Abstract
The MYC proto-oncogene is associated with the pathogenesis of most human neoplasia. Conversely, its experimental inactivation elicits oncogene addiction. While MYC constitutes a formidable therapeutic target, it also plays an essential role in normal physiology, thus creating the need for context--specific targeting strategies. The analysis of post-translational MYC activity modulation yields novel targets for MYC inactivation. Specifically, following regulatory network analysis in human B cells, we identify a novel role of the STK38 kinase as a regulator of MYC activity and a candidate target for abrogating tumorigenesis in MYC addicted lymphoma. We found that STK38 regulates MYC protein stability and turnover in a kinase-activity-dependent manner. STK38 kinase inactivation abrogates apoptosis following B-cell receptor (BCR) activation, while its silencing significantly decreases MYC levels and increases apoptosis. Moreover, STK38 knockdown suppresses growth of MYC addicted tumors in vivo thus providing a novel viable target for treating these malignancies.
Keywords: STK38, MYC post-translational modifications, protein turnover, transcriptional activity, protein-protein interaction
Introduction
Serine-Threonine Kinase 38, STK38/NDR1, is a member of the highly conserved NDR-kinase family expressed in mammalian cells (1). We have previously identified STK38 as a modulator of transcription factor (TF) activity in human B cells using a systems biology approach (2). STK38 emerged as the single most pleiotropic kinase in mature human B cells, affecting 303 TFs including MYC, ETV6, RXRA, IRF5, ARID3A, NME1, FOXJ2, CUX1, FOXM1, TFDP1 and ZNF207 among others. Notably, among kinases that are post-translational modulators of MYC activity, STK38 was ranked within the top 6, with activity comparable to other established MYC modulators, such as CSNK2A1 and HCK (3). Indeed the vast majority of B-cell-specific MYC targets (4) were found to be regulated in a STK38-dependent fashion (p<10−10Fisher Exact Test (FET)) (3).
STK38 mRNA is ubiquitously expressed in most human tissues including peripheral blood lymphocytes (5), where it is generally most expressed. NDR proteins expressed in yeast, plants, flies, worms and protozoa, are involved in the regulation of mitosis, cell morphology, cell proliferation and apoptosis, suggesting diverse and essential roles (1). STK38 function and pathway-related activity is still poorly characterized. STK38 was shown to be involved in regulation of centrosome duplication, whose deregulation may result in chromosomal instability (6). Additionally, STK38 has been described as a repressor of MEKK1/2 activity (7), albeit the mechanism remains unknown. Although knowledge of STK38 in vivo function is limited, biochemical studies are unraveling a complex regulation of its kinase activity. Activation of STK38 requires autophosphorylation at Ser-281, phosphorylation at Thr-444 mediated by a Ca2+- dependent upstream kinase (8), and interaction of the N-terminal domain with accessory proteins like S100B (9) or hMOB1 (10). Inactivation of STK38 kinase by dephosphorylation appears to be mediated by protein phosphatase 2A (PP2A), based on increased STK38 kinase activity following treatment with the PP2A inhibitor okadaic acid (OA) (11).
The MYC oncogene has been implicated in the etiology of most types of human neoplasia (12). When overexpressed, MYC elicits autonomous cell proliferation and growth, fueling tumorigenesis (13). MYC has been implicated in the pathogenesis of many different types of human cancer. Conversely, when MYC expression is suppressed back to physiologic levels in tumor cells, the phenomenon of “oncogene addiction” is elicited (14). Oncogene addiction arises when cancer cells become dependent upon the continued activation of tumor initiating oncogene lesions (15). A variety of possible mechanisms of oncogene addiction have been suggested, including the notion that suppression of oncogenic activity inverts the positive balance between proliferation/survival and apoptotic signals typically observed in tumors, leading to arrest of tumor growth (16). Unfortunately, however, since MYC presides over many essential functions in normal cells, its inactivation would likely be associated with significant toxicity. Thus, the ability to suppress MYC activity in context-specific fashion would be especially valuable as a therapeutic strategy. Indeed, no viable pharmacologic approaches currently exist to target MYC in cancer (17).
Here we show that STK38 regulates MYC activity in vivo critically affecting its ability to maintain a neoplastic phenotype. Mechanistically, STK38 both modulates MYC protein turnover through kinase-activity-dependent superposition of distinct molecular mechanisms as well as mediates signaling from BCR. Its modulatory potential is mediated by complex formation with distinct MYC domains. Knockdown of STK38 protein in vivo significantly suppresses tumor growth in a B-cell lymphoma xenograft mouse model. Thus STK38 inactivation abrogates MYC protein levels and function, suppressing MYC-induced tumorigenesis.
Results
BCR-signaling dependent MYC modulation is mediated by STK38
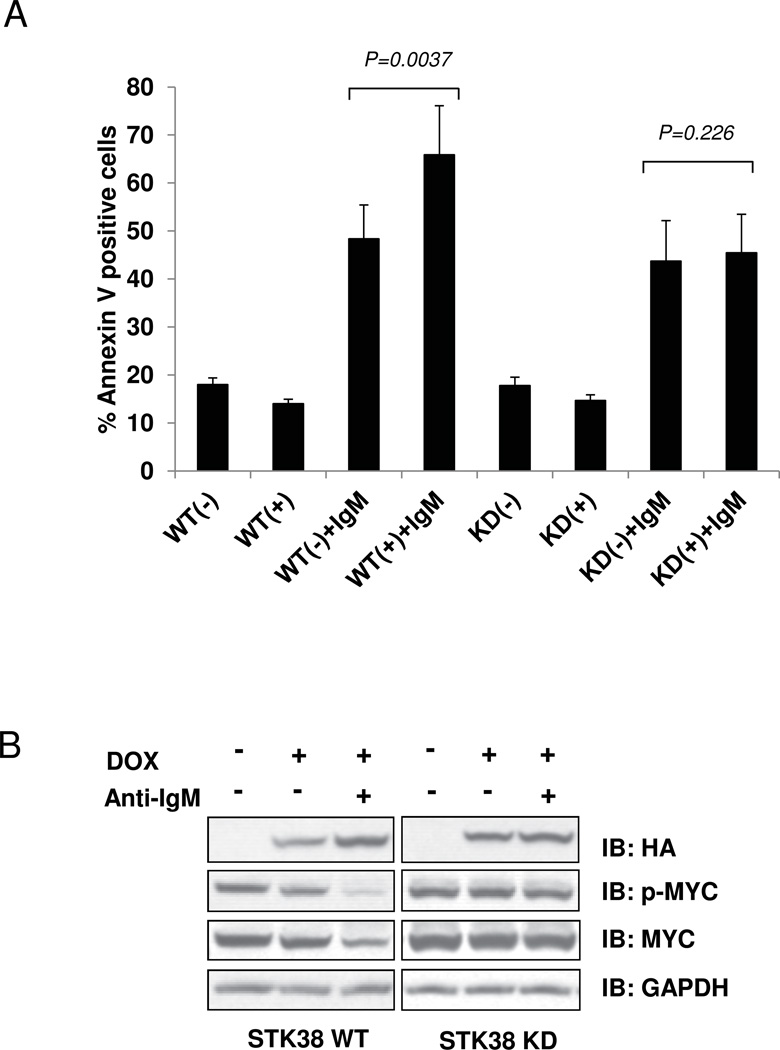
To elucidate the role of STK38-mediated signals in the BCR pathway, we established ST486 Burkitt lymphoma (BL) cell lines conditionally overexpressing either wild-type STK38 (STK38-WT) or its kinase inactive form (STK38-KD), using the inducible Tet system. A single-residue mutation at Lys118 (K118R) in the catalytic site of STK38-KD (18), results in kinase activity reduction (Supplementary Fig. S1). In BL cell lines BCR signal transduction pathway activated by cross-linking of surface IgM with anti-IgM antibodies, induces MYC-dependent growth arrest and apoptosis (19). To evaluate if BCR-mediated apoptosis is regulated by STK38, we crosslinked BCR and analyzed apoptosis by Annexin V binding using flow cytometry. In the absence of BCR signaling, expression of either form of STK38 resulted in slightly decreased apoptotic levels compared with untreated controls (~14% and ~18%, respectively). Consistent with previous observations (19), anti-IgM induced apoptosis in ST486 cells increased significantly (~48%, p =0.01), when compared to untreated cells. Induction of STK38-WT expression further enhanced apoptosis levels (~66%, p =0.0037) while we did not observe significant apoptotic changes in cells expressing STK38-KD upon BCR crosslinking (Fig. 1A).
Figure 1. STK38 kinase mediates anti-IgM induced MYC down regulation and cell apoptosis in ST486 cell line.
STK38-WT and STK38-KD expression in the ST486 cell lines was induced by doxycycline treatment. Cross-linking anti-IgM antibody was performed 24 hours after doxycycline induction. Cells were cultured in the presence of anti-IgM for an additional 6 (A) or 24 hours B; (A) Apoptosis levels measured by flow cytometry using Annexin V binding assay. Bars represent percentage of Annexin V positive cells ±SEM. P-values are estimated by paired t-test analysis. (B) Immunoblot analysis of the total cell extracts showing anti-phosphorylated MYC (p-MYC), anti-HA STK38 fusion proteins (HA), MYC and GAPDH. Results are representative of three independent experiments.
