Abstract
Strategies for the rapid and reliable analysis of microorganisms have been sought to meet national needs in defense, homeland security, space exploration, food and water safety, and clinical diagnosis. Mass spectrometry has long been a candidate technique because it is extremely rapid and can provide highly specific information. It has excellent sensitivity. Molecular and fragment ion masses provide detailed fingerprints, which can also be interpreted. Mass spectrometry is also a broad band method—everything has a mass—and it is automatable. Mass spectrometry is a physiochemical method that is orthogonal and complementary to biochemical and morphological methods used to characterize microorganisms.

Key words: Microorganisms, Proteomics, Detect-to-protect, Detect-to-treat
Perspective
This laboratory’s interest is in strategies that ask broadly, “what is there?” rather than strategies that define a specific target and ask “is it there?” For detection of biological agents in homeland defense, two time frames are particularly relevant: a short “detect to protect,” and a longer “detect to treat,” This Perspective article will address progress in analysis in the short time frame required on the battlefield or by first responders, as well as the longer time frame available for clinical diagnosis and public health.
Most of the literature discussing automatable rapid analysis of unfractionated microorganisms deals with bacteria and bacterial spores [1–9], for example B. anthracis and other species defined as biological warfare agents. Fungi, including yeast, are susceptible to the same strategies [10–12]. Viruses provide a challenging current frontier for unfractionated characterization, since they are hard to lyse, synthesize a smaller selection of characteristic proteins, and are more difficult to culture. Nonetheless, some progress has been reported using mass spectrometry. The molecular masses of abundant capsid proteins and surface glycoproteins are proposed to be characteristic of the virus responsible for severe acute respiratory syndrome (SARS) [13] and of Sindbis, a simulant for the pathogens Venezuelan equine encephalitis and western equine encephalitis viruses [14].
Sample Collection and Preparation
For analyses in the “detect to protect” time frame, microorganisms must be captured on site and analyzed directly. This usually means concentration from air, which provides a relatively clean sample. Since no time is available for culturing, high sensitivity is required. A major challenge associated with collection/enrichment from the atmosphere (e.g., using a particle impactor), is provided by contaminating microorganisms from the environmental background. Direct analysis of bacteria from water and food has been achieved using antibodies, lectins, or functionalized nanoparticles [9].
The “detect to treat” time frame usually allows cell culturing, and standardized cultures form the basis for current applications of mass spectrometers in clinical microbiology laboratories. A current frontier in clinical analysis is direct analysis of physiologic samples, without waiting to culture. I expect that a new window for characterization of viruses will be provided by on-going studies of host–virus interactions.
Although some practitioners speak of “whole cell” or “intact cell” mass spectrometry” to indicate that no fractionation steps are employed, vegetative bacteria cells are, in fact, lysed by exposure to the organic solvents and acids used in MALDI matrices and electrospray, and are thus no longer whole cells. Bacillus spores are usually treated with stronger acid to release characteristic small acid soluble proteins and viruses also require a chemical lysis step. These treatments can be automated. A good overview has been published recently by Franco Basile [15].
Ionization Methods and Biomarker Classes
In Figure 1, spectra are shown of two species of Staphylococcus, profiled in 1975 using electron impact and a double focusing mass spectrometer of Mattauch Herzog geometry [16]. Intact metabolites and lipids were assigned as biomarkers using high resolution measurements of the ions detected.
Figure 1.
