Abstract
Conventional leaching (extraction) methods for gold recovery from electronic waste involve the use of strong acids and pose considerable threat to the environment. The alternative use of bioleaching microbes for gold recovery is non-pollutive and relies on the secretion of a lixiviant or (bio)chemical such as cyanide for extraction of gold from electronic waste. However, widespread industrial use of bioleaching microbes has been constrained by the limited cyanogenic capabilities of lixiviant-producing microorganisms such as Chromobacterium violaceum. Here we show the construction of a metabolically-engineered strain of Chromobacterium violaceum that produces more (70%) cyanide lixiviant and recovers more than twice as much gold from electronic waste compared to wild-type bacteria. Comparative proteome analyses suggested the possibility of further enhancement in cyanogenesis through subsequent metabolic engineering. Our results demonstrated the utility of lixiviant metabolic engineering in the construction of enhanced bioleaching microbes for the bioleaching of precious metals from electronic waste.
The ever increasing demand for electronics, fueled by a rising affluence in societies, coupled with the short lifetime of electronic devices, has resulted in the generation of a torrential electronic waste stream in our modern world1. This electronic waste stream presents a major disposal challenge as electronic waste contains toxic metallics such as lead and mercury. In addition to its inherent toxicity, electronic waste also contains significant amounts of precious metals such as gold (Au). In fact, compared with natural Au ores, the Au content in electronic scrap material (ESM) is significantly higher (10 g to 10 kg Au per ton of ESM2,3 as compared to 0.5 g to 13.5 g Au per ton of natural Au ore3,4), creating an economic driving force for the recycling of electronic waste2,4.
In 2009, the United Nations Environment Program (UNEP) estimated the global electrical and electronic waste (e-waste) generated to be around 40 million tons annually5. The same report projected that by 2020, the waste stream would double in OECD countries and surge up to 500% from 2007 levels in developing nations. Electronic waste shows a higher growth rate than any other category of municipal waste6. E-waste is growing at an alarming rate due to increased consumption of electronic devices and shorter life span of mobile phones, computers, televisions and other electronic devices. Manufacturing mobile phones and personal computers consumes 3% of gold and silver, 13% of palladium and 15% of cobalt mined worldwide each year5.
Au is used in electronics for its excellent resistance to corrosion and high electrical conductivity. Existing processes for recovering Au from electronic waste are either pyrometallurgical (which is energy intensive), or hydrometallurgical (which uses chemical lixiviants such as aqua regia, a mixture of concentrated nitric and hydrochloric acids), and are thus extremely pollutive and not environmentally sustainable4,7. Bioleaching using lixiviant-secreting microbes has been considered as a sustainable alternative in the recovery of precious metals from electronic waste8,9.
In contrast to existing processes, bioleaching, involving microorganisms such as Chromobacterium violaceum, may allow metal recycling in a process analogous to natural biogeochemical cycles, and hence reduces the demand for resources such as ores, energy or space for landfills10. In a process similar to industrial Au cyanidation, cyanogenic microorganisms produce the cyanide lixiviant which then reacts with solid Au to complete the leaching process2,9,11,12. The cyanide lixiviant is derived from the secondary metabolite hydrogen cyanide (HCN) produced from glycine using the enzyme HCN synthase (encoded by the hcnABC operon in cyanogenic C. violaceum)13,14,15; however, the amount of lixiviant metabolite produced is limited (20 mg of cyanide per liter of bacteria culture, approximately 1 × 1016 colony forming units), and tight regulation of this operon under quorum control further restricts its widespread use in industrial Au recovery from electronic waste16,17. Here we report the construction of two metabolically-engineered C. violaceum strains that produced more cyanide lixiviant, and recovered more Au from electronic waste, than wild-type bacteria. We envision the industrial use of metabolically-engineered microorganisms in the recovery of commodity metals (such as Au, Fe and Cu) from electronic waste, the detoxification of toxic metals from waste and landfills, and the recovery and remediation of mine tailings.