Since BCR activation leads to decreased MYC expression (36), we analyzed changes in MYC protein and mRNA levels upon STK38 overexpression. BCR activation by anti-IgM treatment resulted in a reduction of total MYC protein in both control and STK38-WT expressing cells (Fig. 1B and Supplementary Fig. S2A). However, MYC protein reduction was abrogated in cells overexpressing STK38-KD (Fig. 1B). Additionally, qRT-PCR showed that STK38-WT, but not STK38-KD, can affect MYC mRNA levels upon BCR stimulation (Supplementary Fig. S2B).
STK38 affects MYC protein stability in a kinase-activity-dependent way
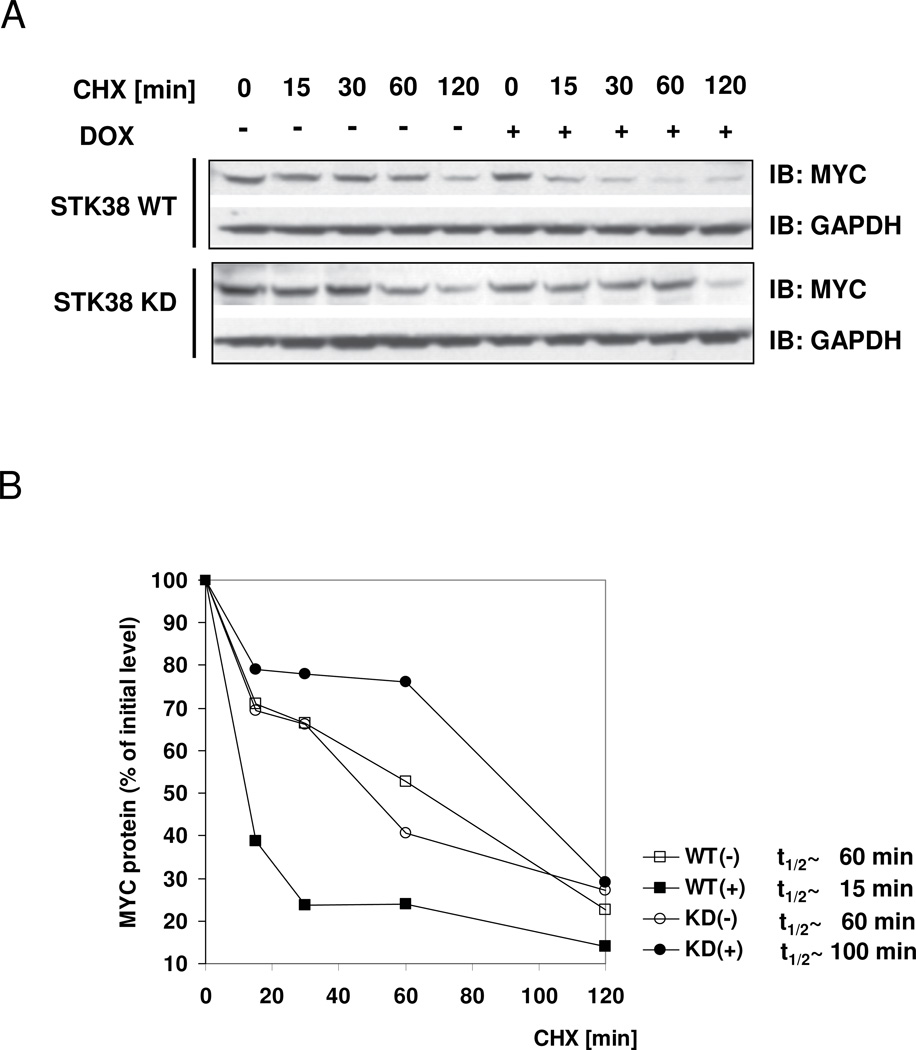
In non-BCR activated ST486 cells, lentiviral-mediated shRNA silencing of endogenous STK38 resulted in significant MYC protein level reduction while MYC mRNA levels were not affected (3). To evaluate whether MYC protein stability could be altered by STK38 kinase activity, we tested MYC protein levels over time in ST486 stable cell lines overexpressing STK38-WT or STK38-KD. STK38-WT overexpression dramatically reduced MYC half-life (4–fold), compared to control un-induced cells (Fig. 2A), while STK38-KD overexpression led to increased MYC half-life (1.7 fold), illustrating an involvement in MYC protein turnover regulation (Fig. 2B). In concordance with the central role of MYC in cell proliferation and survival, decreased MYC protein stability upon STK38 silencing correlated with reduced lymphoma cell survival (Supplementary Fig. S3).
Figure 2. STK38 kinase affects MYC protein stability in ST486 BL cell line.
(A, B) STK38 kinase activity modulates MYC protein turnover. Cells expressing doxycycline-inducible STK38-WT or STK38-KD were incubated with or without doxycycline for 24 hours. Half-life of MYC protein was analyzed by Immunoblotting of total protein lysates harvested at the indicated times after the inhibition of protein synthesis by cyclohexamide (CHX); (A) Immunoblot showing MYC and GAPDH protein levels at different time points; (B) Densitometric analysis of MYC protein levels for each time point compared to the initial MYC protein level. The estimated half-life for each treatment is also shown. Results correspond to a representative experiment out of three.
STK38 interacts with Myc C-terminal domain
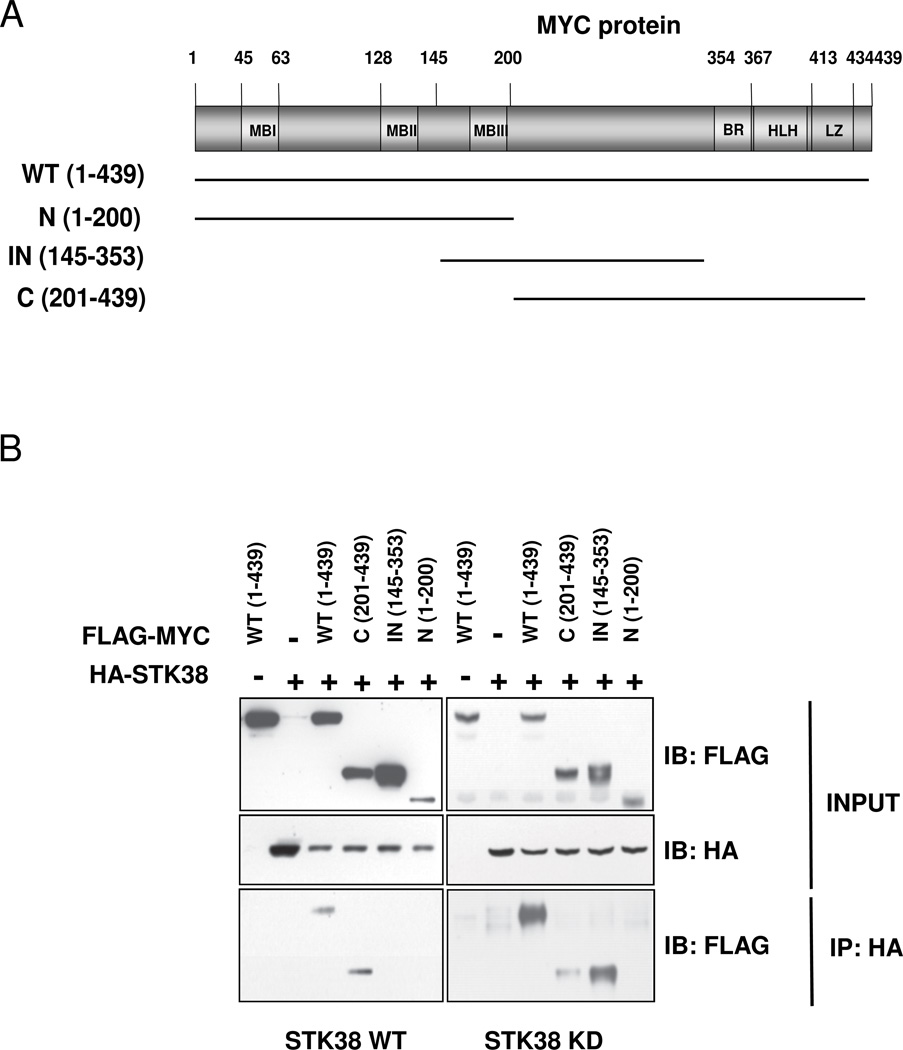
Endogenous MYC and STK38 have been shown to participate in a complex in Ramos B-lymphoma cells (3). To assess which specific MYC domains support STK38 interaction and whether binding is kinase-activity-dependent, we co-transfected HA-tagged expression plasmids for STK38-WT or STK38-KD with Flag-tagged MYC deletion constructs in 293T cells (Fig. 3). Only wild-type and C-terminal domain of MYC protein co-precipitated with STK38-WT indicating a C-terminal domain specific interaction with MYC. In contrast, STK38-KD co-immunoprecipitated with wild-type, internal (145–353 aa), and to a much lesser degree C-terminal (201–439 aa) MYC domains. Surprisingly, STK38-KD bound strongly to the internal domain of MYC that does not appear to bind the STK38-WT, suggesting domain specific binding to MYC is dependent upon STK38 kinase activity. The MYC N-terminus did not interact with either STK38 (Fig. 3B).