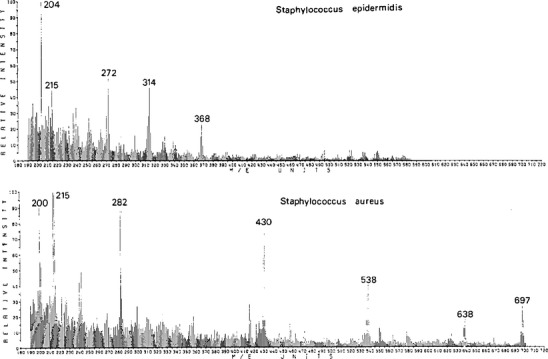
Electron impact mass spectra of Staphylococcus epidermidis and Staphylococcus aureus [16]. Reprinted with permission from the American Chemical Society
Most of the ionization techniques developed for mass spectrometry since 1975 have been applied to profile bacteria, for example, plasma desorption, fast atom bombardment (FAB), laser desorption, matrix assisted laser desorption/ionization (MALDI), electrospray (ESI), desorption electrospray ionization, and laser ablation electrospray ionization. Both anions and cations can be detected and in some cases analysis of anions is preferable [17]. Mass profiles are provided by molecular ions of intact biomarkers. Fatty acids were evaluated early on as biomarkers, but rejected as too dependent on sample history. Advances in desorption ionization (e.g., FAB) favored profiling of polar lipids. Eventually it was concluded that phospholipid and glycolipid biosynthesis are too dependent on growth and storage conditions to provide reliable biomarkers in the field. Non-ribosomal lipopeptides have also been proposed as biomarkers in the mass range below 2000 [18, 19]. However, the most reliable MS biomarkers are considered to be proteins. Demonstrations in 1996 that MALDI [20, 21] and ESI [22] spectra ionized small proteins from unfractionated bacteria and phage pointed the way for protein mass matching and identifications provided by genome-based bioinformatics. In general, m/z values below 20,000 Da and above 4000 Da are used in protein-based analysis. Minimum sensitivity for detecting these mass patterns is generally reported to be in the range of 5000 cells.
Ion Analyzers
The requirement for robustness in field-worthy systems has favored time-of-flight (TOF) analyzers. TOF analyzers are highly compatible with MALDI. This combination is featured in commercial systems sold for clinical diagnostics, reflecting its mass range and simplicity. TOF has been combined with ion mobility for rapid analysis of bacteria [23]. An end-to-end automated system has been developed, based on MALDI and a TOF analyzer, which provides sample preparation and analysis in less than 30 min (e.g., for use in public health laboratories [24]).
Tandem mass spectrometry has been exploited for characterization of biomarkers, initially for phospholipids from unfractionated bacteria [25] and, with the addition of collisionally induced dissociation (CID), for proteolytic peptides generated in situ [26, 27]. A variety of tandem and hybrid analyzers has been employed to access the enhanced identifications provided by peptide sequencing. An ion trap-based MALDI system has been successfully tested that provides automated protein cleavage in situ, CID analysis of the peptide products, and bioinformatic identification of the peptides, proteins and bacterial species [28].
The identification of microorganisms based on top-down analysis of proteins offers high potential for the work of central reference laboratories. Such an approach requires MS/MS with accurate mass measurement of both precursor and product ions, and it has been implemented on FTICR, Orbitrap, and TOF-TOF instruments. Optimally, the analysis also exploits LC-MS. Both ESI and MALDI have been used successfully in top-down analysis of proteins from microorganisms [29–32]. Figure 2 presents CID product ion spectra of a 7260.92 Da protein from Yersinia rohdei eluted from a reverse phase HPLC column into an Orbitrap mass spectrometer [31]. The protein was identified as 50S ribosomal protein L29 using a ProSight search program. In this case, no annotated genome was available for Y. rohdei, a simulant for Y. pestis; however, recognition of proteins with high homology in related species allowed many proteins to be identified, and finally the orphan bacterium to be phylogenetically characterized.
Figure 2.
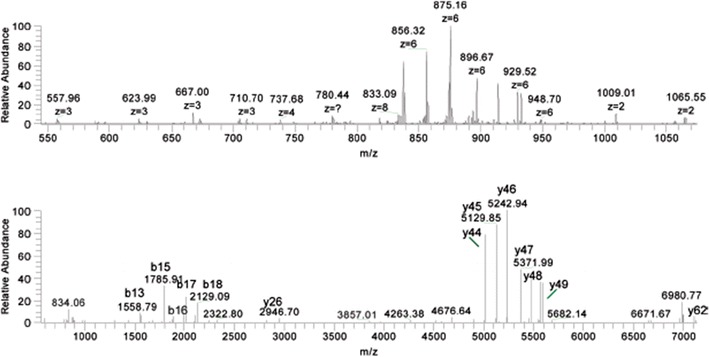
Top: tandem mass spectrum from the precursor ion at m/z 807.80 (+9 charge state, intact mass 7260.92 Da) from Y. rohdei; bottom: the same MS/MS spectrum with all fragment ions converted to zero charge state. Some of the protein fragment ions are indicated [31]. Reprinted with permission from the American Chemical Society
The research team led by Steve Musser at the FDA Center for Food Safety and Applied Nutrition has proposed that the mutations and post-translational modifications characterized by chromatographic fractionation and top-down analysis provide better distinction of strains and serovars than PCR can discern [33]. In Figure 3, protein maps are compared for two serovars of E. coli, which were analyzed using a top-down electrospray strategy [34]. Top-down analysis also provides characterization of protein toxins.