Results
Construction of metabolically-engineered C. violaceum strains
Two metabolically-engineered C. violaceum strains (L-arabinose and IPTG for pBAD and pTAC, respectively) were constructed by site-specific integration of the duplicated hcnABC operon under the transcriptional control of the exogenous promoter pBAD (L-arabinose-inducible) and pTAC (Isopropyl-thiogalactoside/IPTG-inducible), respectively (Fig. 1).
Figure 1. Lixiviant metabolic engineering of Chromobacterium violaceum for enhanced cyanide production and gold recovery.
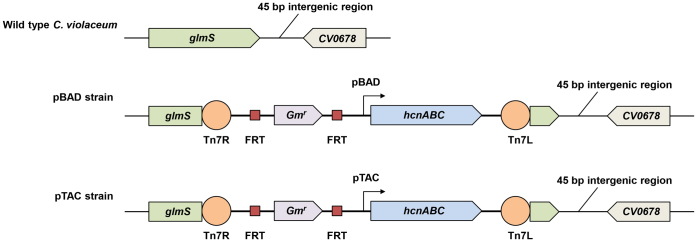
Schematic of genomic region between locus tags CV_0677 and CV_0678 of wild-type and engineered C. violaceum. The duplicated hcnABC operon, with the respective pBAD or pTAC promoter, was inserted into the glmS gene using Tn7-based transposition.
Enhanced cyanide lixiviant production in pBAD and pTAC strains
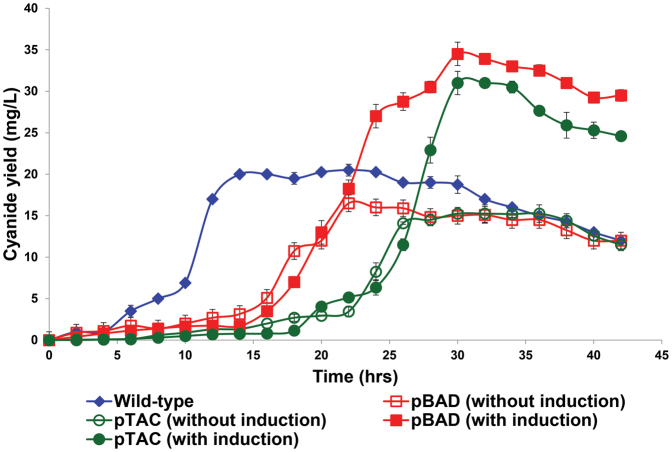
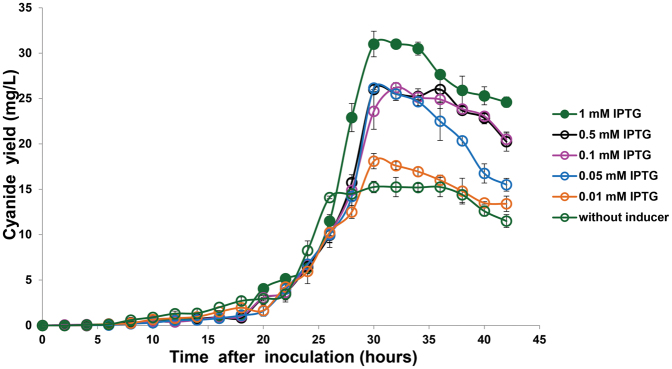
Under optimal inducer concentrations, the metabolically-engineered pBAD strain produced 68% more cyanide lixiviant compared to wild-type bacteria, whilst the pTAC strain produced 51% more cyanide lixiviant than the non-engineered wild-type cyanogenic bacteria (Fig. 2). The differing times for production of cyanide between wild-type and engineered strains of C. violaceum is a result of the differing growth rates and induction times between the strains (Fig. S1). Wild-type C. violaceum was grown in media without antibiotic supplementation, while engineered strains of C. violaceum were grown in media with gentamycin supplementation; the absence or presence of antibiotics in the growth media resulted in the differing growth rates. Both pBAD (Fig. 3) and pTAC (Fig. 4) strains exhibited typical dose-responses (cyanide production) to varying concentrations of the inducer (L-arabinose for pBAD, and IPTC for pTAC, respectively).
Figure 2. Cyanide production profiles of wild-type and engineered Chromobacterium violaceum strains.
pBAD and pTAC strains were induced with 0.002% L-arabinose and 1 mM IPTG, respectively.
