Abstract
Heterorhabditis bacteriophora are entomopathogenic nematodes that have evolved a mutualism with Photorhabdus luminescens bacteria to function as highly virulent insect pathogens. The nematode provides a safe harbor for intestinal symbionts in soil and delivers the symbiotic bacteria into the insect blood. The symbiont provides virulence and toxins, metabolites essential for nematode reproduction, and antibiotic preservation of the insect cadaver. Approximately half of the 21,250 putative protein coding genes identified in the 77 Mbp high quality draft H. bacteriophora genome sequence were novel proteins of unknown function lacking homologs in Caenorhabditis elegans or any other sequenced organisms. Similarly, 317 of the 603 predicted secreted proteins are novel with unknown function in addition to 19 putative peptidases, 9 peptidase inhibitors and 7 C-type lectins that may function in interactions with insect hosts or bacterial symbionts. The 134 proteins contained mariner transposase domains, of which there are none in C. elegans, suggesting an invasion and expansion of mariner transposons in H. bacteriophora. Fewer Kyoto Encyclopedia of Genes and Genomes Orthologies in almost all metabolic categories were detected in the genome compared with 9 other sequenced nematode genomes, which may reflect dependence on the symbiont or insect host for these functions. The H. bacteriophora genome sequence will greatly facilitate genetics, genomics and evolutionary studies to gain fundamental knowledge of nematode parasitism and mutualism. It also elevates the utility of H. bacteriophora as a bridge species between vertebrate parasitic nematodes and the C. elegans model.
Introduction
Nematodes are the most abundant multicellular animals on the planet [1], and exhibit remarkably diverse lifestyles to impact all life [2]. While some nematode parasites harm humans and agriculture, entomopathogenic (i.e., insect-parasitic) nematodes (EPNs) are beneficial in controlling insect pests [3], [4]. Two EPN families, Heterorhabditidae and Steinernematididae, [5], [6] have independently evolved mutual associations with insect pathogenic Photorhabdus and Xenorhabdus bacteria, respectively [7], [8]. A specialized stage of the nematode, analogous to the C. elegans dauer, called the infective juvenile (IJ) harbors the mutualistic bacteria in its intestine while in search of an insect host [9]. Once found, the nematodes penetrate the insect body, sense unknown cue(s) in the hemolymph, and then regurgitate the symbionts [10], [11]. The bacteria grow logarithmically and produce virulence factors and toxins causing rapid insect mortality [12]–[16]. The bacteria produce exoenzymes to degrade the insect tissues and produce unknown metabolites essential for nematode reproduction. Unlike C. elegans and other bacteria-feeding nematodes, H. bacteriophora reproduces only when associated with specific Photorhabdus bacteria both in insects and nutrient rich media [17], [18]. In addition, the H. bacteriophora intestine is more permissive to symbiotic and non-symbiotic Escherichia coli OP50 intestinal bacteria than C. elegans [19]. The bacteria produce potent secondary metabolites that are antibiotics [20] and which deter scavenging arthropods [21], enabling the nematode proliferation to nearly 500,000 IJs from a single infected insect, which then disperse in search of new insect hosts [19], [22].
Heterorhabditis bacteriophora and its mutualistic bacterium Photorhabdus luminescens represent a model system for the study of symbiosis and parasitism [11], [23], [24]. Although mutually dependent in nature, both organisms can be grown, manipulated and re-associated in culture. Heterorhabditis and Photorhabdus have congruent evolutionary lineages, indicating significant coevolution [25]. The bacteria adhere, persist, invade and grow inside nematode cells, breaching the alimentary tract to gain access to the developing IJs in the mother’s body [19]. The IJs select for bacteria that adhere to pharyngeal-intestinal valve cells, possibly invade these cells and exit to grow unattached in the intestinal lumen. It is likely that nematode receptors are exposed on specific cells in developmental stages where the bacteria adhere. For example, a phase variant subpopulation of the bacteria express maternal adhesion (Mad) fimbriae required for adhesion to the maternal intestine and transmission to IJs [26]. More surprisingly, the maternal nematodes select for a M-form phenotypic variant that is avirulent and slow growing compared to the insect pathogenic P form [27]. Visualizing the M-form cells persisting in the posterior intestine among the majority transients enabled the discovery that the P form changed to a small cell morphology (i.e. ∼1/7 vol) of the M form. The optical transparency of the nematodes and differential labelling of transient and persistent bacteria made apparent the mutualistic function of phenotypic variant easily ignored. Furthermore, the genetic tractability of the symbiont and ease of screening revealed the mutable locus and transcription factors required for the P and M form switching [26]. It is unknown why nematodes acquire the M form, which switch genetically back to the P form in fully developed IJs and arm these nematodes for insect infection.
The IJs and bacteria endure cooperatively [27], often for many weeks to months without feeding [28] while in search for their host. Lowering their metabolism through cellular acidification and repressed motility may aid the bacteria to persist in the gut of the IJ [27]. In addition to vectoring the bacteria between insect hosts, the IJs may contribute to immune suppression of the insect hosts [29]. Thus, H. bacteriophora has evolved sophisticated adaptations for bacterial mutualism enabling it to function as an entomopathogen.
The availability of recent data on genome sequences has laid the necessary foundation for the development of this model system. The complete genome of H. bacteriophora strain TT01 symbiont, Photorhabdus luminescens subsp. laumondii TT01, was released in 2003 [30]. Transcriptomic data of H. bacteriophora TT01 and GPS11 recently became available [31]–[33]. Forward genetics by mutagenesis using ethyl methane sulfonate (EMS) was successful [34], [35] [36] and reverse genetics, by gene silencing using RNAi, has been demonstrated in H. bacteriophora [24].
Moreover, techniques for genetic diversity assessment [37], [38], genetic selection [39]–[43], hybridization [44], subtractive amplification [45], [46], transcriptional profiling [47], proteomics [48], [49] and DNA transformation [50] have been achieved. Transformation of the H. bacteriophora germline with the C. elegans heat shock promoter transcriptionally fused to beta-galactosidase [50] and mec-4 (mechanosensitive) promoter transcriptionally fused to GFP [51] suggest that functional analysis of H. bacteriophora genes is possible.
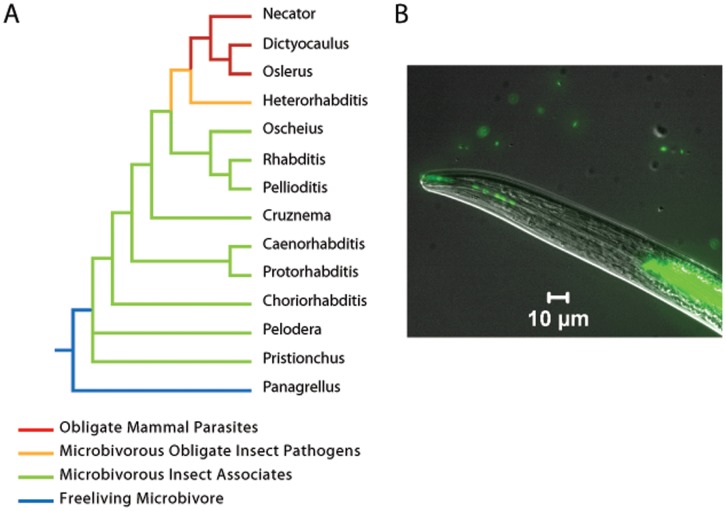
Evolutionarily, Heterorhabditis is a transitional taxon among the Rhabditina. It exhibits ancestral traits shared with its microbivorous ancestors such as C. elegans, but has also evolved parasitism and shares most recent common ancestry with obligate mammalian parasites, such as hookworms and lungworms. Given this phylogenetic position, Heterorhabditis can serve as a sort of “bridge” taxon for exploring the evolutionary changes that free-living microbivores have undergone along the path to obligate parasitism of mammals (Figure 1A). Although this figure is not intended to be comprehensive, it does illustrate the general evolutionary trend from free-living microbivory through facultative and obligate associations with invertebrates, to obligate parasitism of vertebrates: Panagrellus represents a large clade of free-living microbivores, which gave rise to a series of subsequent evolutionary lineages that are non-parasitic associates of invertebrates, followed by Heterorhabditis and its sister taxon, the Strongyloidea (represented by Necator, Dictylocaulus and Oslerus; obligate parasites of vertebrates). According to this scenario, a parsimonious reconstruction of evolutionary history features free-living microbivores giving rise to numerous microbivorous taxa that are facultative or opportunistic associates of invertebrates. However, such facultative and opportunistic conditions gave rise to a clade that evolved obligate parasitism. In Heterorhabditis microbivory (Figure 1B) and association with an invertebrate host were maintained. In contrast, the Strongyloidea have lost microbivory during the evolution of obligate parasitism. However, the entomopathogenic symbiosis can also be viewed as an innovation in parasitism where nematode association with an insect pathogen increases the virulence and fitness of insect infection. The clade containing Dictyocaulus and Oslerus (lungworms; Trichostrongylidae, Metastrongylidae, respectively) has direct lifecycles, being ingested as larvae by their mammalian hosts [52]–[54]. Necator (Hookworms; Ancylostomatidae) penetrate tissue to infect its host. Most of the lungworms require an invertebrate (mollusk) intermediate host. Building on this foundation, the objective of this study was to obtain a high quality genome sequence to facilitate further insights into the mutualistic and parasitic lifestyles of Heterorhabditis. The analysis of H. bacteriophora genome sequence reveals unique features that correspond to the evolution of mutualistic (lover) and parasitic (fighter) aspects of its biology.
Figure 1. Phylogenetic position of Heterorhabditis relative to other notable Rhabditina.
A. At the base of the tree is the free-living microbivorous Panagrellus (Panagrolaimoidea). Lineages in green are semaphoronts of large, diverse clades of microbivorous nematodes whose members associate with invertebrates at some point in their lifecycle, typically via phoresy and/or necromeny [52]–[54]. Heterorhabditis is a transitional taxon, exhibiting ancestral microbivorous traits, but has also evolved obligate pathenogensis and shares most recent common ancestry with obligate mammalian parasites (Strongyloidea; lineages in red). Modified after [127]–[129]. Taxonomy follows the ranking hypotheses and nomenclature of Hodda, 2011 [130]. B. H. bacteriophora nematodes have evolved a mutualism with insect pathogenic P. luminescens bacteria (green) where each partner cooperates to achieve voracious entomopathogenicity. An infective juvenile regurgitating intestinal symbionts (right) out the pharynx is shown. The movement of the nematode head causes slight misalignment of the fluorescent and differential interference micrograph image overlays.