Figure 3.
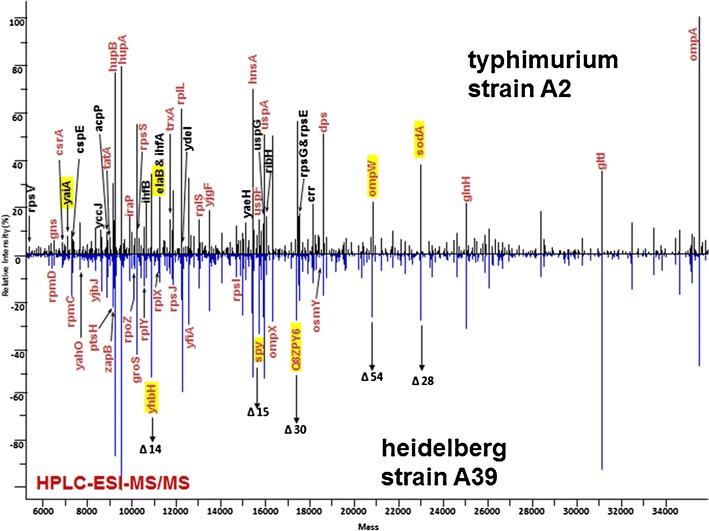
Protein expression profiles from two Salmonella serovars, S. Typhimurium strain A2 and S. Heidelberg strain A39, characterized intact by LC-MS/MS. The masses highlighted in yellow represent cross serovar protein mass shifts. A subset of the proteins is also labeled with gene symbols. Those labeled in red were identified in both serovars [34]
Automated Analysis of Spectra
Software for pattern matching and library searching has been developed or adapted to compare and differentiate mass patterns from different species of bacteria [20, 35, 36]. Machine learning and other strategies have been applied to define core elements in library spectra for more sophisticated automated matching, however, without identification of the biomarkers. Both commercial and public libraries have been assembled for pattern matching, and always, the reproducibility of mass patterns is sensitive to sample culture, preparation, storage, and to the kind of mass spectrometer used [1, 8, 37, 38].
Bioinformatics provides an alternative, interpretive approach to pattern matching, which is not dependent on growth conditions, sample preparation, or instrumentation. All ionization techniques are applicable, and all ionization techniques are supported by the same databases of protein sequences. Furthermore, proteomic or bioinformatic methods have the great advantage that they identify the biomarkers as part of the analysis of the microorganism. Identifications of proteins and microorganisms can be based on molecular masses [24, 39, 40], or on fragmentation in tandem MS measurements of either intact proteins (see remarks above on the top-down approach) or their related peptides, the bottom-up approach [26, 27]. Bioinformatic or proteomic analysis has been proven for bacteria, spores, viruses, fungi, and toxins. Proteomic strategies have been proposed to recognize and characterize genetically engineered bacteria [41], and also to classify bacteria with unsequenced genomes [31, 42]. Strategies based on bioinformatics will be greatly facilitated for analysis of bacteria when the database maintained by the National Center for Biologic Information (NCBI) accomplishes its planned expansion to 100,000 annotated bacteria genomes [43].
Mass Spectrometry and PCR
Significant advantages are offered by mass spectrometry over classical phenotypic and biochemical characterization, which include speed, reliability and cost [7, 36].