Figure 3. Production of cyanide by pBAD strain was dependent on L-arabinose inducer concentrations.
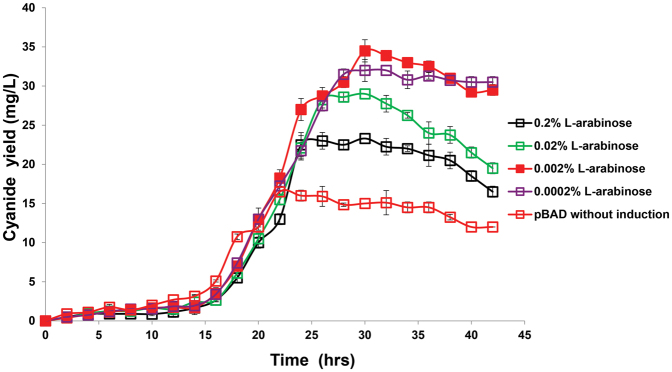
Figure 4. Production of cyanide by pTAC strain was dependent on IPTG inducer concentrations.
Enhanced Au recovery from electronic waste by metabolically-engineered strains
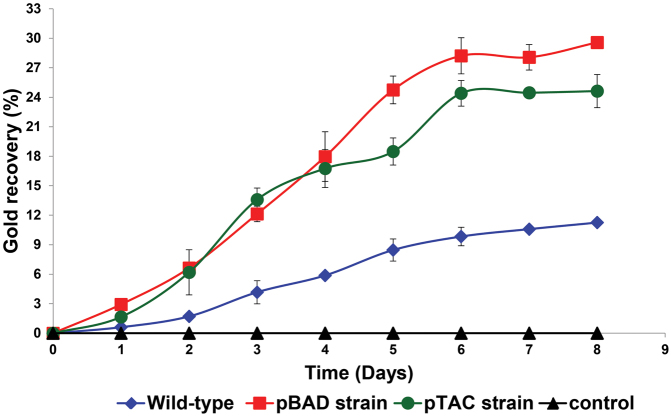
With the expectation that an increase in cyanide lixiviant production would accompany a corresponding increase in bioleaching of Au from electronic waste, the engineered pBAD and pTAC C. violaceum strains were used to recover precious Au from ESM. Bioleaching studies revealed that there were significant increases in the recovery of Au from the ESM electronic waste for both strains of engineered C. violaceum, as compared to wild-type bacteria. After 8 days of bioleaching treatment, an Au recovery of 30% and 25% of the total amount of Au present in the electronic waste, was achieved with the engineered pBAD and pTAC strains, respectively (Fig. 5); using wild-type bacteria under identical bioleaching conditions, an Au recovery of 11% of the total amount of Au present in the ESM was obtained. This represented increases of 170% and 120%, respectively, in Au recovery from ESM by the pBAD and pTAC strains.
Figure 5. Gold recovery profiles of wild-type and engineered Chromobacterium violaceum strains.
Gold recovery from ESM was obtained using pBAD and pTAC strains that were induced with 0.002% L-arabinose and 1 mM IPTC, respectively.
Was cyanide production decoupled from quorum control?
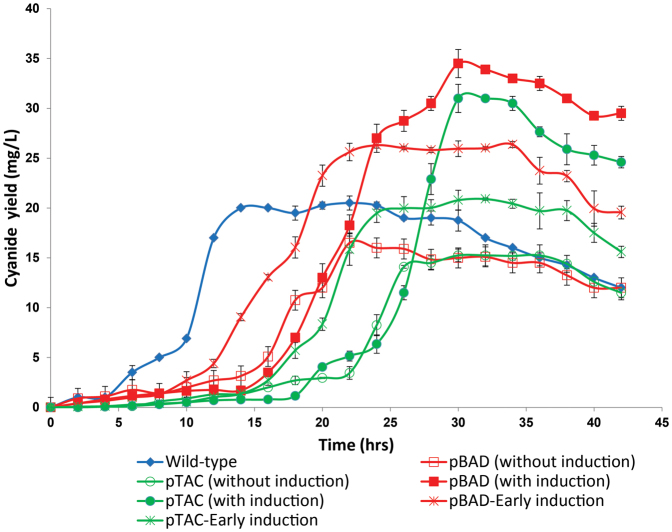
In order to decouple lixiviant production from quorum control, exogenous promoters pBAD and pTAC were used to drive the expression of the hcnABC operon. When early induction (prior to mid-logarithmic phase) was performed, we observed the early production of cyanide lixiviant by the metabolically-engineered bacteria (Fig. 6). These observations, coupled with the observations of tunable, dose-responsive (under varying inducer concentrations) cyanide production, corroborated our expectations that cyanide production in the metabolically-engineered pBAD and pTAC strains have been effectively decoupled from quorum control.