Results and Discussion
A total of 6,845,656 sequencing reads totaling 2,410,251,025 base pairs were obtained from H. bacteriophora genome. After quality trimming and assembly, a draft genome consisting of 1,263 scaffolds totaling 77,007,652 bp was obtained. The size of the scaffolds ranged from 327 to 2,228,510 bp with 166 scaffolds larger than 100 kb. The N50 value of the assembled genome is 312,328 bp. The overall GC content is 32.2%, which is similar to the free-living nematode C. elegans, plant-parasitic nematode M. hapla, and human-parasitic nematode B. malayi (Table 1).
Table 1. Comparison of Heterorhabditis bacteriophora genome with the complete genome of Caenorhabditis elegans (WS220) and the draft genomes of Meloidogyne hapla [132] and Brugia malayi [87].
| C. elegans | H. bacteriophora | M. hapla | B. malayi | |
| Life style | Free living | Insect parasitic | Plant parasitic | Human parasitic |
| Genome size, Mb | 100 | ∼ 80 | 54 | 90–95 |
| Scaffolds | n/a | 1,263 | 1,523 | 8,810 |
| Scaffold N50, bp | n/a | 312,328 | 83,645 | 93,771 |
| Assembled, bp | 100,267,623 | 77,007,652 | 53,578,246 | 70,837,048 |
| Gene models | 21,193 | 21,250 | 14,420 | 11,515 |
| Median exon, bp | 147 | 112 | 145 | 140 |
| Average exon/gene | 6 | 6 | 6 | 7 |
| Median intron, bp | 68 | 125 | 55 | 219 |
| G+C, % | 35.4 | 32.2 | 27.4 | 30.5 |
Protein-coding Genes
The protein-coding genes were predicted using parameters optimized for C. elegans in the ab initio gene prediction programs. In total, 21,250 protein-coding genes were predicted (Table S1). The majority of the predicted protein genes, 11,207, had no significant homolog to C. elegans (WormBase release WS220), whilst 10,043 H. bacteriophora proteins had homologs with an E value cutoff of 1e-5 (Table S2). Of the protein-coding genes that have no homologs in WS220, 9,893 had no significant sequence similarity to known proteins in the GenBank non-redundant database and were hence considered novel.
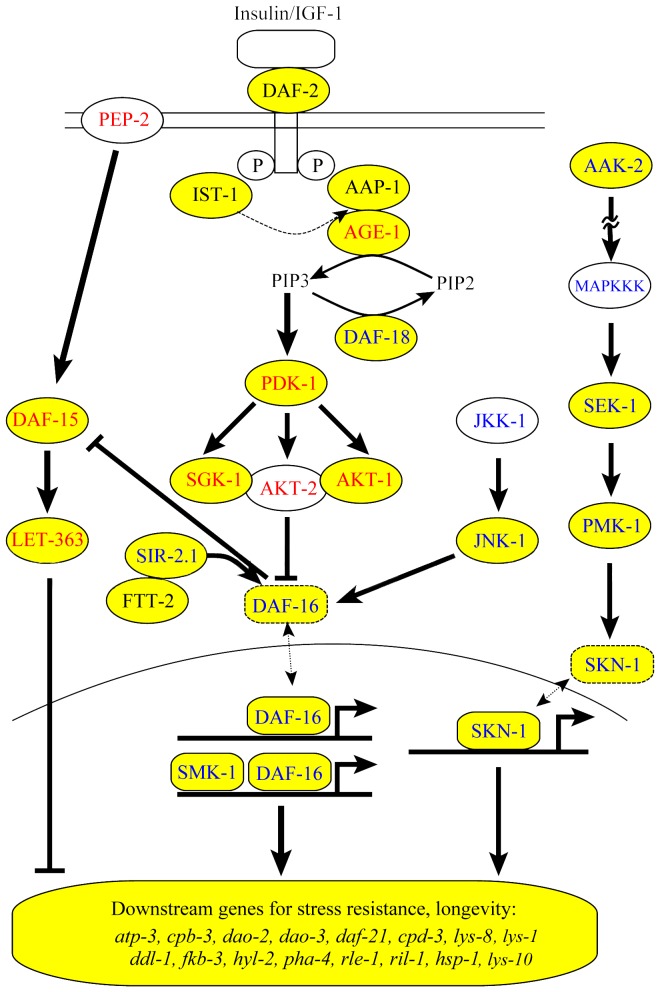
H. bacteriophora and strongylid parasites like hookworms have adapted a developmentally arrested and alternative third larval stage, known as dauer larva in C. elegans, as the infective stage [55]. Entomopathogenic IJs harbor gut symbionts that benefit their insect parasitism [56]. The C. elegans dauer develops under stressful conditions such as overcrowding by sensing dauer and other ascaroside pheromones, signal transduction through insulin and TGF-β pathways and DAF-12 nuclear hormone receptor [57]–[63]. H. bacteriophora produces an ascaroside ethanolamine (C11 EA) derivative that maintains the IJ state at high IJ densities and additional ascarosides [64], [65]. We found that H. bacteriophora has most (19 of 23) of the insulin/IGF-1 signaling pathway genes that are critical for dauer formation and for regulation of longevity, stress resistance and innate immunity in C. elegans (Figure 2). We also found a daf-12 homolog predicted to function in ascaroside transcriptional response [66]. Study of IJ formation and exit from diapause, easily tested in insects like Drosophila melanogaster and assessed by release of intestinal symbionts [10], [11], may lead to new antiparasitic strategies. Increasing IJ longevity and stress resistance may lead to improvements of EPNs for pest control [28], [67], [68].
Figure 2. Genes of insulin/IGF-1 signaling pathway in H. bacteriophora (highlighted in yellow) and C. elegans (all genes).
The genes in red and blue fonts are negative regulator and positive regulator, respectively, of stress resistance, lifespan, and immunity in C. elegan [131].
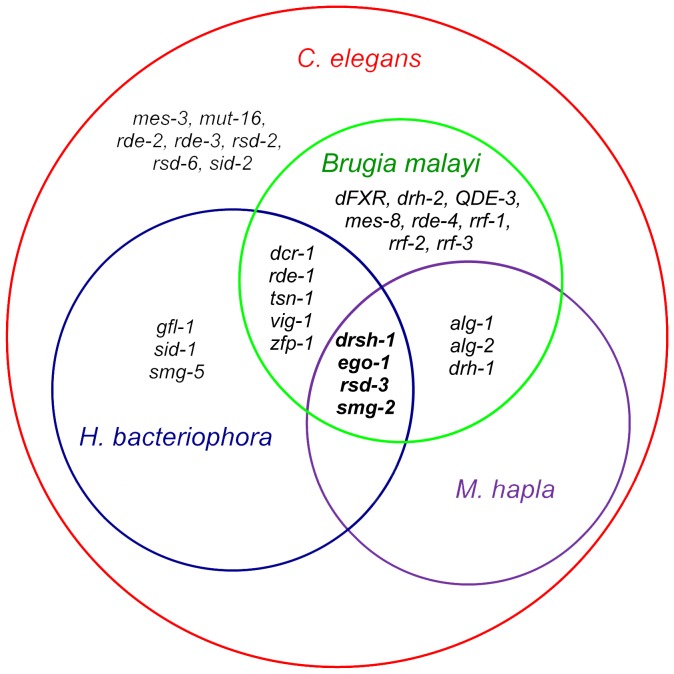
RNA interference (RNAi) is a pathway for gene regulation and powerful tool to manipulate gene expression in functional genomics [69]. RNAi by soaking has been achieved in H. bacteriophora [24]. We detected sid-1 and sid-3 homologs required for systemic RNAi in C. elegans [70], [71] but not a sid-2 homolog required in C. elegans for the uptake of dsRNA in the intestine [72]. Either an Hba- sid-2 homolog was left out of the current H. bacteriophora assembly or another transport mechanism is employed. Although C. elegans efficiently transports environmental DNA, most other related Caenorhabditis species do not [73]. Genes involved in RNA interference in H. bacteriophora, B. malayi, and M. hapla were identified based on sequence similarity to C. elegans gene products (Figure 3). Four genes, drsh-1, ego-1, rsd-3, and smg-2, have been identified in all four nematode species compared. In C. elegans, drsh-1 gene encodes a predicted RNase III-type ribonuclease that is orthologous to Drosha protein in Drosophila and human that is involved in processing primary miRNA transcripts (pri-miRNAs) in the nucleus [74]. ego-1 gene encodes putative RNA-directed RNA polymerase that is required for germline RNAi [75]. smg-2 is involved in non-sense-mediated mRNA decay that selectively and rapidly degrades eukaryotic mRNAs with premature stop codons [76]. rsd-3 is one of four RNA Spreading Defective genes (WormBase). A homolog of dcr-1 DiCer Related endonuclease [77] was detected in H. bacteriophora but not Dcr-1 associated protein rde-4, which is required for RNAi in C. elegans [78]. Since RNAi has been reported for H. bacteriophora [24], B. malayi [79], [80], and M. hapla [81], different mechanisms are possibly employed.
Figure 3. Comparison of genes involved in RNA interference pathway in C. elegans, B. malayi, M. hapla, and H. bacteriophora.
Four genes in bold, drsh-1, ego-1, rsd-3, and smg-2 were identified in all four species. sid-1 gene that is required for systemic RNAi in C. elegans was only identified in C. elegans and H. bacteriophora.