Figure 3. STK38 kinase interacts with MYC C-terminal domain.
(A) Schematic representation of MYC protein domains indicating the fragments used for the experiment shown in panel (B); (B) Interaction of STK38 with different MYC protein fragments. STK38-WT or STK38-KD expression plasmids were co-transfected with the Flag-MYC deletion mutants in 293T cells. Cells were lysed 24 hours post-transfection and immunoprecipitated with anti-HA beads. The precipitated protein complexes were resolved by SDS-PAGE and Immunoblotting with anti-FLAG polyclonal antibody and anti-HA.
STK38 modulates MYC transcriptional activity
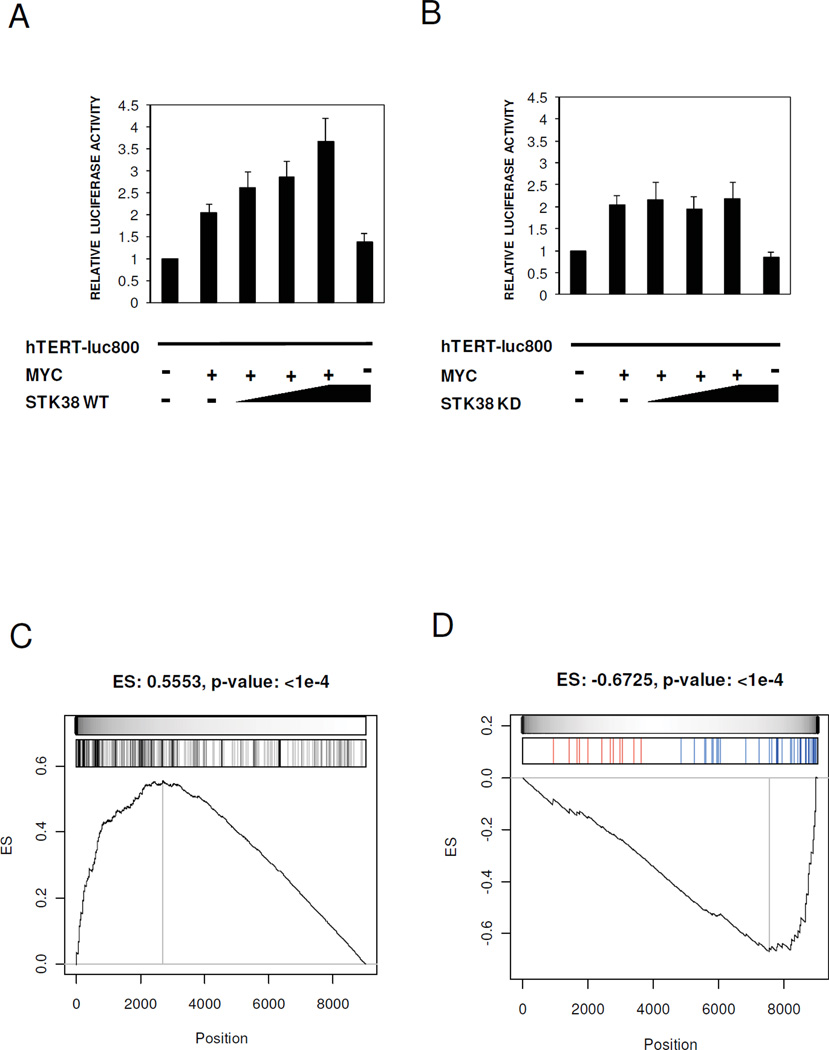
To evaluate whether STK38 kinase activity affects MYC transcriptional activity, we used a MYC human TERT promoter reporter assay (TERTLuc800) (20). U2OS cells were transiently co-transfected with STK38-WT, STK38-KD, MYC and the promoter region of hTERT, and relative luciferase activity was measured. STK38-WT increased MYC-mediated transcriptional activation of the reporter gene in a dose-dependent fashion (Fig. 4A) while STK38-KD did not (Fig. 4B).
Figure 4. STK38 modulates MYC transcriptional activity.
(A, B) The U2OS cells were transiently co-transfected with STK38-WT, STK38-KD, MYC and the promoter region of hTERT. Relative luciferase activity measured in the cells co-expressing STK38-WT (A) or STK38-KD B. Bars represent the mean ±SEM of three independent experiments. (C) GSEA of 258 MYC target genes on the expression profile of ST486 after STK38 knock-down. (D) GSEA of 47 genes known to be transcriptionally activated by MYC. Shown is the enrichment running sum (black line) and its maximum value or enrichment score (ES with associated p-value as headline of each panel). The grey scale bar is proportional to the weight used for each hit (MYC target gene) used by the GSEA algorithm. The bar-code plots indicate the position of MYC target genes on the gene expression profile. The leading edge genes are the group of MYC target genes located to the left (in panel C) and to the right (in panel D) of the grey vertical line. Upregulated and downregulated MYC target genes after STK38 knock-down are shown in red and blue, respectively, in the bar-code plot of panel (D).
To confirm that STK38 is specific for endogenous MYC transcriptional activity in Bcells, we computationally analyzed gene expression profiles of ST486 cells after shRNA mediated STK38 silencing. GSEA demonstrated strong enrichment in MYC regulon B-cell specific MYC target genes) among differentially expressed genes followed by STK38 silencing (Fig. 4C) and in the subset of transcriptionally activated MYC target genes (4) among down-regulated genes after STK38 silencing (p < 10−4Fig. 4D), with 22 out of 47 genes in the leading edge of the GSEA enrichment plot (Supplementary Table S1).
STK38 knockdown suppresses tumor growth in vivo
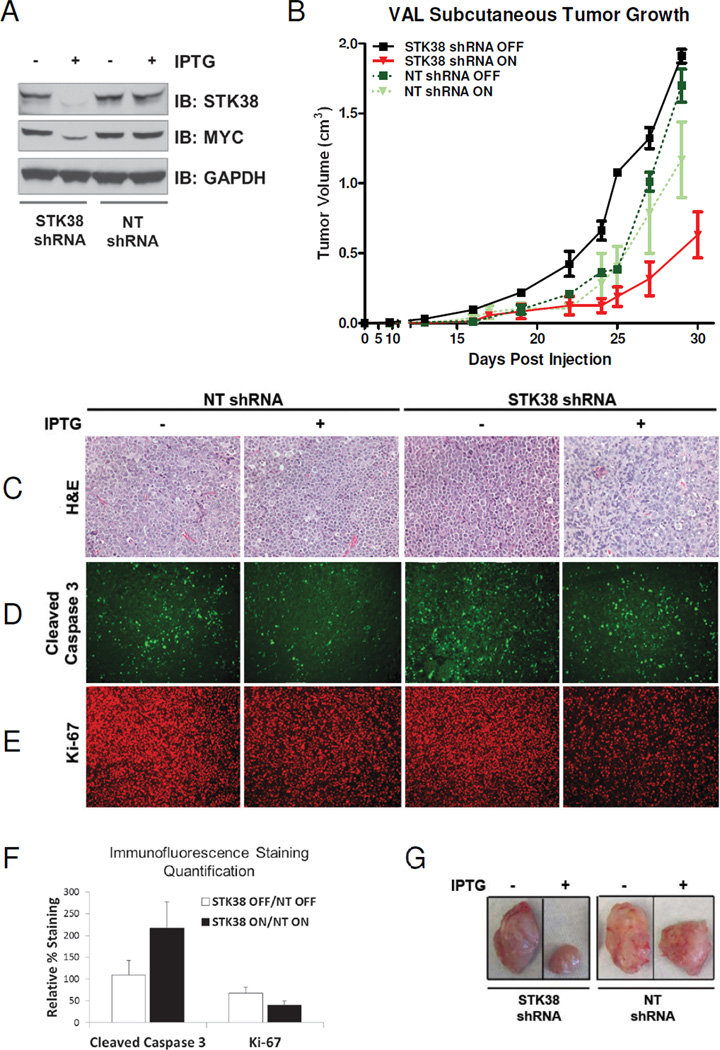
We showed STK38 requirement for cell survival in the ST486 lymphoma line (Supplementary Fig. S3). To evaluate the effect of STK38 knockdown on tumorigenic growth of a MYC-dependent human lymphoma in vivo we introduced xenografts of stable cell lines expressing either control or STK38 shRNA into SCID mice. The VAL cell line, which has a MYC translocation leading to MYC overexpression (21), was engineered to express a shRNA against STK38 (VAL-iSTK38-shRNA) or a non-targeted control (NT) shRNA (VAL-iNT-shRNA) under conditional regulation of IPTG. Conditional knockdown of STK38 induced by an IPTG was confirmed by Immunoblot analysis in vitro Fig. 5A). Induction of STK38 knockdown in vivo with IPTG resulted in an 8 day delay in tumor growth (n=5, p=0.05, Fig. 6B, red line), compared to the STK38 shRNA expressing mice, receiving no IPTG treatment (n=4, Fig. 5B, black line). Expression of the NT shRNA had no significant effect on cells either in the presence or absence of IPTG treatment (n=3, n=3, p=0.61, Fig. 6B, dark and light green dashed lines). Representative tumors excised at day 25 post-injection showed a significant reduction in tumor size in treated versus non-treated tumors for VAL-iSTK38-shRNA cells compared to VAL-iNT-shRNA cells (Fig. 5G). It is noteworthy that resulting tumors that grew from either induced or uninduced tumor groups, including VAL-iSTK38-shRNA tumor cells, expressed STK38 and MYC (Supplementary Fig. S4). Similar results occurred when analogous experiments were done using a second MYC-dependent human cell line, Jurkat T-cell lymphoma, (Supplementary Fig. S5A and B).