Figure 6. Decoupling cyanide production from quorum control.
Cyanide production profiles of wild-type and engineered strains; pBAD and pTAC strains were induced with 0.002% L-arabinose and 1 mM IPTG, respectively.
Comparative proteomics study of cyanogenically-enhanced C. violaceum
Using iTRAQ18 (isobaric Tags for Relative and Absolute Quantitation) labeling of peptide digests of the wild-type and pBAD proteomes, coupled with mass spectrometric analyses, quantitative proteomics maps of the engineered and wild-type strains were obtained (Table S1).
Discussion
Here we report the construction of two metabolically-engineered C. violaceum strains (pBAD and pTAC, respectively) that produced significantly more cyanide lixiviant compared to wild-type bacteria. Lixiviant production was effectively decoupled from quorum control through the use of exogenous promoters. In corroboration with the observed increase in cyanide production by the engineered strains, Au recovery from ESM was significantly increased in the engineered strains. Our efforts in modulating the lixiviant metabolism of C. violaceum demonstrate proof-of-concept that enhanced bioleaching microbes can be constructed as sustainable means of recovering precious metals such as Au from electronic waste.
Wild-type C. violaceum produces cyanide from glycine for short periods at the early stationary phase in its growth. The reliance of lixiviant production on cell population density is quorum-controlled, and is probably co-evolved as a defense mechanism in its bid for niche colonization19. In an effort to obtain suitable strains of C. violaceum for bioleaching of Au from electronic waste, we examined if lixiviant production can be decoupled from quorum control, and if so, whether cyanide production can be increased by modulating the lixiviant metabolic pathways of the organism. Normal cyanide metabolism is maintained by combined actions of the cyanogenic hcnABC operon and the cyanolytic cynTSX operon. We sought to engineer a tightly-regulated and tunable (responsive to varying concentrations of inducer) bioleaching strain by integrating a single copy of the hcnABC operon, under the transcriptional control of the exogenous promoters, into the bacterial genome. This site-specific genomic integration of an inducible cyanogenic operon was performed using Tn7-mediated transposition developed by Schweizer and coworkers20. We inserted a duplicate copy of the hcnABC operon (under the transcriptional control of exogenous promoters pBAD or pTAC, respectively) to decouple cyanogenesis from quorum control.
By design, genomic insertions can be made at specific Tn7 attachment (attTn7) sites located downstream of the highly conserved glmS gene (encoding for glucosamine 6-phosphate synthetase) in bacteria. The glmS gene of wild-type C. violaceum (GI: 34496132) is located upstream of a functionally unassigned gene (GI: 34496133) with a 45 bp intergenic region that is predicted to contain the attTn7 site. However, in contrast to studies with other bacterial species21, genomic integration in C. violaceum did not occur at the intergenic region downstream of the glmS gene; instead, the transposable segment was inserted within the 3′region of the glmS gene (after 1607 bp of the 1830 bp intact glmS gene). Despite disruption of the glmS gene, bacterial fitness in the engineered C. violaceum strains was not compromised (growth curves of wild-type and engineered strains are virtually identical, Fig. S2).