Protein Domains
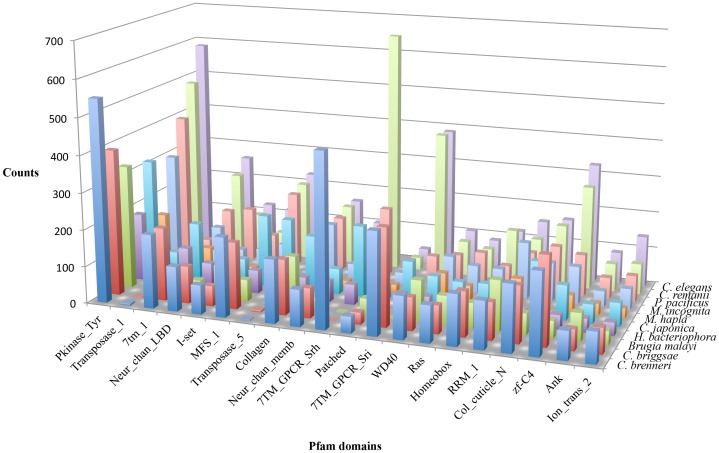
To begin to learn how the more than 10,000 unknown proteins function, we analyzed the proteins for conserved domains. A total of 7,957 Pfam domains with 4,144 different Pfam accessions were predicted using the program HMMER [82] with an E value cutoff of 1e-4. We compared the Pfam domains in H. bacteriophora with other nematodes [59] (Figure 4; Table S3). Based on protein domain information, we identified 82 members of GPCR (G protein coupled receptor) gene family and 24 members of NHR (nuclear hormone receptor) gene family. The domain richness index analysis (see methods) revealed 56 domains in H. bacteriophora that are significantly different from other nematodes. One significantly different richness domain index is the Mariner transposase (PF01359.11), with 138 identified in H. bacteriophora proteins compared to 65 in C. japonica, one each in M. incognita and M. hapla, but none in C. elegans and Brugia malayi. The Mariner transposases have been shown to be sufficient to mediate transposition in vitro in a purified form [83]. The enrichment of Mariner transposase domain is in agreement with the 1,314 predicted Mariner DNA motifs that belong to 23 types (Table 2; Table S4). In contrast, a search with the same parameters returned 844 Mariner DNA motifs that belong to 43 types in C. elegans genome (Table 2; Table S5). More strikingly, 28 types of Mariner DNA motifs are exclusively present in C. elegans genome and 8 types are exclusively present in H. bacteriophora genome. The differences in the number and type of Mariner DNA motifs between H. bacteriophora and C. elegans along with the enrichment of Mariner transposase domains and predicted transposition activity in H. bacteriophora is likely evidence of a past or presently mobile genome.
Figure 4. Comparison of top 20 Pfam domains in H. bacteriophora genome with those in the 10 nematode species in the study.
The top 20 Pfam domains were identified as the ones having the 20 largest number of occurrence in H. bacteriophora genome.
Table 2. Numbers of mariner type motifs in H. bacteriophora and C. elegans genomes.
| Mariner type | Hba | Cel | Mariner type | Hba | Cel | |
| Mariner2_CE | 36 | 93 | Mariner36_CB | 39 | 4 | |
| Mariner3_CE | 18 | 73 | Mariner37_CB | – | 4 | |
| Mariner4_CB | 2 | 1 | Mariner38B_CB | – | 4 | |
| Mariner4_CE | 27 | 8 | Mariner38C_CB | – | 2 | |
| Mariner5_CE | 4 | 68 | Mariner38_CB | – | 1 | |
| Mariner7_CB | – | 180 | Mariner40_CB | – | 11 | |
| Mariner8_CB | – | 6 | Mariner41_CB | – | 2 | |
| Mariner10_CB | – | 3 | Mariner42_CB | 1 | 2 | |
| Mariner12_CB | 1 | 1 | Mariner43_CB | 8 | – | |
| Mariner13_CB | 59 | 9 | Mariner44_CB | – | 1 | |
| Mariner14_CB | 135 | – | Mariner45_CB | – | 6 | |
| Mariner15_CB | 332 | 1 | Mariner47A_CB | – | 14 | |
| Mariner16_CB | 108 | – | Mariner47B_CB | – | 9 | |
| Mariner17_CB | 12 | – | Mariner47_CB | – | 6 | |
| Mariner18_CB | 448 | – | Mariner48_CB | – | 2 | |
| Mariner19_CB | 12 | – | Mariner51_CB | – | 2 | |
| Mariner20_CB | – | 1 | Mariner52_CB | – | 13 | |
| Mariner22_CB | 2 | 4 | Mariner53_CB | – | 94 | |
| Mariner23_CB | – | 1 | Mariner54_CB | – | 17 | |
| Mariner25_CB | – | 1 | Mariner55_CB | 1 | – | |
| Mariner26_CB | 1 | – | Mariner56_CB | – | 1 | |
| Mariner27_CB | 1 | 1 | Mariner60_CB | – | 4 | |
| Mariner28_CB | – | 166 | Mariner61_CB | – | 3 | |
| Mariner31_CB | 1 | 10 | Mariner65_CB | – | 2 | |
| Mariner32_CB | 64 | 1 | Mariner66_CB | – | 4 | |
| Mariner34_CB | 2 | 8 | Total | 1314 | 844 |
Abbreviations: Hba, Heterorhabditis bacteriophora; Cel, Caenorhabditis elegans.
We detected far fewer (9 vs. 133) C-type lectin domain-containing proteins than are present in C. elegans. Homologs of lec-1, lec-2, lec-3, lec-5, lec-6, and lec-12 were detected that function in innate immunity in C. elegans [84]. The reduction in C-lectin domain proteins in H. bacteriophora may be related to the mutualistic relationship with P. luminescens bacteria [19]. Viable symbiotic bacteria are required in the intestine for maternal transmission and in IJs for insect infection. The H. bacteriophora intestine is more permissive to symbiotic bacteria and non-symbiotic E. coli OP50 than C. elegans. Broad-spectrum antibiotics produced by the symbionts likely contribute to defense against pathogenic and saprophyitic microorganisms. H. bacteriophora might also contain a diverse and novel set of innate immune effectors that were not detected by homology to C. elegans.
Non-coding RNA (ncRNA) and Regulatory Elements
A total of 134 potential microRNA (miRNA) genes were identified in H. bacteriophora genome representing 26 different animal microRNA species (Table S6). Other ncRNA include the U1, U2, U3, U4, U5, and U6 small nuclear RNA (snRNA) components of the spliceosome, SL1 involved in trans-splicing (none if 1e-10 cutoff is used), ribonuclease P (RNaseP), and eukaryotic-type signal recognition particle RNA. The number of the non-coding RNAs detected in H. bacteriophora is considerably less than those known to be present in C. elegans (Table S6). For instance, let-7 is absent in the current assembly although its presence and temporal expression were considered to be conserved among animals with bilateral symmetry [85], possibly due an incomplete genome assembly. The ncRNAs have important roles in regulating transcription, translation, and other biological processes.
A total of 254 transfer RNA (tRNA) genes and 1 tRNA pseudogene were predicted in H. bacteriophora genome by tRNAScan-SE (see Table S7) for all 20 standard amino acids, but not the tRNA-Selenocysteine gene. The number of detected tRNA genes in H. bacteriophora is dramatically lower than the 659 tRNA genes and at least 29 tRNA pseudogenes in C. elegans [86]. However, the number of tRNAs are close to those identified in human and plant parasitic nematodes. There are 233 tRNA genes and 26 tRNA pesudogenes identified in the human parasitic B. malayi [87] and 467 tRNA genes, 120 tRNA pseudogenes and 28 other tRNA genes in plant parasitic M. incognita [88].
Microsatellite Repeats
Microsatellites, also known as simple sequence repeats (SSRs), are tandem repeat sequences of 2–6 bp that serve as informative genetic markers to resolve relationships among closely related species because of their high mutation rate [89]. A total of 3,794 microsatellite loci were predicted in 506 contigs of the current draft H. bacteriophora genome (Table S8). Among them, 849 were located in coding regions. Previously, we developed 8 polymorphic microsatellite markers for H. bacteriophora that distinguished a Northeast Ohio population from other populations [90]. These microsatellite markers can serve as useful tools for determining the phylogeographic, demographic and genetic structure of H. bacteriophora populations.
Estimation of Divergence Time between H. bacteriophora and C. elegans
The divergence time between H. bacteriophora and C. elegans was estimated based on a set of 350 orthologs among H. bacteriophora, C. elegans, Anopheles gambiae, and Homo sapiens. Based on the divergence time of 800–1000 MYA between nematodes and insects [91], the estimated divergence time between H. bacteriophora and C. elegans is approximately 86–331 MYA. By contrast, the C. elegans and C. briggsae speciation date was estimated as 78–113 MYA [91]. The large (conservative) discrepancy between the upper and lower bounds are probably most strongly influenced by the sparse taxonomic sample (n = 4), as well as other analytical biases [92].
Characterization of the Secretome
H. bacteriophora secreted proteins are potentially important for parasitic interactions with insects, mutualistic interactions with symbiotic bacteria, immunity to pathogens and in development and reproduction. We detected 753 proteins with predicted signal peptides of which 150 also were predicted to be membrane localized. The 603 potentially secreted proteins (2.8% of total predicted proteins) are similar to the fraction of B. malayi secretome proteins (2.3%), but are less than the free-living nematodes C. elegans (10.1%), C. briggsae (9.4%), C. brenneri (8.9%), C. japonica (6.2%), and C. remanei (8.8%), and the insect-associated P. pacificus (7.4%) when predicted with the same method and criteria. It is also about half of that of plant-parasitic nematodes M. hapla (5.2%) and M. incognita (5.2%). The low number of predicted secreted proteins in parasitic H. bacteriophora and B. malayi could be due to their reliance on mutualistic bacteria for these proteins.
Among the 603 H. bacteriophora secreted proteins, 164 had significant similarity (E value cutoff of 1e-5) to proteins in the SwissProt database (Table S9). Among the remaining 439 secreted proteins, 122 had significant similarity to proteins in the GenBank non-redundant database. The remaining 317 secreted proteins were novel proteins of unknown function. A search of the MEROPS database containing peptidases and peptidase inhibitors revealed the presence of 1 cysteine, 9 serine, and 9 metallo-peptidases and 9 peptidase inhibitors in H. bacteriophora secreted proteins (Table 3). Secreted peptidases have known roles in degrading host tissues for the benefit of parasites [92]. EPNs have been reported to release proteolytic enzymes to aid penetration of the insect gut to reach the hemocoel [93]. Following nematode penetration into the hemocoel, IJ secreted peptides and peptide inhibitors might function to disarm the insect serine proteinase cascade that results in pro-phenoloxidase activation and melanization, the elementary immune defense reaction [94]. However, during subsequent development of the nematode in the host hemocoel, the symbiont secretes peptidases/proteases [13]–[16], [30], which may contribute to such functions. Indeed, the mutualistic bacteria of EPNs also act independently to suppress the insect immune system [29], [95]. Therefore, both partners act synergistically in combating the insect immune system. A peptidase(s) also might function in utilizing symbiont-produced crystalline inclusion proteins (CipA and CipB) that are high in essential amino acid content and required for nematode reproduction [96]. H. bacteriophora also has homologs to C. elegans lysozyme genes lys-1, lys-3–8 and lys-10 that function in bacterial cell lysis and innate immunity [97]. Thus, although similarity suggests common function, it remains to be determined what roles most secreted proteins have in interspecies interactions.