Figure 5. Suppression of STK38 in human B-cell lymphoma can delay tumor progression.
(A) Immunoblot analysis of STK38 protein silencing at 48 hours after IPTG (400 uM) induction of shRNA expression in stable VAL cell lines expressing an IPTG-inducible shRNA against either STK38 or a non-targeted (NT) sequence. (B) Tumor growth of the VAL stable cell lines injected into SCID mice ±SEM. The VAL STK38 shRNA cells, with or without IPTG, are shown as red and black lines, respectively. The VAL NT shRNA cells, with or without IPTG are shown as light and dark green lines, respectively. (C–E) VAL STK38 shRNA cells and NT shRNA cells before and after IPTG induction. (C) H&E staining (D) Immunofluorescence staining for cleaved caspase 3 (E) Immunofluorescence staining for Ki-67. (F) Quantification of the cleaved caspase 3 and Ki-67 staining. Bars represent the percentage of positively stained VAL STK38 shRNA cells with or without IPTG induction normalized to the NT shRNA cells with or without IPTG ±SEM, respectively. (G) Representative tumors excised at Day 25 post-injection.
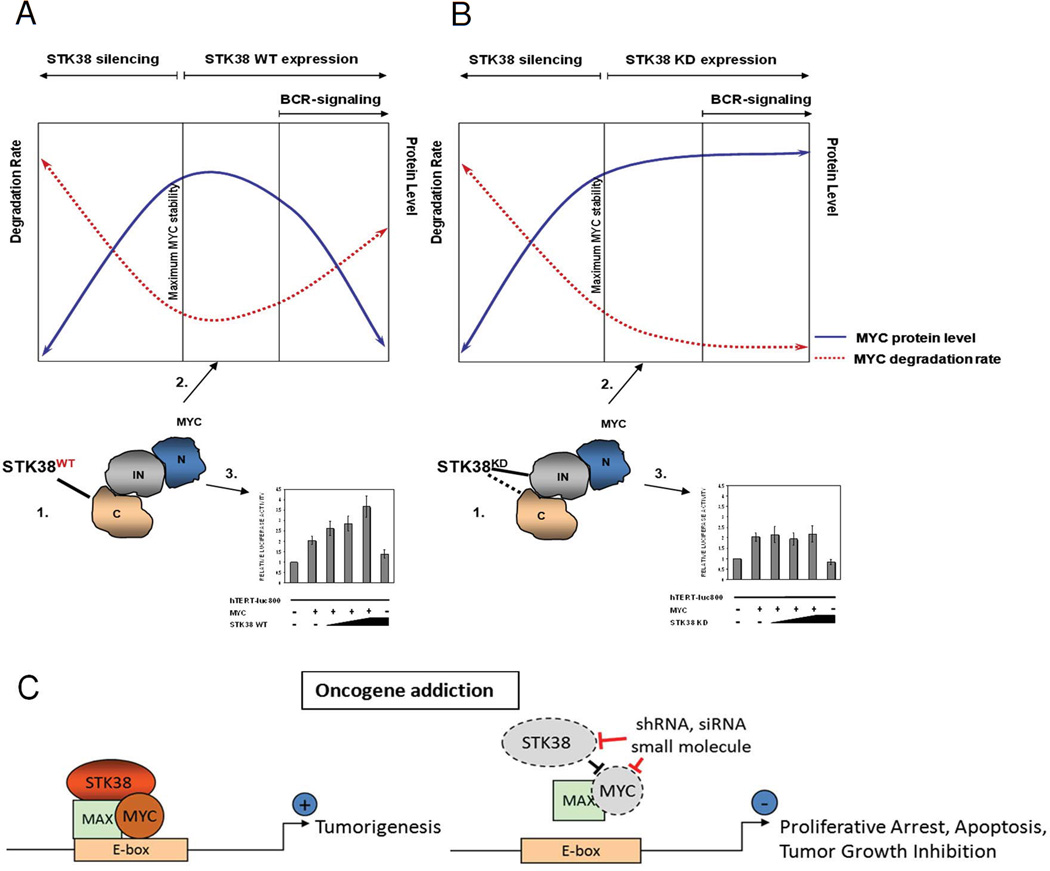
Figure 6. Model of STK38 kinase-activity-dependent modes of MYC modulation.
STK38 kinase affects MYC at several different cellular levels: 1. protein-protein interaction, 2. protein turnover and 3. transcriptional activity. (A) Kinase active STK38 binds to the C-terminus of MYC protein. Overexpression of the kinase increases MYC protein degradation rate and reduces MYC stability. Increased MYC turnover is associated with increased MYC transcriptional activity. Induction of BCR-signaling by anti-IgM crosslinking activates STK38 which causes rapid MYC degradation and induces cell apoptosis; (B) STK38-KD form interacts with MYC through an internal domain of MYC (145–353 aa) and partially C-terminus. Overexpression of STK38-KD reduces MYC protein degradation rate and stabilizes MYC protein. Reduction in MYC protein turnover is associated with lack of effect on transcriptional activity of MYC. BCRsignaling induced by anti-IgM crosslinking is disrupted by mutated inactive STK38 and therefore it cannot affect MYC turnover or transcriptional activity and cells are spared from apoptosis; silencing of endogenous STK38 results in increased MYC degradation rate followed by cell death. (C) Suppression of STK38 elicits oncogene addiction in MYC-driven tumors. STK38 stabilizes MYC protein and regulates MYC transcriptional activity in MYC-addicted tumors (left panel). Pharmacological targeting of STK38 by shRNA, siRNA or small molecule inhibitors, rapidly reduces MYC stability, leading to cell growth arrest and apoptosis (right panel).
To evaluate the mechanism by which suppression of STK38 impedes in vivo tumor growth, we performed in situ immunohistochemistry. Previously, we have shown that MYC suppression in lymphomas is associated with proliferative arrest, apoptosis and senescence (22, 23). Also, the IPTG induced STK38 silencing in VAL cell lines in vitro resulted in increased apoptosis levels compared with control cells (Supplementary Fig. S6). STK38 suppression in vivo was similarly associated with a decrease in proliferative arrest as measured by Ki-67 staining (Fig. 6E and F, p<0.003), and an increase in apoptosis as measured by cleaved caspase 3 staining (Fig. 6D and F, p<0.002).
Discussion
The identification of agents that can elicit oncogene addiction has led to the successful treatment of cancer. The mechanism by which oncogene addiction occurs is not entirely clear, but appears to causally associated with the change in the balance of pro-survival and pro-death signals necessarily evoking tumor regression (16). However, effective small-molecules inhibitors of established oncogenes are rare. Moreover, even if such agents could be identified the broad inhibition of an oncogene that also has normal functions could be highly toxic. Thus, a general strategy for identifying effective agents to target oncogenes and in particular to inhibit them in a manner that would elicit oncogene addiction but minimize toxic effects would be highly useful.
The targeted inactivation of MYC would be potentially useful for the treatment of many cancers. However, TFs are particularly difficult to target. A strategy for their context-specific inactivation would be generally valuable for many TF oncogenes. Moreover, given the pleiotropic regulatory properties of TFs, their systemic inactivation would likely affect many critical cellular functions, resulting in severe toxicity. Furthermore, TFs are often characterized as undruggable targets, due to scarcity of well-defined small-molecule binding pockets in their 3D structure. Thus, availability of methodologies to identify druggable TF modulators whose inhibition will abrogate TF activity in vivo could have important implications in designing targeted therapeutic strategies.
Here, we show that computationally predicted TF modulators, inferred by the MINDy algorithm (24), can provide actionable upstream targets for inactivation of oncogenic TF-regulated programs within a given cellular context, which can abrogate tumorigenesis in vivo .