In attempts to increase the production of the cyanide lixiviant, we used a range of inducer concentrations to obtain dose-responsive profiles of cyanide production. Using L-arabinose as the exogenous inducer, cyanide production was increased above wild-type levels (Fig. 3), with maximal cyanide production (relative to uninduced levels) observed with 0.002% L-arabinose. An analogous dose-responsive profile was obtained with IPTG as the exogenous inducer (Fig. 4), where the addition of 1 mM IPTG resulted in maximal cyanide production, compared to uninduced or wild-type levels. Our observations indicated that the engineered pBAD and pTAC strains exhibited tunable, enhanced cyanide lixiviant production. The observed proto-typical dose-responsive profiles also suggested that lixiviant production in these strains had been decoupled from quorum-control. A comparison of the lixiviant profiles of wild-type versus engineered strains revealed that the pBAD and pTAC strains produced peak concentrations (at 30 hrs after inoculation) of 34.5 mg/L and 31 mg/L of cyanide, respectively (representing significant increases over the wild-type peak concentration of 20 mg/L of cyanide, Fig. 2).
With the expectation that an increase in cyanide lixiviant production would accompany a corresponding increase in bioleaching of Au from electronic waste, the engineered pBAD and pTAC C. violaceum strains were used to recover precious Au from ESM. Bioleaching studies revealed that there were significant increases in the recovery of Au from the ESM electronic waste for both strains of engineered C. violaceum, as compared to wild-type bacteria (Fig. 5). There have been studies on the use of cyanogenic bacteria in the recovery of Au from solid waste10,22,23,24: the reported modest recovery (up to 14.9% of total amount of Au present) corroborate with our observation (11% of total amount of Au present) on the limited utility of wild-type cyanogenic bacteria in bioleaching. In comparison, we were able to achieve Au recovery in excess of 30% of total amount of Au present using engineered C. violaceum. We view these significant increases in Au recovery from electronic waste (over wild-type cyanogenic bacteria) as a heartening possibility of using bioleaching as a sustainable means of recovering precious metals from electronic waste in the future.
To facilitate subsequent engineering efforts, we sought insights into the modulations of the lixiviant metabolic network of the pBAD strain through a comparative proteomics study of the cyanogenically-enhanced variant against the wild-type C. violaceum strain (Fig. 7). We examined if the observed levels of cyanide produced in the engineered strain could be further increased with future metabolic engineering. A logical corollary follows that an increase in cyanide production could be met with modulations in the proteome that could have decreased cyanogenesis, and increased cyanolysis, leading to a limiting “cap” in the amount of cyanide production. Conversely, one could expect the observed increase in cyanogenesis to be within the inherent lixiviant capacity of the cyanogenic bacterium, and minimal modulations in the cyanogenic and cyanolytic pathways of the engineered strain would be observed.
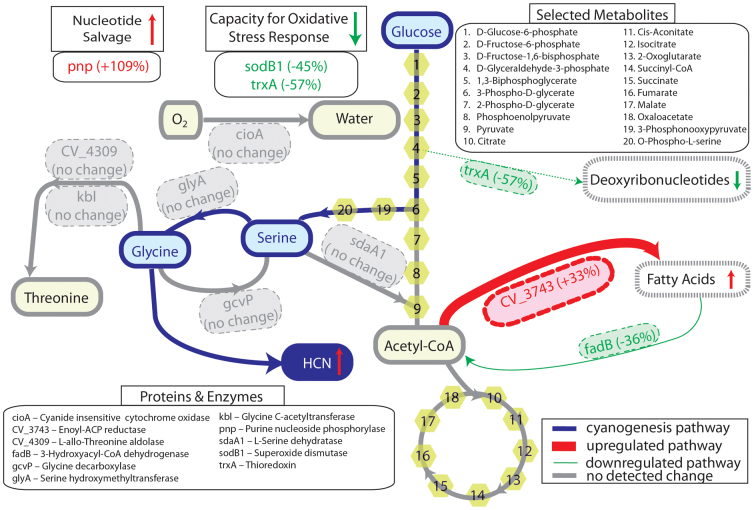
Figure 7. Comparative proteome analyses after increased cyanogenesis in Chromobacterium violaceum.
The cyanogenic pathway is highlighted in blue. Up-regulations are represented in red, and down-regulations are represented in green. The identities of proteins and enzymes detected in the study are shown in Supplementary Table S1.