Table 3. Summary of secreted peptidases and peptidase inhibitors identified in H. bacteriophora.
| Protein name | MEROPS family | Query start-end | MEROPS accession | Hit start-end | E value |
| Cysteine peptidases | |||||
| Hbpro09515 | C46 | 185–256 | MER011696 | 342–415 | 1.90e-07 |
| Metallopeptidases | |||||
| Hbpro17338 | M10A | 122–237 | MER003153 | 200–317 | 3.80e-30 |
| Hbpro04992 | M12A | 92–170 | MER003171 | 124–202 | 1.60e-20 |
| Hbpro10653 | M12A | 94–207 | MER015241 | 216–328 | 6.10e-25 |
| Hbpro13863 | M12A | 319–441 | MER024920 | 128–246 | 2.60e-23 |
| Hbpro15592 | M12A | 133–320 | MER002349 | 134–320 | 1.10e-91 |
| Hbpro15986 | M12A | 145–326 | MER001107 | 94–261 | 1.60e-37 |
| Hbpro20263 | M12A | 70–253 | MER001593 | 60–237 | 2.10e-41 |
| Hbpro11857 | M12B | 104–164 | MER002292 | 347–417 | 2.00e-25 |
| Hbpro13918 | M13 | 37–327 | MER002350 | 78–370 | 1.60e-90 |
| Serine peptidases | |||||
| Hbpro01274 | S01A | 9–76 | MER099499 | 26–91 | 2.80e-06 |
| Hbpro17402 | S08A | 795–954 | MER134526 | 298–451 | 8.30e-05 |
| Hbpro16490 | S08B | 203–296 | MER001610 | 179–272 | 1.20e-52 |
| Hbpro11245 | S09X | 17–347 | MER037861 | 26–353 | 1.20e-36 |
| Hbpro11940 | S09X | 34–208 | MER037861 | 29–207 | 4.70e-32 |
| Hbpro20894 | S10 | 35–61 | MER000430 | 39–65 | 3.80e-07 |
| Hbpro12626 | S28 | 59–214 | MER162965 | 54–211 | 3.50e-34 |
| Hbpro14365 | S28 | 130–242 | MER171698 | 102–212 | 2.00e-40 |
| Hbpro12626 | S37 | 59–207 | MER001350 | 62–194 | 1.20e-05 |
| Peptidase inhibitors | |||||
| Hbpro11626 | I02 | 386–432 | MER018193 | 250–296 | 4.50e-13 |
| Hbpro12168 | I02 | 20–51 | MER022808 | 669–700 | 2.90e-09 |
| Hbpro15022 | I02 | 282–333 | MER092785 | 4–53 | 3.10e-07 |
| Hbpro17931 | I02 | 18–71 | MER020231 | 5–56 | 1.50e-11 |
| Hbpro06248 | I08 | 21–81 | MER017818 | 10–63 | 2.50e-05 |
| Hbpro11583 | I17 | 117–167 | MER019417 | 27–69 | 4.90e-06 |
| Hbpro20975 | I21 | 49–128 | MER016218 | 70–155 | 5.60e-13 |
| Hbpro11626 | I31 | 325–375 | MER020813 | 331–379 | 6.10e-08 |
| Hbpro19310 | I51 | 120–189 | MER029866 | 66–135 | 1.70e-26 |
Gene Ontology Enrichment
The predicted Gene Ontology of H. bacteriophora proteins was compared to those of the proteins from the other nine sequenced nematode genomes (Table S10). A striking difference is the significant enrichment of DNA metabolic process (GO:0006259), DNA recombination (GO:0006310), DNA-mediated transposition (GO:0006313), DNA integration (GO:0015074), transposition (GO:0032196) and transposase activity (GO:0004803) in H. bacteriophora compared to other nematodes, with the exception of C. japonica. These observations are in agreement with the enrichment of mariner transposase domain in H. bacteriophora discussed above.
Metabolic Pathway Comparison
The KEGG (Kyoto Encyclopedia of Genes and Genome) pathways were predicted for H. bacteriophora and other 9 nematode species for which full genome sequence information is available and the numbers of genes in each pathway are summarized in Table S11. The genes and KEGG orthology (KO) in the metabolic pathways were compared to assess whether there is enrichment or reduction in the H. bacteriophora genome compared to other select nematode genomes (Table 4). H. bacteriophora has fewer KOs compared to the free-living nematode C. elegans in almost all metabolic categories, which is compatible with previous observations that parasitic nematodes seem to undergo reductive genome evolution [98]. However, H. bacteriophora has substantially more proteins (48 in total) in the KO groups of glycan biosynthesis and metabolism (Table S12). Glycans are generally found attached to proteins as in glycoproteins and proteoglycans on the exterior surface of cells and play important roles in proper protein folding and cell-cell interactions [99]. At the enzyme level, H. bacteriophora has 17 (out of 23) enzymes in common with C. elegans (19 enzymes in total). Interestingly, C. elegans, B. malayi and M. hapla have only one isoform (isoform 1) of [heparan sulfate]-glucosamine 3-sulfotransferase (3-OST), whereas H. bacteriophora has three isoforms, isoform 1, 2 and 3. The enzyme 3-OST is involved in biosynthesis of glycan structure and different isoforms have been demonstrated to have different substrate specificities depending on the saccharide structures around the modified glucosamine residue [100]. The presence of the two additional isoforms of 3-OST enzyme together with other H. bacteriophora-specific enzymes involved in glycan biosynthesis and metabolism suggests that H. bacteriophora is well evolved to thrive in different environments where different metabolic substrates are available during its life cycle.
Table 4. Genes and KEGG orthology (KO) in metabolic pathways in selected nematode species with different life styles.
| Pathway | KOs in KEGG reference pathway | Insect-parasitic H. bacteriophora | Human parasitic B. malayi | Plant-parasitic M. hapla | Free-living C. elegans | ||||
| Genes | KOs | Genes | KOs | Genes | KOs | Genes | KOs | ||
| Metabolism | 2,258 | 1,224 | 631 | 1,694 | 623 | 971 | 572 | 2,508 | 790 |
| Amino acid metabolism | 484 | 199 | 113 | 196 | 93 | 174 | 102 | 470 | 152 |
| Biosynthesis of other secondary metabolites | 55 | 46 | 22 | 62 | 21 | 32 | 13 | 108 | 25 |
| Carbohydrate metabolism | 550 | 238 | 145 | 448 | 165 | 207 | 140 | 540 | 199 |
| Energy metabolism | 408 | 124 | 46 | 215 | 51 | 93 | 47 | 207 | 58 |
| Glycan biosynthesis and metabolism | 160 | 75 | 48 | 97 | 36 | 43 | 33 | 66 | 35 |
| Lipid metabolism | 325 | 159 | 85 | 200 | 82 | 137 | 77 | 350 | 99 |
| Metabolism of cofactors and vitamins | 301 | 104 | 50 | 130 | 52 | 68 | 40 | 187 | 58 |
| Metabolism of other amino acids | 126 | 67 | 42 | 55 | 33 | 48 | 36 | 156 | 52 |
| Nucleotide metabolism | 174 | 159 | 49 | 223 | 59 | 104 | 50 | 240 | 68 |
| Xenobiotics biodegradation and metabolism | 178 | 53 | 31 | 68 | 31 | 65 | 34 | 184 | 44 |
Orthologs
The orthologous sequences among H. bacteriophora, C. elegans, C. briggsae, C. japonica, C. remanei, C. brenneri, Brugia malayi, Meloidogyne hapla, M. incognita, and Pristionchus pacificus were identified using the orthoMCL program [101] on the predicted protein sequences from the genomes. In total, we identified 183 orthologs among these species (Table S13). Based on the Gene Ontology information of C. elegans genes in the ortholog sets, most of these orthologs are essential in C. elegans and annotated to biological processes such as reproduction (number of orthologs: 50), growth (36), regulation of growth (47), regulation of biological process (61), and larval development (45). Genome sequences of other nematodes, including Bursaphelenchus xylophilus [102], Trichinella spiralis [98], and Ascaris suum [103], are not included in the analysis because trophic categories represented by these nematodes are already included in the current study.
H. bacteriophora is useful for Comparisons of Rapidly Evolving Protein Domains
Some proteins that are conserved from human to C. elegans have domains that are evolving too rapidly to analyze by the large evolutionary distance comparison. One example is the carboxyl terminal tail of EGF-receptor, called LET-23 in nematodes. A three-species comparison of elegans-briggsae-japonica has a C-terminus that is too conserved to be informative (being 65% identical), but addition of H. bacteriophora in a 4-way comparison highlights the tyrosines and PDZ-binding domain that have been shown to be functional in LET-23 [104], [105], with only 26% identified across the four species (Figure 5).
Figure 5. H. bacteriophora informs C. elegans protein structure function.
Multiple alignment of the EGF-receptor (LET-23) carboxyl tail of Caenorhabditis elegans, briggsae and japonica with H. bacteriophora. 3-way, alignment of the three Caenorhabditis proteins; 4-way, alignment of three Caenorhabditis proteins with Hba-LET-23. *, identity; :, strong similarity; ., weak similarity. Red and green highlight the parts of the protein that have been demonstrated to be important in signaling and localization, respectively. Numbers represent the length of the predicted proteins.
Conclusions
H. bacteriophora is an entomopathogenic nematode, which is mutually associated with symbiotic bacteria to function as an insect parasite. The high quality draft genome sequence revolutionizes our knowledge and genetic tractability to understand nematode fundamental processes of gut mutualism and insect parasitism. H. bacteriophora is well-known of symbiosis compared to the C. elegans and thus represent a simple and tractable model of animal-bacteria gut symbiosis. The genome sequence along with RNAi gene silencing methodology provides a powerful reverse genetic approach to probe the functions of signaling pathways and transcription factors in symbiosis as well as insect parasitism. The H. bacteriophora genome sequence along with some sequences from other H. bacteriophora strains (e.g. GPS11) allow single nucleotide polymorphisms (SNPs) to be identified which can be used in mapping. For example, nematode mutations can be mapped to SNPs and identified by genome resequencing and their function validated by RNAi. In addition, H. bacteriophora cis- and untranslated regulatory elements can be identified and used to facilitate expression of transgenes. These approaches can be used to learn how the nematode associates with symbiotic bacteria, what is the basis for dependency of these nematodes on symbiotic bacteria for reproduction and how do nematodes function as parasites? Therefore, the H. bacteriophora TT01 genome facilitates both basic and applied research on entomopathogenic nematodes.