Although strategies exist by which mass spectrometry can be used for rapid characterization of small RNA and DNA fragments [8], protein analysis is the significant product of three decades of development of mass spectrometry for rapid analysis of microorganisms. The comparison of MS-based protein analysis with PCR is not straight-forward. On the one hand, amplification renders 16S rRNA gene PCR more sensitive. A target bacterium and a target gene fragment must be predefined for PCR, whereas MS-based protein identification (by either mass patterns or bioinformatics) is broad-based. Recent advances in PCR technology have rendered it almost as rapid as proteomic analysis of an uncultured sample (~30 min); however, sample processing is more complex for PCR and effects of contaminants are more challenging to overcome in a harsh environment. Protein toxins such as ricin cannot, of course, be identified by DNA-based methods.
Next-Gen Challenges
For the next decade, I see many exciting opportunities and challenges for the “development of rapid methods employing mass spectrometry to detect, characterize, and differentiate intact microorganisms.”1 These include:
continued miniaturization of mass spectrometers (e.g., for bedside applications [44–46])
elimination of the need to culture in clinical microbiology [47]
expansion of genome/protein databases for bacteria, fungi, and viruses
rapid analysis by MS of single microbial cells
development of physical or chemical methods to break viral coats so that MS-based strategies can be readily applied to viruses
definition of host protein changes in infection, particularly viral, to allow rapid characterization of infectious agents in patients
development of meta-strategies for rapid analysis of the human microbiome and its changes [48]
Acknowledgments
I extend special thanks to my students and collaborators in this area through the last 25 years, and also to colleagues in other institutions and countries who have made critical contributions to this field. In particular, I acknowledge and express my gratitude to my husband Professor Robert J. Cotter (deceased Nov. 12, 2012) for his rigorous scientific contributions, his insightful creativity, and his personal support and encouragement. Collaborating institutions in microorganism research through three decades included the Johns Hopkins University Applied Physics Laboratory and Scientific Engineering Systems Inc. Financial support was provided by DARPA, HSARPA, DHS, DOD, NIHGMS, the German Science Foundation, and the ARCS Foundation.
Footnotes
From the citation for the 2012 ASMS award to Catherine Fenselau for Outstanding Contribution in Mass Spectrometry.
References
- 1.Fenselau C, Demirev P. Characterization of intact microorganisms by MALDI mass spectrometry. Mass Spectrom. Rev. 2001;20:157–171. doi: 10.1002/mas.10004. [DOI] [PubMed] [Google Scholar]
- 2.Demirev P, Fenselau C. Mass spectrometry for rapid characterization of microorganisms. Annu. Rev. Anal. Chem. 2008;1:71–93. doi: 10.1146/annurev.anchem.1.031207.112838. [DOI] [PubMed] [Google Scholar]
- 3.Demirev P, Fenselau C. Mass spectrometry in biodefense. J. Mass Spectrom. 2008;43:1441–1457. doi: 10.1002/jms.1474. [DOI] [PubMed] [Google Scholar]
- 4.Fenselau C, editor. Mass Spectrometry for the Characterization of Microorganisms. Washington, DC: American Chemical Society; 1994. pp. 1–240. [Google Scholar]
- 5.Fenselau C, Demirev P, editors. Rapid Characterization of Microorganisms by Mass Spectrometry. Washington, DC: American Chemical Society; 2011. pp. 1–237. [Google Scholar]
- 6.Banoub, J., El Essassi, M. (eds.): Detection of Biological Agents for the Prevention of Bioterrorism. pp. 1–12. Springer Press, Berlin (2011)