Our results provide novel mechanistic insight into post-transcriptional regulation of MYC. We identify a new strategy for therapeutic targeting MYC in hematopoietic malignancies. Our results illustrate that STK38 is important in post-translational regulation of MYC’s function and may be a novel therapeutic target. The characterization of post-translational mechanisms for control of MYC’s transcriptional activity by STK38 suggest a more general role for its homeostatic regulation of normal cell physiology that, when deregulated, can contribute to tumorigenesis. Previous literature has correlated changes in STK38 expression with tumorigenesis. Some reports suggest that upregulation of STK38 is associated with progressive breast ductal carcinoma in situ (25) and melanoma (26). Conversely, significant downregulation of STK38 mRNA was shown in gastric cancer compared with normal gastric mucosa (27) and B-cell lymphoma compared with normal B-cells (28). Therefore, the mechanism by which STK38 deregulation contributes to tumorigenesis appears to be context dependent.
We show that STK38 plays a critical role in transduction of BCR signals in human Bcells and can modulate MYC activity by distinct kinase-activity-dependent mechanisms, including: (a) interaction with distinct MYC protein domains, (b) regulation of MYC protein turnover and (c) modulation of MYC transcriptional activity, independent of MYC protein levels. Given the range and significance of the various mechanisms by which STK38 modulates MYC activity, we will discuss them individually.
Post-translational modifications of MYC, including phosphorylation, ubiquitination, and acetylation affect protein stability and are critical effectors of MYC transcriptional activity (29). Despite their significance, understanding the signal transduction cascades affecting MYC post-translational modification is still sparse. Previously, we demonstrated that STK38 kinase mediates MYC phosphorylation (3). In this report we elucidate the kinase-dependent nature of STK38-MYC interaction. STK38 binds to the MYC C-terminal region (201–439 aa) which includes the basic DNA-binding helix-loop-helix-leucine zipper (b/HLH/LZ) domain (30). Proteins known to interact with MYC in this region, including Max, BRCA1, AP-2, YY1, TFII and Miz-1, are all involved in regulation of MYC transcriptional activity (31).
We directly demonstrate that STK38 regulates MYC’s transcriptional activity using a TERT-reporter in U2OS cells. Furthermore, GSEA analysis of STK38 silencing in ST486 cells line showed that STK38 regulates MYC transcriptional activity. Conversely, the kinase inactive form of STK38 had no effect on MYC transcriptional activity. This could be explained by its interaction with the internal MYC domain (145–353 aa) with a region called “MYC box III” (MbIII) previously shown to be indispensable for MYC transcriptional repression of target genes (32) and its degradation (33). Binding of kinase inactive STK38 to this region could interfere with both processes. Although, a transactivation domain (TAD) located in the MYC N-terminus is known to be involved in MYC transcriptional activity (34) it did not interact with STK38 in our co-immunoprecipitation assay. Future investigation will determine whether regulation of MYC transcriptional activity by STK38 is due to differential binding of the kinase to MYC protein or engagement of other co-activating molecules. Recent studies have demonstrated post-translational regulation of MYC is, in part, mediated through coordination of regulatory proteins into complexes containing multi-domain scaffolding proteins (35). Although speculative, it is worth considering that STK38 may be participating in the multi-protein degradation complex involved in MYC regulation.
Our data suggest that MYC turnover is regulated by STK38 activity, with active STK38 decreasing protein levels while inactive STK38 extends MYC half-life. The significant increase in MYC turnover following STK38 overexpression, as well as silencing, suggests that this kinase could provide a therapeutic target for MYC activity modulation in MYC-dependent tumors. This is provocative, since the direct pharmacological targeting of MYC is challenging. Others have reported that overexpression of active STK38 in 293T cells impaired MYC ubiquitination, although STK38 could not compete with FBW7 ubiquitin ligase for MYC binding (36). Our results indicate that overexpression of active STK38 in B-cell context does not interfere with MYC degradation in contrast to inactive STK38. Whether reduction in MYC turnover in the presence of inactive STK38 results from inefficient proteasomal degradation due to either “protective” binding to the MYC internal domain or phosphorylation-dependent modulation of its ubiquitination, must be defined.
We identified the inverse correlation between MYC half-life and activity following increased expression of STK38-WT. Similar findings have been documented for other proteins (37), using chimeric transcriptional activators (38). Increased activity of the transcriptional activator correlated with enhanced proteasome-mediated degradation, and was one possible mechanism of modulation of intracellular levels of regulatory proteins. A similar mechanism was reported for ubiquitin ligase SKP2, which was shown to increase MYC transcriptional activity while decreasing its protein stability (39).
We speculate that the multilayered regulation by STK38 can be explained by a number of regulatory mechanisms effecting intrinsically disordered proteins (IDPs) such as MYC (40–42). Native unstructured regions present in many eukaryotic proteins (43) are involved in transcriptional regulation and signaling events and their levels and accessibility must be tightly controlled. Unstructured regions of IDPs are susceptible to ubiquitin-independent degradation and post-translational modifications which allow precise regulation (44). Such regulation can be mediated through binding to other proteins that effectively protect the IDP from degradation or mediate the IDP phosphorylation state (45). In that context, inactive STK38 could play a role of a “nanny” (45) by binding to the internal domain of MYC and protecting it from degradation. On the other hand, wild type structure of phosphorylated form of STK38 could favor binding to C-terminus of MYC allowing its full functionality.
We have shown that STK38 is a key effector of BCR-signaling-mediated, MYC-dependent apoptosis and proliferative arrest in human B-cells in concordance with our previous results (2). Depending on its kinase activity, STK38 can either stabilize or destabilize MYC protein levels and affect MYC transcriptional activity. Operating within a complex post-translational regulatory landscape, STK38 overexpression may have opposite effects depending on its kinase activity. This is consistent with previously reported dual-role proteins, like p38 or JNK, which could contribute to both anti- and pro-oncogenic processes (46). Similar view on the dual function of NDR1/2 kinases was discussed broadly in (47).
STK38’s function in specific signaling pathways is poorly defined. So far it has only been characterized as a negative regulator of MEKK1/2, a member of MAPK signaling pathway by exogenous expression in 293T cells (7) and a regulator of p21 stability in cell cycle progression (36). We have defined a role of STK38 in BCR signaling. Our results illustrate that STK38 is a critical effector of BCR-mediated signals modulating MYC protein activity and turnover, as well as cell survival, suggesting that this kinase plays a prominent role in the BCR pathway. BCR activation mediated apoptosis was completely abrogated in cells expressing the kinase inactive form, STK38-KD. STK38 mediated regulation of apoptosis was demonstrated by others in HeLa cells, where transient overexpression of wild-type STK38 enhanced release of apoptotic markers (18).
Concordant with the notion of oncogene addiction, the targeted inactivation of critical genes in oncogenic pathways that are directly responsible for the ability of an oncogene to maintain survival over death signaling would in turn lead to the regression of cancer (15). Hence, targeting these co-dependent genes could elicit oncogene addiction identical to the suppression of the oncogene (48). Therefore, we hypothesized that targeting a key regulator of MYC protein expression would lead to abrogation of MYC induced tumors. In vivo mouse xenograft models demonstrated that silencing STK38 leads to delayed tumor formation and slowed tumor growth. At least two mechanisms underlie this process; decreased proliferation and increased apoptosis. These intrinsic mechanisms aid in the tumor regression observed in MYC inactivation models (23). It appears that balance between pro-death and pro-life is shifted toward pro-death in tumors in which STK38 function is removed. Although, in STK38 knockdown tumors an increase in mass after 25 days of growth was observed, it is noteworthy that both STK38 and MYC protein levels were increased as well, suggesting that a subpopulation of cells within the tumor eventually found a way to circumvent IPTG-inducible loss of STK38. Although, we cannot rule out the possibility that the IPTG induction was not efficient in all cells within the tumor mass, it’s interesting to speculate that this observation indicates some bypass mechanism similar to those observed in the reversal of MYC tumor regression (49).
Our results identify a new general strategy for the identification of key regulators of oncogenes that in turn can serve as therapeutic targets to elicit oncogene addiction. Specifically, we demonstrate that STK38 critically regulates the MYC protein and its activity (Fig. 6), at several cellular levels; 1) protein-protein interaction, 2) protein turnover, and 3) transcriptional activity. We show that STK38 is crucial for survival of the ability of MYC-induced lymphomas (15). Hence, suppression of STK38 elicits oncogene addiction in MYC-addicted tumors. Thus, through the combined computational and experimental characterization of effectors of TF activity, we can isolate key upstream regulators of oncogenes, and in particular a TF such as MYC, that are critically required for these oncogenes to maintain a neoplastic phenotype. Therefore, this is a new paradigm for eliciting oncogene addiction, through the suppression of genes that are critical and co-dependent regulators of oncogenes, hence functioning as surrogate and more readily druggable therapeutic targets.
