to Figure 1E ). (C,D) Overexpressing aPKCCAAX-DN in scrib1+ab clones restrains clonal tissue overgrowth and GFP specks are observed, consistent with cells undergoing cell death (C). Larvae still enter an extended larval stage of development, however, tumour overgrowth (D) is reduced compared to scrib1+ab tumours (eg. Figure 8F ). Yellow scale bar = 50 µm.
(JPG)
wtsX1+ab tumours retain Dll expression, but do not express Dan. ey-FLP induced eye/antennal disc clones marked by GFP (white, or green in merges). The cell fate markers Dll and Dan are shown in white (magenta when overlaid with GFP in merged images). F-actin (white) shows cell morphology. GFP (panels A–D), F-actin (panels A′–D′), Dll (panels A″,B″), Dan (panels C″,D″), GFP/Dll merges (panels A″′,B″′) and GFP/Dan merges (panels C″′,D′″). (A) wtsX1 clones exhibit the normal pattern of Dll in the eye/antennal disc. (B) wtsX1+ab clones retain Dll expression, often resulting in ectopic domains of Dll-expressing tissue (B, arrowhead), similar to ab expressing clones, or scrib1+ab tumours. (C) wtsX1 clones exhibit the normal pattern of Dan in the eye/antennal disc. (D) wtsX1+ab clones do not express Dan in the antenna, and the overgrowths in the eye disc are also characterised by a loss of Dan (D, arrowhead). Yellow scale bar = 50 µm.
(JPG)
Live cell imaging of scrib −+ab tumours. Movie taken at 2 minute intervals over a 20 hour period at 10× magnification. scrib1+ab tumour cells are highly motile, moving between, and over, the brain lobes. Note that, although motile hemocytes have been reported be associated with the tumours, and may be associated with tumour cell engulfment, the intensity and distribution of GFP fluorescence within the motile cells is not consistent with them being phagocytic hemocytes.
(ASF)
Live cell imaging of scrib −+ab+bskDN tumours. Movie taken at 2 minute intervals over a 20 hour period at 10× magnification. scrib1+ab+bskDN tumour cells are non invasive, and the overall tumour mass, whilst growing, remains compact.
(ASF)
Transcription factor matrices enriched amongst peak sequences associated with the promoter regions or introns of potential target genes.
(DOC)