- 7.Havlicek V., Lemr K., Schug K.A. Current trends in microbial diagnostics based on mass spectrometry. Anal. Chem. 2013;85:790–797. doi: 10.1021/ac3031866. [DOI] [PubMed] [Google Scholar]
- 8.Sauer S, Kliem M. Mass spectrometry tools for the classification and identification of bacteria. Nat. Rev. Microbiol. 2010;8:74–82. doi: 10.1038/nrmicro2243. [DOI] [PubMed] [Google Scholar]
- 9.Ho YP, Reddy PM. Advances in mass spectrometry for the identification of pathogens. Mass Spectrom. Rev. 2010;30:1203–1224. doi: 10.1002/mas.20320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amiri-Eliasi B, Fenselau C. Characterization of protein biomarkers desorbed by MALDI from whole fungal cells. Anal. Chem. 2001;73:5228–5231. doi: 10.1021/ac010651t. [DOI] [PubMed] [Google Scholar]
- 11.Padliya ND, Garrett WM, Campbell KB, Tabb DL, Cooper B. Tandem mass spectrometry for the detection of plant pathogenic fungi and the effects of database composition on protein inferences. Proteomics. 2007;7:3932–3942. doi: 10.1002/pmic.200700419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hettick JM, Green BJ, Buskirk AD, Slaven JE, Kashon ML, Beezhold DH. Discrimination of fungi by MALDI-TOF mass spectrometry. In: Fenselau C, Demirev P, editors. Rapid Characterization of Microorganisms by Mass Spectrometry. Washington, DC: American Chemical Society; 2011. p. 35. [Google Scholar]
- 13.Krokhin O, Li Y, Andonov A, Feldmann H, Flick R, Jones S, Stroeher U, Bastien N, Dasuri KV, Cheng K, Simonsen JN, Perreault H, Wilkins J, Ens W, Plummer F, Standing KG. Mass spectrometric characterization of proteins from the SARS virus: a preliminary report. Mol. Cell. Proteomics. 2003;2:346–356. doi: 10.1074/mcp.M300048-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim YJ, Freas A, Fenselau C. Rapid analysis of viral glycoproteins by MALDI-TOF mass spectrometry. Anal. Chem. 2001;73:1544–1548. doi: 10.1021/ac001171p. [DOI] [PubMed] [Google Scholar]
- 15.Basile F. Rapid sample preparation for microorganism analysis by mass spectrometry. In: Fenselau C, Demirev P, editors. Rapid Characterization of Microorganisms by Mass Spectrometry. Washington, DC: American Chemical Society; 2011. p. 5. [Google Scholar]
- 16.Anhalt JP, Fenselau C. Identification of bacteria using mass spectrometry. Anal. Chem. 1975;47:219–225. doi: 10.1021/ac60352a007. [DOI] [Google Scholar]
- 17.Heller DN, Cotter RJ, Fenselau C, Uy OM. Profiling of bacteria by fast atom bombardment mass spectrometry. Anal. Chem. 1987;59:2806–2809. doi: 10.1021/ac00150a018. [DOI] [PubMed] [Google Scholar]
- 18.Hathout Y., Ho Y., Ryzhov V., Demirev P., Fenselau C. Kurstakins: a new class of lipopeptides isolated from Bacillus thuringiensis. J. Nat. Prod. 2000;63:1492–1496. doi: 10.1021/np000169q. [DOI] [PubMed] [Google Scholar]
- 19.Price NPJ, Rooney AP, Swezey JL, Perry E, Cohan FM. Mass spectrometric analysis of lipopeptides from Bacillus strains isolated from diverse geographical locations. FEMS Microbiol. Lett. 2007;271:83–89. doi: 10.1111/j.1574-6968.2007.00702.x. [DOI] [PubMed] [Google Scholar]
- 20.Holland RD, Wilkes JG, Rafii F, Sustherland JR, Persos CC, Voorhees K, Lay JO. Rapid identification of intact whole bacteria based on spectral patterns using matrix-assisted laser desorption/ionization with time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 1996;10:1227–1232. doi: 10.1002/(SICI)1097-0231(19960731)10:10<1227::AID-RCM659>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 21.Krishnamurthy T, Ross PL. Rapid identification of bacteria by direct matrix-assisted laser desorption/ionization mass spectrometric analysis of whole cells. Rapid Commun. Mass Spectrom. 1996;10:1992–1996. doi: 10.1002/(SICI)1097-0231(199612)10:15<1992::AID-RCM789>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 22.Despeyroux D, Phillpotts R, Watts P. Electrospray mass spectrometry for detection and characterization of purified cricket paralysis virus (CrPV) Rapid Commun. Mass Spectrom. 1996;10:937–941. doi: 10.1002/(SICI)1097-0231(19960610)10:8<937::AID-RCM598>3.0.CO;2-9. [DOI] [Google Scholar]