Materials and Methods
Mice
For xenograft tumorigenesis assays, 3–4 week old Fox Chase SCID mice (C.B-17 SCID) were obtained from Charles River Laboratory (Willmington, MA). Animals were kept under standard laboratory conditions and allowed free access to food and water. All experiments were performed with appropriate institutional animal care and use committee approval. Mice were then euthanized via CO2 and secondary method, according to APLAC guidelines at the designated experimental endpoint.
Cell lines, Antibodies and Reagents
293T, HeLa and U2OS cells were cultured in DMEM (Invitrogen) ST486 and VAL cells in IMDM, and Jurkat in RPMI 1640 (Invitrogen) supplemented with 10% FBS (Invitrogen) and 1% penicillin-streptomycin (Cellgro). Stable cell lines ST486 were selected with Geneticin (Invitrogen). Expression of STK38 in stably transduced ST486 cells was induced with 1 µg/ml Doxycycline (Invitrogen). Stable VAL cell lines expressing inducible shRNAs were selected with Puromycin (MP Biomedicals) and induced by IPTG (isopropyl β-D-1-thiogalactopyranoside) (SIGMA). All cell lines were provided by R. Dalla-Favera (Columbia University). Anti-STK38 monoclonal antibody was purchased from Abnova (Taiwan); polyclonal anti-STK38, anti-MYC (9E10), (N262) and FITC conjugate, anti-GAPDH and anti-HA from SantaCruz; anti-β-Actin, anti-FLAG, anti-Tubulin, CHX and OA from Sigma; anti-p-MYC (T58/S62) was purchased from Cell Signaling Technology.
Constructs, Primers, Transfection and Lenti-viral transduction
Human STK38 coding sequence was obtained by RT-PCR from HeLa total RNA using the primers: Forward 5’-CGCAATTGCAATGACAGGCTCAACACC-3’ and Reverse 5’-GCCTCGAGCTATTTTGCTGCTTTCATGTAGG-3’ containing restriction sites for MfeI and XhoI, respectively. PCR product was cloned in EcoRI/XhoI sites of pCMV-HA (Clontech) to obtain the pCMV-HA-STK38-WT expression vector. The STK38 kinase-dead (KD) mutant K118R was generated on the pCMV-HA-STK38-WT vector by site-directed mutagenesis (Genewiz, South Plainfield NJ). HA-STK38-WT and KD were obtained by PCR from pCMV-HA constructs with primers: Forward 5’-GCGGTACCCACCATGTACCCATACGATGTTC-3’ and Reverse 5’-GTACCTCGAGCTATTTTGCTGC-3’ and subcloned in KpnI/XhoI sites of pEN_TTmcs (ATCC MBA-255). The TRE_HA-STK38 inducible cassette was transferred to the lentiviral vector pSLIK_Neo (ATCC MBA-235) by LR gateway recombination (Invitrogen). MYC, WT(1–439), and deletion mutants C(201–439), IN(145–353) and N(1–200), were obtained by PCR from pcDNA3-MYC-HA (50) and cloned in BamHI/EcoRI sites of pCMV-Tag2A vector (Stratagene) to obtain the FLAG tagged MYC fragments. Forward primer for WT and N-fragment: 5’-ATCTTAGGATCCCATGCCCCTCAACGTTA-3’; Reverse primer for N-fragment: 5’-ATTCGGAATTCTTAGTTGAGAGGGTAGGGG-3’; Forward and Reverse primers for IN-fragment: 5’-ATCTTAGGATCCCGTCTCAGAGAAGCTGGC-3’, 5’-ATTCTGAATTCTTAATTCTCCTCGGTGTCC-3’; Forward primer for C-fragment: 5’-ATCTTAGGATCCCGACAGCAGCTCGCCCAA-3’; Reverse primer for WT and C-fragment: 5’-ATTCTGAATTCTTACGCACAAGAGTTCCGT-3’. Primers used for qPCR: MYC, Forward 5’-ATGTCAAGAGGCGAACACAC-3’ and Reverse 5’-GAGCTTTTGCTCCTCTGCTT-3’; GAPDH, Forward 5’-CACCCAGAAGACTGTGGATGGC-3’ and Reverse 5’-GTTCAGCTCAGGGATGACCTTGC-3’. Transient transfections were performed using Lipofectamine (Invitrogen) and FuGENE (Roche) according to the manufacturer’s protocol. Silencing STK38 shRNA clone TRCN0000010215 and control NT shRNA (SHC002) cloned into pLKO.1-puro or pLKO-puro-IPTG-3xLacO lenti-viral vectors (Sigma) were co-transfected with pCMV-Δ8.9 packaging vector and pCMV-VSV-G envelope plasmid according to the manufacturer’s protocol. ST486 cells were infected with viral particles by centrifugation at 1 200 rpm for 90 min, in the presence of 8 µg/ml of Polybrene (Chemicon). Inducible stable ST486 cell lines expressing STK38 WT and KD proteins were generated by lenti-viral transduction with pSLIK-Neo-STK38 constructs.
Real-Time PCR
The cDNA preparation and real-time polymerase chain reaction (qPCR) were performed using the FastLane Cell cDNA and QuantiTech Kits, respectively, according to the manufacturer’s protocol (Qiagen). qPCR reactions were run with SYBR Green (Qiagen) master mix using an Applied Biosystems 7300 thermal cycler.
Sample processing for microarray
Total RNA was extracted with Trizol (Invitrogen) and purified by RNeasy kit (Qiagen). 5μg of total RNA were processed following manufacturer instructions (Affymetrix, 701025 Rev.6), and 15ug of fragmented and biotin-labeled cRNA were hybridized to HG-U95Av2 microarrays (Affymetrix). All microarray data have been submitted to Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo, GSE20229).
Gene reporter assay
U2OS cells were transiently co-transfected with expression vectors encoding MYC, TERTLuc800 (Wu et al., 1999) and STK38-WT or STK38-KD. Relative luciferase activity was measured 48 hours post-transfection using Dual-Luciferase Reporter Assay System (Promega).
Immunoprecipitation and Immunoblotting analysis
Immunoprecipitation using anti-HA agarose or control IgG-agarose was performed according to the manufacturer’s protocol. Immunoprecipitated proteins were denatured with 2x SDS Sample Buffer (Invitrogen), resolved by SDS-PAGE and analyzed by Immunoblotting. Protein lysates were prepared by lysing cells in RIPA buffer (Upstate) supplemented with protease inhibitor cocktail (Roche). Proteins were resolved by SDS-PAGE, transferred to nitrocellulose and analyzed by standard Immunoblotting procedures.
Kinase activity assay
The 293T cells transfected with HA-tagged STK38-WT, STK38-KD and control vectors were washed in PBS and lysed in Cell Lysis Buffer (Cell Signaling Technology) supplemented with protease inhibitor cocktail (Roche). Cell lysates were pre-cleared for 1 hr at 4°C with Sepharose-G (Zymed). Supernatants were incubated with anti-HA agarose overnight at 4°C with rotation. Immune complexes were washed three times with cell lysis buffer followed by three washings with Kinase Buffer (Cell Signaling Technology). For the kinase assay, immunoprecipitates were incubated with 1 mM of synthetic STK38 peptide substrate (KKRNRRLSVA) (Invitrogen) and 10 µM. ATP in 50 µl of kinase buffer at 30°C for 1 hr. Kinase activity was measured by quantitation of the remaining amount of ATP using Kinase-Glo Luminescent Kinase Assay Platform (Promega) (Supplementary Fig. 1).
Flow Cytometry assays
Apoptosis levels in ST486 and VAL cells were measured using Annexin V binding kit (BD Pharmingen). Annexin V fluorescence intensity was measured and analyzed using FACSCalibur flow cytometer and CellQuest version 3.3 software (BD Biosciences). Statistical significance was estimated by paired t-student test. Correlation between mean and variance was reduced by log-transformation of the data.
Xenograft Tumorigenesis assay
Tumorigenic potential of MYC-dependent B-cell lymphoma cells with STK38 suppressed was assessed utilizing xenografts. VAL cells, bearing the t(8:14) chromosomal rearrangement causing MYC overexpression, were infected with lentivirus driving stable expression of either IPTG-inducible STK38 shRNA (VAL-iSTK38-shRNA) or IPTG-inducible non-targeted control (VAL-iNT-shRNA). Jurkat cells, acute lymphoblastic leukemia T-cells known to overexpress MYC, were infected with lentivirus to drive stable expression of either IPTG-inducible STK38 shRNA (Jurkat-iSTK38-shRNA) or IPTG-inducible non-targeted control (Jurkat-iNT-shRNA). VAL-iSTK38-shRNA, VAL-iNT-shRNA, Jurkat-iSTK38-shRNA, or Jurkat-iNT-shRNA cells were injected subcutaneously into right flanks of SCID mice (10 million cells/mouse). Mice were monitored and tumors tracked via caliper measurements. Tumor volume was determined by the formula: tumor volume = ½(large diameter)*(small diameter2. Mice were then either untreated or treated with a combination of 14 µM IPTG in their drinking water and 200 µL IPTG (0.53 mmol) via intraperitoneal (IP) injection every other day from the time of injection. Mice from all treatment groups were euthanized when tumors from untreated mice reached maximum tumor burden (~1.75cm3). Tumors were excised and tissue archived.