Materials and Methods
Nematode Culture
An inbred line, M31e, self-fertilized 13 times, of H. bacteriophora TT01 strain originally isolated from Trinidad and Tobago [106] and kindly provided by Dr. Ann Burnell (NUI-Maynooth, Ireland), was thawed from cryopreserved stocks [24]. Axenic IJs were obtained by culturing the nematodes on strain P. temperata TRN16 that do not colonize IJs [26]. High molecular weight DNA was purified from first and second larval stages harvested from lawns of TRN16 grown previously for 18 h at 28°C on NA+chol (4 g nutrient agar, 1 g sodium pyruvate, 10 g agarose per liter with 2 ml 5 mg/ml cholesterol added after autoclaving). On average, 275 IJs were added to 100 mm lawns for efficient egg laying. Nematodes were washed off the lawns after 82–86 h with 10 ml of Ringer’s containing 0.1% triton X-100. Bacteria were removed by washing on a 10 µm pore nylon filter and hermaphrodites removed by retention on a 30 µm filter. Eggs were surface sterilized with 1% commercial bleach (Chlorox®), washed 3X in Ringer’s solution and allowed to hatch in Ringer’s solution containing 100 µg/ml carbenicillin, 50 µg/ml streptomycin, 30 µg/ml kanamycin and 10 µg/ml gentamicin overnight. A contaminant of Stenotrophomonas maltophilia, likely originating from a contaminated Ringer’s solution, was inadvertently sequenced along with the nematode. Approximately 3×106 L1 nematodes were harvested from 1,000 cultures.
Isolation of RNA
Nematode mRNA was isolated from mixed (L1–L4), adult and IJ stages grown on TRN16. The nematodes were obtained from the cultures with Ringer’s solution and bacteria removed by 3 washes with 15× Ringer’s solution in a 15 ml conical tube and centrifugation for 5 min at 2,000 rpm. The nematodes were frozen in liquid nitrogen, then Trizol reagent (Life Technologies) was added and incubated at 65°C. The IJs were freeze-thawed 3× in liquid nitrogen and at 65°C, before RNA was purified per manufacturer’s instructions. Polyadenylated RNA was purified using oligo(dT) cellulose columns, MicroPoly(A)Purist Kit (Life Technologies).
cDNA Library Construction and Sequencing
The integrity of the mRNA was validated using the Bioanalyzer 2100 (Agilent Technologies) and yield determined via Nanodrop (Thermo Scientific). Two different methods were used for library construction:
The CloneMiner cDNA Library Construction Kit (Life Technologies) was utilized to generate non-radiolabeled cDNA according to the manufacturer’s specifications. A Biotin-attB2-Oligo(dT) primer was hybridized to mRNA. First strand cDNA was synthesized via SuperScript II Reverse Transcriptase. DNA polymerase I was utilized to generate the second strand of cDNA. attB1 adapters were ligated to the 5′ end of the cDNA. The cDNA was purified by column fractionation to remove residual adapters. Through site-specific recombination, attB-flanked cDNA was cloned directly into the pDONR-222 vector (Life Technologies). The ligations were transformed using the ElectroMax DH10B cells (Life Technologies). The transformed cells were spread on LB plates containing 50 µg/mL kanamycin.
mRNA was used as the template for cDNA library construction using the Accuscript HF Reverse Transcriptase Kit (Agilent Technologies) and SMART primers (Life Technologies). PCR cycle optimization was performed to determine the threshold cycle number to minimally amplify full length cDNA products using the SMART primers and Clontech Advantage-HF 2 polymerase Mix (Clontech/Takara Bio). Library normalization was accomplished by using the Trimmer kit (Evrogen). PCR cycle optimization was performed with normalized cDNA to determine the threshold cycle number using the SMART primers and Clontech Advantage-HF 2 polymerase Mix previously mentioned. Finally, 5′ and 3′ adapter excision was performed by restriction exonuclease digestion using MmeI. The excised adapters were removed utilizing AMPure paramagnetic beads (Agencourt, Beckman Coulter Genomics). Two kinds of libraries were prepared for sequencing on ABI3730 and Roche/454 platforms.
For libraries intended for sequencing on ABI3730 platform, the final cDNA product was nebulized, end repaired (Lucingen), and size selected from a 0.8% SeaKem agarose TAE gel. The fraction was purified according to the manufacturer’s instructions in the QIAquick Gel Extraction (Qiagen) protocol and ligated into the pSMART HC-Kan vector system (Lucigen). Ligations were transformed using E. coli cells (Lucingen). The transformed cells were spread onto LB plates containing 50 µg/mL kanamycin.
A 454 fragment library was constructed using GS DNA Library Preparation Kit (Roche) with the cDNA as outlined in the manufacturer’s protocol. Five microgram of cDNA was fragmented via nebulization. Fragmented cDNA was size selected with an AMPure bead (Agencourt, Beckman Coulter Genomics) cleanup, removing fragments less than 300 bp. The cDNA was end polished and ligated to 454 Titanium library adapters utilizing reagents from the Titanium General Library Kit (Roche). An AMPure (Agencourt) bead cleanup was performed to remove library adapter dimers and cDNA fragments less than 400 bp in length. The 454 library was immobilized with Strepavidin beads (-Roche) and single stranded with Sodium Hydroxide. The single stranded library was quantitated by a Quant-iT single stranded DNA assay using the Qubit fluorometer (Life Technologies) and the integrity validated using the BioAnalyzer 2100 (Agilent Technologies). The library fragments were immobilized onto DNA capture beads utilizing clonal amplification kits (Roche). The captured DNA library was emulsified and subjected to PCR in order to amplify the DNA template. The emulsion was chemically broken and the beads containing the DNA were recovered, washed, and enriched utilizing bead recovery reagents (Roche). The DNA library beads were loaded onto a PicoTiterPlate device and sequenced on the Genome Sequencer instrument using the GS FLX Titanium Sequencing Kit XLR70 (Roche).
Genomic Library Construction and Sequencing
High molecular weight genomic DNA was isolated using a protocol kindly provided by Erich Schwartz, which was based on that of Andrew Fire’s lab with slight modifications from the R. Waterston lab and K. Kiontke [107]. The integrity of the genomic DNA was verified by comparing the intensity of H. bacteriophora to serial dilutions of lambda standards of known concentration on a 1.8% agarose gel stained with ethidium bromide. The yield was determined by a high sensitivity Quant-iT double stranded DNA assay using a Qubit fluorometer (Life Technologies). A 454 Titanium fragment library was constructed with 5 µg of genomic DNA as outlined in the manufacturer’s protocol. The genomic DNA was fragmented via nebulization and run on a 0.8% GTG Seakem agarose gel (Lonza) with ethidium bromide in 1× TAE buffer for a size selection of 500–800 bp. Fragmented DNA was isolated from the agarose gel using the QiaQuick Gel Extraction Kit (Qiagen). The size selected DNA was end polished and ligated to 454 Titanium library adapters utilizing reagents from the Titanium General Library Kit (Roche). An AMPure (Agencourt) bead cleanup was performed to remove library adapter dimers and DNA fragments less than 400 bp in length. The 454 library was immobilized with Strepavidin beads (Roche) and single stranded with sodium hydroxide. The single stranded library was quantitated by a Quant-iT single stranded DNA assay using a Qubit fluorometer (Life Technologies) and the integrity validated using the BioAnalyzer 2100 (Agilent Technologies). The library fragments were immobilized onto DNA capture beads utilizing clonal amplification kits. The captured DNA library was emulsified and subjected to PCR in order to amplify the DNA template. The emulsion was chemically broken and the beads containing the DNA were recovered, washed, and enriched utilizing bead recovery reagents. The DNA library beads were loaded onto a PicoTiterPlate device and sequenced on the Genome Sequencer instrument using the GS FLX Titanium Sequencing Kit XLR70 (Roche).
Genome Assembly
The genome sequences from fragments, 3 kb insert from plasmid libraries and end sequencing of bacterial artificial clone libraries were generated at an estimated 26-fold sequence coverage. All sequenced reads were attempted in de novo assembly using the Celera assembler v. 6.0. The assembly was submitted to GenBank genome database under accession number ACKM00000000.
Genome Annotation
The scaffolds were masked for repeats using RepeatMasker version 3.3 [108]. Transfer RNA coding genes were predicted using tRNAscan-SE [109]. To identify microRNA, other non-coding RNA, and regulatory elements, Rfam [110] covariance models were searched using Inferno program [111], [112] with an E value cutoff of 1e-8 after adjusting to the size of the genome. Protein-coding genes were predicted with gene prediction programs of SNAP [113], AUGUSTUS [114]–[116], GlimmerHMM [117], and GeneMark [118]. The results were integrated with other evidence, including the mapping results of ESTs generated by cDNA sequencing with sim4 and sequence similarity to proteins in GenBank non-redundant (nr) database and WormBase WS220 release, by JIGSAW program [119] with linear combiner option. Gene models with in-frame stop codons were considered erroneous and therefore removed. Protein domains in the predicted protein-coding genes were predicted by searching Pfam [120] using the HMMER program [82] with an E value threshold of 1e-4. For comparison, the same prediction parameters were used to predict Pfam domains in other nematodes. A domain richness index for each domain in each nematode was calculated by dividing the number of that domain with the total number of protein sequences in that nematode species. The program T statistics was used to compare the domain richness indices among nematodes. H. bacteriophora protein sequences were assigned Gene Ontology terms by the Blast2GO program [121] based on the BLASTp results against the SwissProt database with an E value cutoff of 1e-10. The orthologous sequences among H. bacteriophora, C. elegans, C. briggsae, C. japonica, C. remanei, C. brenneri, Brugia malayi, Meloidogyne hapla, M. incognita, and Pristionchus pacificus were identified using the orthoMCL program [101] on the predicted protein sequences from the genomes. H. bacteriophora protease/peptidases were predicted based on sequence similarity search of the sequences in MEROPS database Release 9.5 [122].
Estimation of Divergence Time between H. bacteriophora and C. elegans
We obtained a set of 350 orthologs common to H. bacteriophora, C. elegans, Anopheles gambiae (AgamP3.4 release from VectorBase), and Homo sapiens (Ensembl release 55) based on the prediction results of orthoMCL [101]. For each ortholog set, the protein sequences were aligned using ClustalW2 [123], followed by reverse translation to their original transcript sequences that were obtained from the same respective databases as the protein sequences. After conversion to PHYLIP format, the alignments were used to estimate genetic distances among the taxa using the DNADIST program in PHYLIP (PHYLogeny Inference Package; [124]. A phylogenetic tree was then built using the PHYLIP neighbor-joining algorithm NEIGHBOR with human as the outgroup taxon. The sequence alignment and the rooted neighbor-joining tree were used to estimate divergence times using the MCMCTREE program in PAML (Phylogenetic Analysis by Maximum Likelihood [125]). We used 800–1000 MYA (million years ago) as the divergence time of nematodes and insects [91].