Cyanide production through the actions of HCN synthase can be decreased by decreasing the availability of the substrate glycine; consequently, glycine flux away from cyanogenesis can be independently achieved through the actions of enzymes such as serine hydroxymethyltransferase17 (glyA, in the biosynthesis of serine from glycine), glycine decarboxylase25 (gcvP, in the glycine cleavage system), serine dehydratase26 (sdaA1, in serine catabolism), threonine aldolase and glycine C-acetyltransferase27 (CV_4309 and kbl, respectively, in the biosynthesis of threonine). Our comparative proteomics study revealed that upon an increase in cyanide production in the engineered strain, there were no significant changes in the protein levels of these enzymes (directly and indirectly) associated with the cyanolytic pathways (Fig. 7). Cyanolysis can also be directly achieved through the actions of cyanase28; our study did not detect significant changes in the levels of cyanase in the engineered strain, compared to wild-type C. violaceum. In addition, we did not observe any significant changes in the protein levels of enzymes associated with glycine biosynthesis, and hence, cyanogenesis.
Cyanogenesis in bacteria and plants is associated with virulence and defense12; in the context of C. violaceum, our comparative proteomics study revealed significant modulations in metabolic pathways associated with metabolic dormancy and energy conservation. Taken together, an increase in nucleotide salvage (represented by a decrease in the enzyme purine nucleoside phosphorylase29/pnp), a decrease in deoxyribonucleotide biosynthesis (represented by a decrease in thioredoxin/trxA levels, the essential cofactor for ribonucleotide reductase30), a decrease in the capacity for oxidative stress response31 (represented by decreases in superoxide dismutase/sodB1 and thioredoxin/trxA levels), and an increase in fatty acid biosynthesis32 (represented by an increase in enoyl-ACP reductase/CV_3743 levels, an essential enzyme in the biosynthesis of saturated straight-chain fatty acids; and a decrease in 3-hydroxyacyl-CoA dehydrogenase/fadB levels, an enzyme associated with β-oxidation of fatty acids) suggested that the increased levels of cyanide production was within the cyanolytic capacity of the engineered strain. Our hypothesis was further corroborated by the observation that there was no significant change in protein levels of cioA33, a cyanide-insensitive terminal cytochrome oxidase in the respiratory electron transport chain of C. violaceum; if cyanide levels were to exceed the inherent cyanolytic capacity of the bacterium, the expectation would be a significant up-regulation of cioA, so that aerobic respiration (in particular, cytochrome c oxidase) would not be inhibited.
We have demonstrated that lixiviant metabolism in C. violaceum can be engineered for enhanced cyanide production; a decoupling of cyanogenesis from quorum control resulted in a significant increase in cyanide production, and correspondingly, an increase in Au recovery from electronic waste. Comparative proteomics analyses suggested that further increase in cyanogenesis is possible, and our results highlight the utility of lixiviant metabolic engineering in the construction of next-generation bioleaching microbes for the recovery of precious metals such as Au from electronic waste.
Methods
Construction of Chromobacterium violaceum strains
Wild-type C. violaceum ATCC 12472 was obtained from ATCC. The hcnABC operon was PCR amplified from the genomic DNA isolated from C. violaceum (ATCC 12472) using Platinum Pfx DNA polymerase (Invitrogen) and the primer sets hcnABC-XbaI-F/hcnABC-XhoI-R (Table S2). Exogenous inducible promoters pBAD and pTac were cloned upstream of the hcnABC operon. The pBAD and pTac promoters were PCR amplified from pBAD33 and pET15b vectors, respectively. After PCR amplification, the respective promoters were cloned upstream of the hcnABC operon into pUC18-mini-Tn7T-Gm vectors. The Tn7-based genomic transposition method as described by Choi and Schweizer20 was used for genomic integration of the duplicated hcnABC operon into the genome of C. violaceum. Genomic integration was confirmed by DNA sequencing of the insertion sites using the primer sets P-Tn7L and P-0678 (Table S2).