Immunohistochemistry and Immunofluorescence assays
Tumor tissues excised from xenografts were formalin fixed and embedded in paraffin. Paraffin embedded tumor sections were deparaffinized by successive incubations in xylene, 95% ethanol, 90% ethanol, 70% ethanol followed by PBS. Epitopes were unmasked by steaming in DAKO antigen retrieval solution for 45 minutes and rinsed twice in PBS. One section was stained with Hematoxylin and Eosin (H&E) while other sections were incubated overnight at 4°C with 1:100 anti-Ki-67 (Invitrogen) or 1:100 anti-cleaved caspase 3 (BD Biosciences) antibody, washed, and incubated with anti-mouse or anti-rabbit secondary antibody, respectively. Images were obtained on a Nikon microscope and analyzed using Metamorph software (Meta Imaging Series).
Data analysis
Expression data were normalized with the gcrma algorithm implemented in the gcrma R-system package v2.18 (Bioconductor). The original Gene Set Enrichment Analysis (GSEA) (51) was implemented in R-system and nominal p-values were computed by permuting genes uniformly at random 10 000 times.
We used the MINDy (Modulator Inference by Network Dynamics) algorithm (3), to dissect the genome-wide TF activity post-translational modulatory network in human B-cells. MINDy tests whether the conditional mutual information (CMI), I[TF ; t | M ], between a TF and a target t as a function of a modulator M is non-constant. In that case, M is inferred as a candidate post-translational modulator of the TF . In this work we define the signalome as the compendium of 781 signaling proteins (SP) annotated as protein kinases (421 genes), phosphatases (113 genes) or cell surface receptors (295 genes) in the Gene Ontology (GO) (52) and represented in a 254 tumor and normal human B-cell samples gene expression profile (28). SP-TF interactions were inferred by assessing whether one or more TF-target interactions were modulated by the SP using the MINDy algorithm at the genome-wide statistical significance level of 5% (2). In this work, we refer to the complete set of TF-targets modulated by a SP as the SP's regulon while the set of all TFs modulated by a SP is called the SP's modulon.
Supplementary Material
Acknowledgments
We thank Riccardo Dalla-Favera for critical review of this manuscript and Masumichi Saito for supplying MYC deletion mutant expression vectors (both Columbia University). This work was supported by the NCI (R01CA109755), the NIAID (R01AI066116) and the NCBC NIH Roadmap Initiative (U54CA121852) (AC), and Burroughs Welcome Fund Career Award, the Damon Runyon Foundation Lilly Clinical Investigator Award, NIH (R01CA089305, R01CA105102), National Cancer Institute's In-vivo Cellular and Molecular Imaging Center (P50CA114747), Integrative Cancer Biology Program (1P20CA112973), NIH/NCI (P01CA034233), the Leukemia and Lymphoma Society Translational Research grant number R6223-07 (D.W.F.), and the American Cancer Society Fellowship PF-10-237-01-TBG (SJA).
Grant Support: This work was supported by the NCI (R01CA109755), the NIAID (R01AI066116) and the NCBC NIH Roadmap Initiative (U54CA121852) (AC), and Burroughs Welcome Fund Career Award, the Damon Runyon Foundation Lilly Clinical Investigator Award, NIH (R01CA089305, R01CA105102), National Cancer Institute's In-vivo Cellular and Molecular Imaging Center (P50CA114747), Integrative Cancer Biology Program (1P20CA112973), NIH/NCI (P01CA034233), the Leukemia and Lymphoma Society Translational Research grant number R6223-07 (D.W.F.), and the American Cancer Society Fellowship PF-10-237-01-TBG (SJA).
Footnotes
Conflict of Interest
The authors disclose no potential conflicts of interest.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc).
References
- 1.Hergovich A, Schmitz D, Hemmings BA. The human tumour suppressor LATS1 is activated by human MOB1 at the membrane. Biochemical and biophysical research communications. 2006 Jun 23;345(1):50–58. doi: 10.1016/j.bbrc.2006.03.244. [DOI] [PubMed] [Google Scholar]
- 2.Wang K, Alvarez MJ, Bisikirska BC, Linding R, Basso K, Dalla Favera R, et al. Dissecting the interface between signaling and transcriptional regulation in human B cells. Pacific Symposium on Biocomputing. 2009:264–275. [PMC free article] [PubMed] [Google Scholar]
- 3.Wang K, Saito M, Bisikirska BC, Alvarez MJ, Lim WK, Rajbhandari P, et al. Genome-wide identification of post-translational modulators of transcription factor activity in human B cells. Nature biotechnology. 2009 Sep;27(9):829–839. doi: 10.1038/nbt.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zeller KI, Jegga AG, Aronow BJ, O'Donnell KA, Dang CV. An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome biology. 2003;4(10):R69. doi: 10.1186/gb-2003-4-10-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Millward T, Cron P, Hemmings BA. Molecular cloning and characterization of a conserved nuclear serine(threonine) protein kinase. Proceedings of the National Academy of Sciences of the United States of America. 1995 May 23;92(11):5022–5026. doi: 10.1073/pnas.92.11.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hergovich A, Lamla S, Nigg EA, Hemmings BA. Centrosome-associated NDR kinase regulates centrosome duplication. Molecular cell. 2007 Feb 23;25(4):625–634. doi: 10.1016/j.molcel.2007.01.020. [DOI] [PubMed] [Google Scholar]
- 7.Enomoto A, Kido N, Ito M, Morita A, Matsumoto Y, Takamatsu N, et al. Negative regulation of MEKK1/2 signaling by serine-threonine kinase 38 (STK38) Oncogene. 2008 Mar 20;27(13):1930–1938. doi: 10.1038/sj.onc.1210828. [DOI] [PubMed] [Google Scholar]
- 8.Tamaskovic R, Bichsel SJ, Rogniaux H, Stegert MR, Hemmings BA. Mechanism of Ca2+-mediated regulation of NDR protein kinase through autophosphorylation and phosphorylation by an upstream kinase. The Journal of biological chemistry. 2003 Feb 28;278(9):6710–6718. doi: 10.1074/jbc.M210590200. [DOI] [PubMed] [Google Scholar]
- 9.Bhattacharya S, Large E, Heizmann CW, Hemmings B, Chazin WJ. Structure of the Ca2+/S100B/NDR kinase peptide complex: insights into S100 target specificity and activation of the kinase. Biochemistry. 2003 Dec 16;42(49):14416–14426. doi: 10.1021/bi035089a. [DOI] [PubMed] [Google Scholar]
- 10.Devroe E, Erdjument-Bromage H, Tempst P, Silver PA. Human Mob proteins regulate the NDR1 and NDR2 serine-threonine kinases. The Journal of biological chemistry. 2004 Jun 4;279(23):24444–24451. doi: 10.1074/jbc.M401999200. [DOI] [PubMed] [Google Scholar]
- 11.Millward TA, Hess D, Hemmings BA. Ndr protein kinase is regulated by phosphorylation on two conserved sequence motifs. The Journal of biological chemistry. 1999 Nov 26;274(48):33847–33850. doi: 10.1074/jbc.274.48.33847. [DOI] [PubMed] [Google Scholar]
- 12.Marcu KB, Bossone SA, Patel AJ. myc function and regulation. Annu Rev Biochem. 1992;61:809–860. doi: 10.1146/annurev.bi.61.070192.004113. [DOI] [PubMed] [Google Scholar]
- 13.Bouchard C, Staller P, Eilers M. Control of cell proliferation by Myc. Trends Cell Biol. 1998 May;8(5):202–206. doi: 10.1016/s0962-8924(98)01251-3. [DOI] [PubMed] [Google Scholar]
- 14.Felsher DW. Reversibility of oncogene-induced cancer. Curr Opin Genet Dev. 2004 Feb;14(1):37–42. doi: 10.1016/j.gde.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 15.Weinstein IB. Cancer. Addiction to oncogenes--the Achilles heal of cancer. Science, New York, NY. 2002 Jul 5;297(5578):63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- 16.Tran PT, Bendapudi PK, Lin HJ, Choi P, Koh S, Chen J, et al. Survival and death signals can predict tumor response to therapy after oncogene inactivation. Sci Transl Med. 2011 Oct 5;3(103):103ra99. doi: 10.1126/scitranslmed.3002018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Felsher DW. Oncogenes as therapeutic targets. Semin Cancer Biol. 2004 Feb;14(1):1. doi: 10.1016/j.semcancer.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 18.Vichalkovski A, Gresko E, Cornils H, Hergovich A, Schmitz D, Hemmings BA. NDR kinase is activated by RASSF1A/MST1 in response to Fas receptor stimulation and promotes apoptosis. Curr Biol. 2008 Dec 9;18(23):1889–1895. doi: 10.1016/j.cub.2008.10.060. [DOI] [PubMed] [Google Scholar]