Gene Ontology Enrichment and Metabolic Pathway Comparison
H. bacteriophora protein sequences were assigned Gene Ontology (GO) terms by the Blast2GO program [121] based on the BLASTP results against SwissProt database with an E value cutoff of 1e-10. In comparison, proteins from the other 9 nematode genomes underwent the same analysis using the same programs and databases. The pair-wise GO enrichment using H. bacteriophora sequences as the reference was done using the GOSSIP program [126]. The KEGG (Kyoto Encyclopedia of Genes and Genome) Ontologies (KO) in the metabolic pathways were assigned using Blast2GO program [121] for the four nematode species being compared.
Ethics Statement
This study did not involve any human or vertebrate subjects.
Supporting Information
Predicted gene models in Heterorhabditis bacteriophora genome.
(XLSX)
Sequence similarity of conceptually translated H. bacteriohora proteins to C. elegans proteins in Wormbase release W220.
(XLSX)
Comparison of Pfam domains predicted in the proteins of 10 nematode species in this study.
(XLSX)
Predicted mariner DNA motifs in H. bacteriophora genome.
(XLSX)
Predicted mariner DNA motifs in C. elegans genome using the same parameters as the ones used to generate results in Table S4.
(XLSX)
Predicted non-protein-coding RNA in H. bacteriohora genome.
(XLSX)
Predicted tRNA genes in H. bacteriophora genome.
(XLSX)
Predicted microsatellite loci in H. bacteriophora genome.
(XLSX)
Predicted secretome in H. bacteriophora genome.
(XLSX)
Comparison of gene ontology terms that were assigned to genes in the 10 nemtode species included in this study.
(XLSX)
Comparison of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways predicted in the 10 nematode species included in this study.
(XLSX)
Comparison of the metabolism pathways in KEGG Ontologies in the 10 nematode species included in this study.
(XLSX)
The list of the IDs of orthologous sequences in the 10 nematode species included in this study.
(XLSX)
Acknowledgments
We thank the members of the EPN and EPB International Genome Sequence Consortium for support and the Washington University School of Medicine’s Genome Institute, St. Louis, MO, USA for sequencing the genome.
Funding Statement
This project was supported by grants from the United States Department of Agriculture (www.usda.gov) and National Science Foundation (www.nsf.gov) Microbial Genome Sequence Program under the grant #2006-35600-16665. P.W.S. is an investigator with the Howard Hughes Medical Institute (www.hhmi.org) with reference number of 047–101. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bongers T, Ferris H (1999) Nematode community structure as a bioindicator in environmental monitoring. Trends in Ecology and Evolution 14: 224–228. [DOI] [PubMed] [Google Scholar]
- 2. Baldwin JG, Nadler SA, Adams BJ (2004) Evolution of plant parasitism among nematodes. Annu Rev Phytopathol 42: 83–105. [DOI] [PubMed] [Google Scholar]
- 3.Gaugler R (2002) Entomopathogenic nematology. Wallingford, UK: CABI Publishing.
- 4.Grewal PS, Ehlers RU, Shapiro-Ilan DI (2005) Nematodes as biocontrol agents. Wallingford, UK: CABI Publishing. 480 p.
- 5.De Ley P, Blaxter M (2002) Systematic position and phylogeny. In: Lee DL, editor. The Biology of Nematodes. London, UK: Taylor and Francis. 1–30.
- 6. Blaxter ML, De Ley P, Garey JR, Liu LX, Scheldeman P, et al. (1998) A molecular evolutionary framework for the phylum Nematoda. Nature 392: 71–75. [DOI] [PubMed] [Google Scholar]
- 7. Adams BJ, Fodor A, Koppenhöfer HS, Stackebrandt E, Patricia Stock S, et al. (2006) Biodiversity and systematics of nematode–bacterium entomopathogens. Biological Control 37: 32–49. [Google Scholar]
- 8. Boemare N (2002) Interactions between the partners of the entomopathogenic bacterium nematode complexes, Sterinernema-Xenorhabdus and Heterorhabditis-Photorhabdus . Nematology 4: 601–603. [Google Scholar]
- 9. Grewal PS, Lewis EE, Gaugler R (1997) Response of infective stage parasites (Nematoda: Steinernematidae) to volatile cues from infected hosts. J Chem Ecol 23: 503–515. [Google Scholar]
- 10. Ciche TA, Ensign JC (2003) For the insect pathogen Photorhabdus luminescens, which end of a nematode is out? Appl Environ Microbiol 69: 1890–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hallem EA, Rengarajan M, Ciche TA, Sternberg PW (2007) Nematodes, bacteria, and flies: a tripartite model for nematode parasitism. Curr Biol 17: 898–904. [DOI] [PubMed] [Google Scholar]
- 12. ffrench-Constant R, Waterfield N, Daborn P, Joyce S, Bennett H, et al. (2003) Photorhabdus: towards a functional genomic analysis of a symbiont and pathogen. FEMS Microbiol Rev 26: 433–456. [DOI] [PubMed] [Google Scholar]
- 13. An R, Sreevatsan S, Grewal PS (2009) Comparative in vivo gene expression of the closely related bacteria Photorhabdus temperata and Xenorhabdus koppenhoeferi upon infection of the same insect host, Rhizotrogus majalis. BMC Genomics 10: 433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. ffrench-Constant RH, Bowen DJ (2000) Novel insecticidal toxins from nematode-symbiotic bacteria. Cell Mol Life Sci 57: 828–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. ffrench-Constant RH, Waterfield NR (2006) Ground control for insect pests. Nat Biotechnol 24: 660–661. [DOI] [PubMed] [Google Scholar]
- 16. Goodrich-Blair H, Clarke DJ (2007) Mutualism and pathogenesis in Xenorhabdus and Photorhabdus: two roads to the same destination. Mol Microbiol 64: 260–268. [DOI] [PubMed] [Google Scholar]
- 17. Ciche TA, Bintrim SB, Horswill AR, Ensign JC (2001) A Phosphopantetheinyl transferase homolog is essential for Photorhabdus luminescens to support growth and reproduction of the entomopathogenic nematode Heterorhabditis bacteriophora. J Bacteriol 183: 3117–3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Han R, Ehlers RU (2000) Pathogenicity, development, and reproduction of Heterorhabditis bacteriophora and Steinernema carpocapsae under axenic in vivo conditions. J Invertebr Pathol 75: 55–58. [DOI] [PubMed] [Google Scholar]
- 19. Ciche TA, Kim KS, Kaufmann-Daszczuk B, Nguyen KC, Hall DH (2008) Cell Invasion and Matricide during Photorhabdus luminescens Transmission by Heterorhabditis bacteriophora Nematodes. Appl Environ Microbiol 74: 2275–2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Webster JM, Chen G, Hu K, Li J (2002) Bacterial metabolites. In: Gaugler R, editor. Entomopathogenic nematology. Wallingford, UK: CABI Publishing. 99–114.
- 21. Zhou X, Kaya HK, Heungens K, Goodrich-Blair H (2002) Response of ants to a deterrent factor(s) produced by the symbiotic bacteria of entomopathogenic nematodes. Appl Environ Microbiol 68: 6202–6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Grewal PS, Selvan S, Gaugler R (1994) Thermal adaptation of entomopathogenic nematodes: niche breadth for infection, establishment, and reproduction. Journal of Thermal Biology 19: 245–253. [Google Scholar]
- 23. Ruby EG (2008) Symbiotic conversations are revealed under genetic interrogation. Nat Rev Microbiol 6: 752–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ciche TA, Sternberg PW (2007) Postembryonic RNAi in Heterorhabditis bacteriophora: a nematode insect parasite and host for insect pathogenic symbionts. BMC Dev Biol 7: 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Maneesakorn P, An R, Daneshvar H, Taylor K, Bai X, et al. (2011) Phylogenetic and cophylogenetic relationships of entomopathogenic nematodes (Heterorhabditis: Rhabditida) and their symbiotic bacteria (Photorhabdus: Enterobacteriaceae). Mol Phylogenet Evol 59: 271–280. [DOI] [PubMed] [Google Scholar]
- 26.Somvanshi VS, Kaufmann-Daszczuk B, Kim KS, Mallon S, Ciche TA (2010) Photorhabdus phase variants express a novel fimbrial locus, mad, essential for symbiosis. Mol Microbiol in press. [DOI] [PubMed]
- 27. Somvanshi VS, Sloup RE, Crawford JM, Martin AR, Heidt AJ, et al. (2012) A single promoter inversion switches Photorhabdus between pathogenic and mutualistic states. Science 337: 88–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grewal PS, Bai X, Jagdale GB (2011) Longevity and stress tolerance of entomopathogenic nematodes. In: Perry RN, Wharton D, editors. Molecular and Physiological Basis of Nematodes Survival. Wallingford, UK: CABI Publishing. 157–181.