- 23.Hayes JM, Anderson LC, Schultz JA, Ugarov M, Egan TF, Lewis EK, Womack V, Woods AS, Jackson SN, Hauge RH, Kittrell C, Ripley S, Murray KK. Matrix assisted laser desorption ionization ion mobility time-of-flight mass spectrometry of bacteria. In: Fenselau C, Demirev P, editors. Rapid Characterization of Microorganisms by Mass Spectrometry. Washington, DC: American Chemical Society; 2011. p. 143. [Google Scholar]
- 24.Hagan NA, Lin JS, Antoine MD, Cornish TJ, Quizon RS, Collins BF, Feldman AB, Demirev P. MALDI mass spectrometry for rapid detection and characterization of biological threats. In: Fenselau C, Demirev P, editors. Rapid Characterization of Microorganisms by Mass Spectrometry. Washington, DC: American Chemical Society; 2011. p. 211. [Google Scholar]
- 25.Heller DN, Murphy CM, Cotter RJ, Fenselau C, Uy OM. Constant neutral loss scanning for the characterization of bacterial phospholipids desorbed by fast atom bombardment. Anal. Chem. 1988;60:2787–2791. doi: 10.1021/ac00175a029. [DOI] [PubMed] [Google Scholar]
- 26.Warscheid B, Jackson K, Sutton C, Fenselau C. MALDI analysis of Bacilli in spore mixtures by applying a quadrupole ion trap time of flight tandem mass spectrometer. Anal. Chem. 2003;75:5608–5617. doi: 10.1021/ac0344081. [DOI] [PubMed] [Google Scholar]
- 27.Dworzanski JP, Snyder AP, Chen R, Zhang H, Wishart D, Li L. Identification of bacteria using tandem mass spectrometry combined with a proteome database and statistical scoring. Anal. Chem. 2004;76:2355–2366. doi: 10.1021/ac0349781. [DOI] [PubMed] [Google Scholar]
- 28.Oktem B, Sundaram AK, Razumovskaya J, Gudlavalleti SK, Saul TD, Doroshenko V. Rapid detection and identification of aerosolized biological materials, toxins, and microorganisms by an AP-MALDI-MS-based system. In: Fenselau C, Demirev P, editors. Rapid Characterization of Microorganisms by Mass Spectrometry. Washington, DC: American Chemical Society; 2011. p. 197. [Google Scholar]
- 29.Demirev PA, Feldman AB, Kowalski P, Lin JS. Top-down proteomics for rapid identification of intact microorganisms. Anal. Chem. 2005;77:7455–7561. doi: 10.1021/ac051419g. [DOI] [PubMed] [Google Scholar]
- 30.Ferguson JT, Wenger CD, Metcalf WW, Kelleher NL. Top-down proteomics reveals novel protein forms expressed in Methanosarcina acetivorans. J. Am. Soc. Mass Spectrom. 2009;20:1743–1750. doi: 10.1016/j.jasms.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wynne C, Fenselau C, Demirev PA, Edwards N. Top-down identification of protein biomarkers in bacteria with unsequenced genomes. Anal. Chem. 2009;81:9633–9642. doi: 10.1021/ac9016677. [DOI] [PubMed] [Google Scholar]
- 32.Fagerquist CK. Rapid identification of food-borne pathogens by top-down proteomics using MALDI-TOF/TOF mass spectrometry. In: Fenselau C, Demirev P, editors. Rapid Characterization of Microorganisms by Mass Spectrometry. Washington, DC: American Chemical Society; 2011. p. 99. [Google Scholar]
- 33.Williams TL, Monday SR, Edelson-Mammel S, Buchanon R, Musser SM. A top-down proteomics approach for differentiating thermal resistant strains of Enterobacter sakazakii. Proteomics. 2005;5:4161–4169. doi: 10.1002/pmic.200401263. [DOI] [PubMed] [Google Scholar]
- 34.McFarland, M.A., Andrzejewski, D., Callahan, J.H., Musser, S.M.: Private communication, (2012)
- 35.Platt J, Uy OM, Heller DN, Cotter RJ, Fenselau C. Computer based library matching and multivariant analysis of desorption mass spectra of microorganisms. Anal. Chem. 1988;60:1415–1419. doi: 10.1021/ac00165a014. [DOI] [PubMed] [Google Scholar]