- 19.Kaptein JS, Lin CK, Wang CL, Nguyen TT, Kalunta CI, Park E, et al. Anti- IgM-mediated regulation of c-myc and its possible relationship to apoptosis. The Journal of biological chemistry. 1996 Aug 2;271(31):18875–18884. doi: 10.1074/jbc.271.31.18875. [DOI] [PubMed] [Google Scholar]
- 20.Wu KJ, Grandori C, Amacker M, Simon-Vermot N, Polack A, Lingner J, et al. Direct activation of TERT transcription by c-MYC. Nature genetics. 1999 Feb;21(2):220–224. doi: 10.1038/6010. [DOI] [PubMed] [Google Scholar]
- 21.Bonnefoy-Berard N, Genestier L, Flacher M, Rouault JP, Lizard G, Mutin M, et al. Apoptosis induced by polyclonal antilymphocyte globulins in human B-cell lines. Blood. 1994 Feb 15;83(4):1051–1059. [PubMed] [Google Scholar]
- 22.Wu CH, van Riggelen J, Yetil A, Fan AC, Bachireddy P, Felsher DW. Cellular senescence is an important mechanism of tumor regression upon c-Myc inactivation. Proceedings of the National Academy of Sciences of the United States of America. 2007 Aug 7;104(32):13028–13033. doi: 10.1073/pnas.0701953104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Felsher DW, Bishop JM. Reversible tumorigenesis by MYC in hematopoietic lineages. Molecular cell. 1999 Aug;4(2):199–207. doi: 10.1016/s1097-2765(00)80367-6. [DOI] [PubMed] [Google Scholar]
- 24.Bansal M, Califano A. Genome-wide dissection of posttranscriptional and posttranslational interactions. Methods Mol Biol. 2012;786:131–149. doi: 10.1007/978-1-61779-292-2_8. [DOI] [PubMed] [Google Scholar]
- 25.Adeyinka A, Emberley E, Niu Y, Snell L, Murphy LC, Sowter H, et al. Analysis of gene expression in ductal carcinoma in situ of the breast. Clin Cancer Res. 2002 Dec;8(12):3788–3795. [PubMed] [Google Scholar]
- 26.Millward TA, Heizmann CW, Schafer BW, Hemmings BA. Calcium regulation of Ndr protein kinase mediated by S100 calcium-binding proteins. The EMBO journal. 1998 Oct 15;17(20):5913–5922. doi: 10.1093/emboj/17.20.5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cui DX, Zhang L, Yan XJ, Zhang LX, Xu JR, Guo YH, et al. A microarray-based gastric carcinoma prewarning system. World J Gastroenterol. 2005 Mar 7;11(9):1273–1282. doi: 10.3748/wjg.v11.i9.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, Califano A. Reverse engineering of regulatory networks in human B cells. Nature genetics. 2005 Apr;37(4):382–390. doi: 10.1038/ng1532. [DOI] [PubMed] [Google Scholar]
- 29.Hann SR. Role of post-translational modifications in regulating c-Myc proteolysis, transcriptional activity and biological function. Semin Cancer Biol. 2006 Aug;16(4):288–302. doi: 10.1016/j.semcancer.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 30.Blackwell TK, Kretzner L, Blackwood EM, Eisenman RN, Weintraub H. Sequence-specific DNA binding by the c-Myc protein. Science, New York, NY. 1990 Nov 23;250(4984):1149–1151. doi: 10.1126/science.2251503. [DOI] [PubMed] [Google Scholar]
- 31.Sakamuro D, Prendergast GC. New Myc-interacting proteins: a second Myc network emerges. Oncogene. 1999 May 13;18(19):2942–2954. doi: 10.1038/sj.onc.1202725. [DOI] [PubMed] [Google Scholar]
- 32.Herbst A, Hemann MT, Tworkowski KA, Salghetti SE, Lowe SW, Tansey WP. A conserved element in Myc that negatively regulates its proapoptotic activity. EMBO reports. 2005 Feb;6(2):177–183. doi: 10.1038/sj.embor.7400333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herbst A, Salghetti SE, Kim SY, Tansey WP. Multiple cell-type-specific elements regulate Myc protein stability. Oncogene. 2004 May 6;23(21):3863–3871. doi: 10.1038/sj.onc.1207492. [DOI] [PubMed] [Google Scholar]
- 34.Kato GJ, Barrett J, Villa-Garcia M, Dang CV. An amino-terminal c-myc domain required for neoplastic transformation activates transcription. Mol Cell Biol. 1990 Nov;10(11):5914–5920. doi: 10.1128/mcb.10.11.5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arnold HK, Zhang X, Daniel CJ, Tibbitts D, Escamilla-Powers J, Farrell A, et al. The Axin1 scaffold protein promotes formation of a degradation complex for c-Myc. The EMBO journal. 2009 Mar 4;28(5):500–512. doi: 10.1038/emboj.2008.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cornils H, Kohler RS, Hergovich A, Hemmings BA. Human NDR kinases control G(1)/S cell cycle transition by directly regulating p21 stability. Mol Cell Biol. 2011 Apr;31(7):1382–1395. doi: 10.1128/MCB.01216-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mooney LM, Whitmarsh AJ. Docking interactions in the c-Jun N-terminal kinase pathway. The Journal of biological chemistry. 2004 Mar 19;279(12):11843–11152. doi: 10.1074/jbc.M311841200. [DOI] [PubMed] [Google Scholar]
- 38.Molinari E, Gilman M, Natesan S. Proteasome-mediated degradation of transcriptional activators correlates with activation domain potency in vivo. The EMBO journal. 1999 Nov 15;18(22):6439–6447. doi: 10.1093/emboj/18.22.6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim SY, Herbst A, Tworkowski KA, Salghetti SE, Tansey WP. Skp2 regulates Myc protein stability and activity. Molecular cell. 2003 May;11(5):1177–1188. doi: 10.1016/s1097-2765(03)00173-4. [DOI] [PubMed] [Google Scholar]
- 40.Babu MM, van der Lee R, de Groot NS, Gsponer J. Intrinsically disordered proteins: regulation and disease. Curr Opin Struct Biol. 2011 Jun;21(3):432–440. doi: 10.1016/j.sbi.2011.03.011. [DOI] [PubMed] [Google Scholar]
- 41.Nair SK, Burley SK. X-ray structures of Myc-Max and Mad-Max recognizing DNA. Molecular bases of regulation by proto-oncogenic transcription factors. Cell. 2003 Jan 24;112(2):193–205. doi: 10.1016/s0092-8674(02)01284-9. [DOI] [PubMed] [Google Scholar]
- 42.Follis AV, Hammoudeh DI, Wang H, Prochownik EV, Metallo SJ. Structural rationale for the coupled binding and unfolding of the c-Myc oncoprotein by small molecules. Chem Biol. 2008 Nov 24;15(11):1149–1155. doi: 10.1016/j.chembiol.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 43.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004 Mar 26;337(3):635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 44.Tsvetkov P, Reuven N, Prives C, Shaul Y. Susceptibility of p53 unstructured N terminus to 20 S proteasomal degradation programs the stress response. The Journal of biological chemistry. 2009 Sep 25;284(39):26234–26242. doi: 10.1074/jbc.M109.040493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsvetkov P, Reuven N, Shaul Y. The nanny model for IDPs. Nat Chem Biol. 2009 Nov;5(11):778–781. doi: 10.1038/nchembio.233. [DOI] [PubMed] [Google Scholar]
- 46.Engelberg D. Stress-activated protein kinases-tumor suppressors or tumor initiators? Semin Cancer Biol. 2004 Aug;14(4):271–282. doi: 10.1016/j.semcancer.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 47.Cornils H, Kohler RS, Hergovich A, Hemmings BA. Downstream of human NDR kinases: impacting on c-myc and p21 protein stability to control cell cycle progression. Cell Cycle. 2011 Jun 15;10(12):1897–1904. doi: 10.4161/cc.10.12.15826. [DOI] [PubMed] [Google Scholar]
- 48.Jain M, Arvanitis C, Chu K, Dewey W, Leonhardt E, Trinh M, et al. Sustained loss of a neoplastic phenotype by brief inactivation of MYC. Science, New York, NY. 2002 Jul 5;297(5578):102–104. doi: 10.1126/science.1071489. [DOI] [PubMed] [Google Scholar]
- 49.Karlsson A, Giuriato S, Tang F, Fung-Weier J, Levan G, Felsher DW. Genomically complex lymphomas undergo sustained tumor regression upon MYC 24 inactivation unless they acquire novel chromosomal translocations. Blood. 2003 Apr 1;101(7):2797–2803. doi: 10.1182/blood-2002-10-3091. [DOI] [PubMed] [Google Scholar]
- 50.Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M, et al. Non-transcriptional control of DNA replication by c-Myc. Nature. 2007 Jul 26;448(7152):445–451. doi: 10.1038/nature05953. [DOI] [PubMed] [Google Scholar]
- 51.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005 Oct 25;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature genetics. 2000 May;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.