- 29. Eleftherianos I, ffrench-Constant RH, Clarke DJ, Dowling AJ, Reynolds SE (2010) Dissecting the immune response to the entomopathogen Photorhabdus. Trends Microbiol 18: 552–560. [DOI] [PubMed] [Google Scholar]
- 30. Duchaud E, Rusniok C, Frangeul L, Buchrieser C, Givaudan A, et al. (2003) The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens . Nat Biotechnol 21: 1307–1313. [DOI] [PubMed] [Google Scholar]
- 31. Bai X, Adams BJ, Ciche TA, Clifton S, Gaugler R, et al. (2009) Transcriptomic analysis of the entomopathogenic nematode Heterorhabditis bacteriophora TTO1. BMC Genomics 10: 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bai X, Grewal PS, Hogenhout SA, Adams BJ, Ciche TA, et al. (2007) Expressed sequence tag analysis of gene representation in insect parasitic nematode Heterorhabditis bacteriophora. J Parasitol 93: 1343–1349. [DOI] [PubMed] [Google Scholar]
- 33. Sandhu SK, Jagdale GB, Hogenhout SA, Grewal PS (2006) Comparative analysis of the expressed genome of the infective juvenile entomopathogenic nematode, Heterorhabditis bacteriophora . Mol Biochem Parasitol 145: 239–244. [DOI] [PubMed] [Google Scholar]
- 34. Rahimi FR, McGuire TR, Gaugler R (1993) Morphological mutant in the entomopathogenic nematode, Heterorhabditis bacteriophora. J Hered 84: 475–478. [DOI] [PubMed] [Google Scholar]
- 35. Zioni Cohen-Nissan S, Glazer I, Segal D (1992) Phenotypic and Genetic Analysis of a Mutant of Heterorhabditis bacteriophora Strain HP88. J Nematol 24: 359–364. [PMC free article] [PubMed] [Google Scholar]
- 36. O’Leary SA, Burnell AM (1997) The isolation of mutants of Heterorhabditis megidis (Strain UK211) with increased desiccation tolerance. Fundam Appl Nematol 20: 197–205. [Google Scholar]
- 37. Hashmi G, Gaugler R (1998) Genetic diversity in insect-parasitic nematodes (Rhabditida: heterorhabditidae). J Invertebr Pathol 72: 185–189. [DOI] [PubMed] [Google Scholar]
- 38. Jagdale GB, Saeb AT, Somasekhar N, Grewal PS (2006) Genetic variation and relationships between isolates and species of the entomopathogenic nematode genus Heterorhabditis deciphered through isozyme profiles. J Parasitol 92: 509–516. [DOI] [PubMed] [Google Scholar]
- 39. Gaugler R, Campbell JF, McGuire TR (1989) Selection for host-finding in Steinernema feltiae . J Invertebr Pathol 54: 363–372. [Google Scholar]
- 40. Glazer I, Gaugler R, Segal D (1991) Genetics of the Nematode Heterorhabditis bacteriophora Strain HP88: The Diversity of Beneficial Traits. J Nematol 23: 324–333. [PMC free article] [PubMed] [Google Scholar]
- 41. Grewal PS, Gaugler R, Wang YI (1996) Enhanced cold tolerance of the entomopathogenic nematode Steinernema feltiae through genetic selection. Annals of Applied Biology 129: 335–341. [Google Scholar]
- 42. Grewal PS, Gaugler R, Shupe C (1996) Rapid Changes in Thermal Sensitivity of Entomopathogenic Nematodes in Response to Selection at Temperature Extremes. J Invertebr Pathol 68: 65–73. [DOI] [PubMed] [Google Scholar]
- 43. Segal D, Glazer I (2000) Genetics for improving biological control agents: the case of entomopathogenic nematodes. Crop Protection 19: 685–689. [Google Scholar]
- 44. Shapiro-Ilan DI, Glazer I, Segal D (1997) Genetic diversity in wild and laboratory populations of Heterorhabditis bacteriophora as determined by RAPD-PCR analysis. Fundam Appl Nematol 20: 581–585. [Google Scholar]
- 45. Bai X, Grewal PS (2007) Identification of two down-regulated genes in entomopathogenic nematode Heterorhabditis bacteriophora infective juveniles upon contact with insect hemolymph. Mol Biochem Parasitol 156: 162–166. [DOI] [PubMed] [Google Scholar]
- 46. Gal TZ, Glazer I, Koltai H (2003) Differential gene expression during desiccation stress in the insect-killing nematode Steinernema feltiae IS-6. J Parasitol 89: 761–766. [DOI] [PubMed] [Google Scholar]
- 47. Adhikari BN, Lin CY, Bai X, Ciche TA, Grewal PS, et al. (2009) Transcriptional profiling of trait deterioration in the insect pathogenic nematode Heterorhabditis bacteriophora. BMC Genomics 10: 609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chen S, Glazer I, Gollop N, Cash P, Argo E, et al. (2006) Proteomic analysis of the entomopathogenic nematode Steinernema feltiae IS-6 IJs under evaporative and osmotic stresses. Mol Biochem Parasitol 145: 195–204. [DOI] [PubMed] [Google Scholar]
- 49. Gal TZ, Glazer I, Sherman A, Koltai H (2005) Protein interaction of nucleosome assembly protein 1 and casein kinase 2 during desiccation response in the insect-killing nematode Steinernema feltiae IS-6. J Parasitol 91: 691–693. [DOI] [PubMed] [Google Scholar]
- 50. Hashmi S, Hashmi G, Gaugler R (1995) Genetic transformation of an entomopathogenic nematode by microinjection. J Invertebr Pathol 66: 293–296. [DOI] [PubMed] [Google Scholar]
- 51. Hashmi S, Hatab MAA, Gaugler R (1997) GFP: green fluorescent protein a versatile gene marker for entomopathogenic nematodes. Fundam Appl Nematol 20: 323–327. [Google Scholar]
- 52. Dietrich LE, Teal TK, Price-Whelan A, Newman DK (2008) Redox-active antibiotics control gene expression and community behavior in divergent bacteria. Science 321: 1203–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Grewal PS, Grewal SK, Tan L, Adams BJ (2003) Parasitism of molluscs by nematodes: types of associations and evolutionary trends. J Nematol 35: 146–156. [PMC free article] [PubMed] [Google Scholar]
- 54. Kiontke KC, Felix MA, Ailion M, Rockman MV, Braendle C, et al. (2011) A phylogeny and molecular barcodes for Caenorhabditis, with numerous new species from rotting fruits. BMC Evol Biol 11: 339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Anderson RC (1984) The origins of zooparasitic nematodes. Canadian Journal of Zoology 62: 317–328. [Google Scholar]
- 56. Dillman AR, Chaston JM, Adams BJ, Ciche TA, Goodrich-Blair H, et al. (2012) An entomopathogenic nematode by any other name. PLoS Pathog 8: e1002527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Golden JW, Riddle DL (1982) A pheromone influences larval development in the nematode Caenorhabditis elegans. Science 218: 578–580. [DOI] [PubMed] [Google Scholar]
- 58. Butcher RA, Fujita M, Schroeder FC, Clardy J (2007) Small-molecule pheromones that control dauer development in Caenorhabditis elegans. Nat Chem Biol 3: 420–422. [DOI] [PubMed] [Google Scholar]
- 59. Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G (1997) daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science 277: 942–946. [DOI] [PubMed] [Google Scholar]
- 60. Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, et al. (1997) The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature 389: 994–999. [DOI] [PubMed] [Google Scholar]
- 61. Estevez M, Attisano L, Wrana JL, Albert PS, Massague J, et al. (1993) The daf-4 gene encodes a bone morphogenetic protein receptor controlling C. elegans dauer larva development. Nature 365: 644–649. [DOI] [PubMed] [Google Scholar]
- 62. Antebi A, Culotti JG, Hedgecock EM (1998) daf-12 regulates developmental age and the dauer alternative in Caenorhabditis elegans. Development 125: 1191–1205. [DOI] [PubMed] [Google Scholar]
- 63. Schaedel ON, Gerisch B, Antebi A, Sternberg PW (2012) Hormonal signal amplification mediates environmental conditions during development and controls an irreversible commitment to adulthood. PLoS Biol 10: e1001306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Noguez JH, Conner ES, Zhou Y, Ciche TA, Ragains JR, et al. (2012) A novel ascaroside controls the parasitic life cycle of the entomopathogenic nematode Heterorhabditis bacteriophora. ACS Chem Biol 7: 961–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Choe A, von Reuss SH, Kogan D, Gasser RB, Platzer EG, et al. (2012) Ascaroside signaling is widely conserved among nematodes. Curr Biol 22: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Motola DL, Cummins CL, Rottiers V, Sharma KK, Li T, et al. (2006) Identification of ligands for DAF-12 that govern dauer formation and reproduction in C. elegans. Cell 124: 1209–1223. [DOI] [PubMed] [Google Scholar]
- 67. Grewal PS, Wang X, Taylor RA (2002) Dauer juvenile longevity and stress tolerance in natural populations of entomopathogenic nematodes: is there a relationship? Int J Parasitol 32: 717–725. [DOI] [PubMed] [Google Scholar]
- 68.Grewal PS (2002) Formulation and application technology. In: Gaugler R, editor. Entomopathogenic Nematology. Wallingford, UK: CABI Publishing. 265–287.