- 36.van Belkum A, Welker M, Erhard M, Chatellier S. Biomedical mass spectrometry in today’s and tomorrow’s clinical microbiology laboratories. J. Clin. Microbiol. 2012;50:1513–1517. doi: 10.1128/JCM.00420-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wunschel SC, Jarman KH, Petersen CE, Valentine NB, Wahl KL, Schauki D, Jackman J, Nelson CP, White VE. Bacterial analysis by MALDI-TOF mass spectrometry: an inter-laboratory comparison. J. Am. Soc. Mass Spectrom. 2005;16:456–462. doi: 10.1016/j.jasms.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 38.Mellmann A, Bimet F, Bizet C, Borovskaya AD, Drake RR, Eigner U, Fahr AM, He Y, Ilina EN, Kostrzewa M, Maier T, Mancinelli L, Moussaoui W, Prevost G, Putignani L, Seachord CL, Tang YW, Darmsen D. High interlaboratory reproducibility of matrix-assisted laser desorption ionization-time of flight mass spectrometry-based species identification of nonfermenting bacteria. J. Clin. Microbiol. 2009;47:3732–3734. doi: 10.1128/JCM.00921-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Demirev P, Ho YP, Ryzhov V, Fenselau C. Microorganism identification by mass spectrometry and protein database searches. Anal. Chem. 1999;71:2732–2738. doi: 10.1021/ac990165u. [DOI] [PubMed] [Google Scholar]
- 40.Fenselau C, Russell S, Swatkoski S, Edwards N. Proteomic strategies for rapid characterization of micro-organisms. Eur. J. Mass Spectrom. 2007;13:35–39. doi: 10.1255/ejms.845. [DOI] [PubMed] [Google Scholar]
- 41.Russell SC. Rapid profiling of recombinant protein expression from crude cell cultures by matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) In: Fenselau C, Demirev P, editors. Rapid Characterization of Microorganisms by Mass Spectrometry. Washington, DC: American Chemical Society; 2011. p. 61. [Google Scholar]
- 42.Edwards N, Wynne C, Dhabaria A, Fenselau C. Top-down protein analysis and phylogenetic characterization of unsequenced bacteria. In: Fenselau C, Demirev P, editors. Rapid Characterization of Microorganisms by Mass Spectrometry. Washington, DC: American Chemical Society; 2011. p. 121. [Google Scholar]
- 43.Available at: http://100Kgenome.vetmed.ucdavis.edu/ Accessed 6 Jan 2013
- 44.Cotter R.J. High energy collisions on tandem time-of-flight mass spectrometers. J. Am. Soc. Mass Spectrom. 2013;24:657–674. doi: 10.1007/s13361-012-0518-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ouyang Z, Cooks RG. Miniature mass spectrometers. Annu. Rev. Anal. Chem. 2009;2:187–214. doi: 10.1146/annurev-anchem-060908-155229. [DOI] [PubMed] [Google Scholar]
- 46.Frank M, Gard EE, Tobias HJ, Adams KL, Bogan MJ, Coffee KR, Farquar GR, Fergenson DP, Martin SI, Pitesky M, Riot VJ, Srivastava A, Steele PT, Williams AM. Single-particle aerosol mass spectrometry (SPAMS) for high-throughput and rapid analysis of biological aerosols and single cells. In: Fenselau C, Demirev P, editors. Rapid Characterization of Microorganisms by Mass Spectrometry. Washington, DC: American Chemical Society; 2011. p. 161. [Google Scholar]
- 47.Hsieh S, Tseng C, Lee Y, Kuo A, Sun C, Lin Y, Chen J. Highly efficient classification and identification of human pathogenic bacteria by MALDI-TOF MS. Mol. Cell. Proteomics. 2008;7:448–456. doi: 10.1074/mcp.M700339-MCP200. [DOI] [PubMed] [Google Scholar]
- 48.Mahowald MA, Rey FE, Seedorf H, Turnbaugh PJ, Fulton RS, Wollam A, Shah N, Wang C, Magrini V, Wilson RK, Cantarel BL, Coutinho PM, Henrissat B, Crock LW, Russell A, Verberkmoes NC, Hettich RL, Gordon JI. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc. Natl. Acad. Sci. U. S. A. 2009;106:5859–5864. doi: 10.1073/pnas.0901529106. [DOI] [PMC free article] [PubMed] [Google Scholar]