- 69. Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, et al. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811. [DOI] [PubMed] [Google Scholar]
- 70. Jose AM, Kim YA, Leal-Ekman S, Hunter CP (2012) Conserved tyrosine kinase promotes the import of silencing RNA into Caenorhabditis elegans cells. Proc Natl Acad Sci U S A 109: 14520–14525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Winston WM, Molodowitch C, Hunter CP (2002) Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science 295: 2456–2459. [DOI] [PubMed] [Google Scholar]
- 72. Winston WM, Sutherlin M, Wright AJ, Feinberg EH, Hunter CP (2007) Caenorhabditis elegans SID-2 is required for environmental RNA interference. Proc Natl Acad Sci U S A 104: 10565–10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Nuez I, Felix MA (2012) Evolution of susceptibility to ingested double-stranded RNAs in Caenorhabditis nematodes. PLoS One 7: e29811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ (2004) Processing of primary microRNAs by the Microprocessor complex. Nature 432: 231–235. [DOI] [PubMed] [Google Scholar]
- 75. Vought VE, Ohmachi M, Lee MH, Maine EM (2005) EGO-1, a putative RNA-directed RNA polymerase, promotes germline proliferation in parallel with GLP-1/notch signaling and regulates the spatial organization of nuclear pore complexes and germline P granules in Caenorhabditis elegans. Genetics 170: 1121–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kim JK, Gabel HW, Kamath RS, Tewari M, Pasquinelli A, et al. (2005) Functional genomic analysis of RNA interference in C. elegans. Science 308: 1164–1167. [DOI] [PubMed] [Google Scholar]
- 77. Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, et al. (2001) Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106: 23–34. [DOI] [PubMed] [Google Scholar]
- 78. Tabara H, Yigit E, Siomi H, Mello CC (2002) The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell 109: 861–871. [DOI] [PubMed] [Google Scholar]
- 79. Aboobaker AA, Blaxter ML (2003) Use of RNA interference to investigate gene function in the human filarial nematode parasite Brugia malayi. Mol Biochem Parasitol 129: 41–51. [DOI] [PubMed] [Google Scholar]
- 80. Song C, Gallup JM, Day TA, Bartholomay LC, Kimber MJ (2010) Development of an in vivo RNAi protocol to investigate gene function in the filarial nematode, Brugia malayi. PLoS Pathog 6: e1001239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Huang G, Allen R, Davis EL, Baum TJ, Hussey RS (2006) Engineering broad root-knot resistance in transgenic plants by RNAi silencing of a conserved and essential root-knot nematode parasitism gene. Proc Natl Acad Sci U S A 103: 14302–14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Eddy SR (1998) Profile hidden Markov models. Bioinformatics 14: 755–763. [DOI] [PubMed] [Google Scholar]
- 83. Lampe DJ, Churchill ME, Robertson HM (1996) A purified mariner transposase is sufficient to mediate transposition in vitro. EMBO J 15: 5470–5479. [PMC free article] [PubMed] [Google Scholar]
- 84. Schulenburg H, Hoeppner MP, Weiner J, 3rd, Bornberg-Bauer E (2008) Specificity of the innate immune system and diversity of C-type lectin domain (CTLD) proteins in the nematode Caenorhabditis elegans. Immunobiology 213: 237–250. [DOI] [PubMed] [Google Scholar]
- 85. Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, et al. (2000) Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 408: 86–89. [DOI] [PubMed] [Google Scholar]
- 86. The C. elegans Sequencing Consortium (1998) Genome Sequence of the Nematode C. elegans: A Platform for Investigating Biology. Science 282: 2012–2018. [DOI] [PubMed] [Google Scholar]
- 87. Ghedin E, Wang S, Spiro D, Caler E, Zhao Q, et al. (2007) Draft genome of the filarial nematode parasite Brugia malayi. Science 317: 1756–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Abad P, Gouzy J, Aury J-M, Castagnone-Sereno P, Danchin EGJ, et al. (2008) Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nature Biotechnology 26: 909–915. [DOI] [PubMed] [Google Scholar]
- 89. Bowcock AM, Ruiz-Linares A, Tomfohrde J, Minch E, Kidd JR, et al. (1994) High resolution of human evolutionary trees with polymorphic microsatellites. Nature 368: 455–457. [DOI] [PubMed] [Google Scholar]
- 90. Bai X, Saeb ATM, Michel A, Grewal PS (2009) Isolation and characterization of microsatellite loci in the entomopathogenic nematodeHeterorhabditis bacteriophora. Molecular Ecology Resources 9: 207–209. [DOI] [PubMed] [Google Scholar]
- 91. Stein LD, Bao Z, Blasiar D, Blumenthal T, Brent MR, et al. (2003) The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. PLoS Biol 1: E45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. McKerrow JH, Caffrey C, Kelly B, Loke P, Sajid M (2006) Proteases in parasitic diseases. Annu Rev Pathol 1: 497–536. [DOI] [PubMed] [Google Scholar]
- 93. AbuHatab M, Selvan S, Gaugler R (1995) Role of Proteases in Penetration of Insect Gut by the Entomopathogenic Nematode Steinernema glaseri (Nematoda: Steinernematidae). Journal of Invertebrate Pathology 66: 125–130. [Google Scholar]
- 94. Cerenius L, Soderhall K (2004) The prophenoloxidase-activating system in invertebrates. Immunol Rev 198: 116–126. [DOI] [PubMed] [Google Scholar]
- 95. Crawford JM, Portmann C, Zhang X, Roeffaers MB, Clardy J (2012) Small molecule perimeter defense in entomopathogenic bacteria. Proc Natl Acad Sci U S A 109: 10821–10826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Bintrim SB, Ensign JC (1998) Insertional inactivation of genes encoding the crystalline inclusion proteins of Photorhabdus luminescens results in mutants with pleiotropic phenotypes. J Bacteriol 180: 1261–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Mallo GV, Kurz CL, Couillault C, Pujol N, Granjeaud S, et al. (2002) Inducible antibacterial defense system in C. elegans. Curr Biol 12: 1209–1214. [DOI] [PubMed] [Google Scholar]
- 98. Mitreva M, Jasmer DP, Zarlenga DS, Wang Z, Abubucker S, et al. (2011) The draft genome of the parasitic nematode Trichinella spiralis. Nat Genet 43: 228–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Varki A, Cummings R, Esko J, Freeze H, Hart G, et al.. (1999) Essentials of glycobiology. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [PubMed]
- 100. Liu J, Shworak NW, Sinay P, Schwartz JJ, Zhang L, et al. (1999) Expression of heparan sulfate D-glucosaminyl 3-O-sulfotransferase isoforms reveals novel substrate specificities. J Biol Chem 274: 5185–5192. [DOI] [PubMed] [Google Scholar]
- 101. Li L, Stoeckert CJ Jr, Roos DS (2003) OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13: 2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Kikuchi T, Cotton JA, Dalzell JJ, Hasegawa K, Kanzaki N, et al. (2011) Genomic insights into the origin of parasitism in the emerging plant pathogen Bursaphelenchus xylophilus. PLoS Pathog 7: e1002219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Jex AR, Liu S, Li B, Young ND, Hall RS, et al. (2011) Ascaris suum draft genome. Nature 479: 529–533. [DOI] [PubMed] [Google Scholar]
- 104. Simske JS, Kaech SM, Harp SA, Kim SK (1996) LET-23 receptor localization by the cell junction protein LIN-7 during C. elegans vulval induction. Cell 85: 195–204. [DOI] [PubMed] [Google Scholar]
- 105. Lesa GM, Sternberg PW (1997) Positive and negative tissue-specific signaling by a nematode epidermal growth factor receptor. Mol Biol Cell 8: 779–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Constant P, Marchay L, Fischer-Le-Saux M, Briand-Panoma S, Mauleon H (1998) Natural occurrence of entomopathogenic nematodes (Rhabditida: Steinernematidae and Heterorhabditidae) in Guadeloupe islands. Fundam Appl Nematol 21: 667–672. [Google Scholar]
- 107. Mortazavi A, Schwarz EM, Williams B, Schaeffer L, Antoshechkin I, et al. (2010) Scaffolding a Caenorhabditis nematode genome with RNA-seq. Genome Res 20: 1740–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Smit AFA, Hubley R, Green P (1996–2010) RepeatMasker Open-3.0 Available: http://www.repeatmasker.org.
- 109. Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25: 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, et al. (2005) Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res 33: D121–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Eddy SR, Durbin R (1994) RNA sequence analysis using covariance models. Nucleic Acids Res 22: 2079–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Nawrocki EP, Eddy SR (2007) Query-dependent banding (QDB) for faster RNA similarity searches. PLoS Comput Biol 3: e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Korf I (2004) Gene finding in novel genomes. BMC Bioinformatics 5: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Stanke M, Morgenstern B (2005) AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res 33: W465–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Stanke M, Schoffmann O, Morgenstern B, Waack S (2006) Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics 7: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Stanke M, Steinkamp R, Waack S, Morgenstern B (2004) AUGUSTUS: a web server for gene finding in eukaryotes. Nucleic Acids Res 32: W309–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Majoros WH, Pertea M, Salzberg SL (2004) TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics 20: 2878–2879. [DOI] [PubMed] [Google Scholar]
- 118. Lukashin AV, Borodovsky M (1998) GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res 26: 1107–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Allen JE, Salzberg SL (2005) JIGSAW: integration of multiple sources of evidence for gene prediction. Bioinformatics 21: 3596–3603. [DOI] [PubMed] [Google Scholar]
- 120. Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, et al. (2008) The Pfam protein families database. Nucleic Acids Res 36: D281–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, et al. (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676. [DOI] [PubMed] [Google Scholar]
- 122. Rawlings ND, Barrett AJ, Bateman A (2010) MEROPS: the peptidase database. Nucleic Acids Res 38: D227–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- 124.Felsenstein J (2005) PHYLIP (Phylogeny Inference Package) version 3.6. Distributed by the author Department of Genome Sciences, University of Washington, Seattle.
- 125. Yang Z (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24: 1586–1591. [DOI] [PubMed] [Google Scholar]
- 126. Bluthgen N, Brand K, Cajavec B, Swat M, Herzel H, et al. (2005) Biological profiling of gene groups utilizing Gene Ontology. Genome Informatics 16: 106–115. [PubMed] [Google Scholar]
- 127.Adams BJ, Peat SM, Dillman AR (2007) Phylogeny and Evolution. In: Nguyen KB, Hunt DJ, editors. A monograph of the nematodes in the families Sterinernematidae & Heterorhabditidae. Leiden, The Netherlands: Brill.
- 128. Elsworth B, Wasmuth J, Blaxter M (2011) NEMBASE4: the nematode transcriptome resource. Int J Parasitol 41: 881–894. [DOI] [PubMed] [Google Scholar]
- 129.Kiontke K, Fitch DHA (2005) The phylogenetic relationships of Caenorhabditis and other rhabditids. In: Girard LR, editor. WormBaook: The C. elegans Research Community. [DOI] [PMC free article] [PubMed]
- 130. Hodda M (2011) Phylum Nematoda Cobb 1932. Zootaxa 3148: 63–95. [Google Scholar]
- 131. Baumeister R, Schaffitzel E, Hertweck M (2006) Endocrine signaling in Caenorhabditis elegans controls stress response and longevity. J Endocrinol 190: 191–202. [DOI] [PubMed] [Google Scholar]
- 132. Opperman CH, Bird DM, Williamson VM, Rokhsar DS, Burke M, et al. (2008) Sequence and genetic map of Meloidogyne hapla: A compact nematode genome for plant parasitism. Proceedings of the National Academy of Sciences 105: 14802–14807. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Predicted gene models in Heterorhabditis bacteriophora genome.
(XLSX)
Sequence similarity of conceptually translated H. bacteriohora proteins to C. elegans proteins in Wormbase release W220.
(XLSX)
Comparison of Pfam domains predicted in the proteins of 10 nematode species in this study.
(XLSX)
Predicted mariner DNA motifs in H. bacteriophora genome.
(XLSX)
Predicted mariner DNA motifs in C. elegans genome using the same parameters as the ones used to generate results in Table S4.
(XLSX)
Predicted non-protein-coding RNA in H. bacteriohora genome.
(XLSX)
Predicted tRNA genes in H. bacteriophora genome.
(XLSX)
Predicted microsatellite loci in H. bacteriophora genome.
(XLSX)
Predicted secretome in H. bacteriophora genome.
(XLSX)
Comparison of gene ontology terms that were assigned to genes in the 10 nemtode species included in this study.
(XLSX)
Comparison of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways predicted in the 10 nematode species included in this study.
(XLSX)
Comparison of the metabolism pathways in KEGG Ontologies in the 10 nematode species included in this study.
(XLSX)
The list of the IDs of orthologous sequences in the 10 nematode species included in this study.
(XLSX)