Electronic scrap material (ESM)
The electronic waste, ESM, used in this study was supplied by Cimelia Resource Recovery (Singapore). The grey dust-like ESM was generated by shredding and other mechanical separation processes during mechanical recycling of ESM parts such as printed circuit boards. ESM used in this study were of particle size less than 100 μm. We used the same batch of ESM for all our bioleaching experiments. To check on the uniformity in the composition of gold, we conducted acid digestion experiments after every few batches of bioleaching runs. The initial amount of gold present in the ESM is 0.24 ± 0.04 mg/g of ESM. Gold content in this ESM from printed circuit board scrap was higher than those reported by Ilyas et al, Liang et al and Xiang et al34,35,36 and was comparable with ESM reported by Hageluken37.
Metal analyses
ESM samples with weight 1.000 ± 0.005 g were added in 250 mL Erlenmeyer flasks. Each sample was subjected to aqua regia [mixture of concentrated nitric acid (69% m/v) and hydrochloric acid (37% m/v) at 1:3 ratio] leaching, and the resulting residue was sent for Inductively-Coupled Plasma Mass Spectrometry (ICP-MS) metal-analysis using an Agilent 7500a ICP-MS spectrometer. The residue was dried and weighed to determine the mass of the solubilized metal.
Growth and induction of bacteria cultures
Wild-type and engineered C. violaceum were inoculated and grown in LB media from single colonies. The engineered pBAD and pTAC strains were grown in LB supplemented with gentamycin at 15 μg/mL. From an overnight confluent culture (1 × 107 cfu/mL), a 1% (v/v) inoculum was used to inoculate 100 mL of LB medium in a 250 mL Erlenmeyer flask; the culture was incubated at 30°C, with shaking at 170 rpm; and for the engineered strains respective inducers (0.0002%, 0.002%, 0.02% and 0.2% L-arabinose for pBAD; 0.01 mM, 0.05 mM, 0.1 mM, 0.5 mM and 1 mM IPTG for pTAC strains, respectively) were added to the cultures during mid-log phase (12 hrs). To demonstrate decoupling of cyanide production from quorum control for the engineered strains, respective inducers were added to the cultures pre mid-log phase (6 hrs).
Cyanide analyses
Free cyanide present in samples was measured using a cyanide electrode and an Ion Selective Electrode (ISE) meter (Thermoscientific Orion). Six-point calibrations (0, 0.5, 1, 5, 10, 25 ppm) were performed prior to cyanide measurements. Dilutions were done to fit the measurements within the dynamic range of the calibrated electrode when cyanide concentrations were above 25 ppm.
Bioleaching studies
Bioleaching of ESM was carried out with wild-type and engineered C. violaceum in 250 mL Erlenmeyer flasks with 100 mL of Luria-Bertani (LB) at a pulp density of 0.5% (w/v). A two-step bioleaching process was performed during Au bioleaching of ESM: bacteria were initially cultured in LB in the absence of ESM; sterilized ESM was then added to the bacterial cultures upon cyanide production. The cultures were incubated at 30°C, with shaking at 170 rpm, over eight days after ESM addition. Samples were taken daily, with debris removed by centrifugation (11,000 g for 15 mins), and cyanide and metal analyses were performed as described prior. The cyanide produced over eight days after ESM addition is shown in Fig. S3.
Comparative proteome analyses
Proteome analyses were performed using three biological replicates each of wild-type and engineered C. violaceum; a technical replicate for each biological replicate was also performed. The growth of wild-type and pBAD strains were performed as previously described. For the engineered pBAD strain, 0.002% L-arabinose was used to induce the bacteria cultures at mid-log phase. After 16 hours post-induction (corresponding to maximal cyanide production), the cells were harvested by centrifugation, washed with M9 minimal media, and resuspended in lysis buffer38 (8 M urea, 25 mM TEAB, 2% Triton X-100, 0.1% SDS, 1× protease inhibitor cocktail and 50 μg/mL of DNase and RNase). The cells were lysed by sonication, lysates were clarified by centrifugation, and the supernatants were collected and subjected to iTRAQ labeling.
For iTRAQ labeling, 50 μg of proteins were reduced and alkylated prior to trypsin digestion; the resulting peptides were then lyophilized and reconstituted before labeling, according to the manufacturer's instructions (AB SCIEX). Protein identification and relative iTRAQ quantitation were performed using an AB SCIEX TripleTOF 5600 mass spectrometer, with ProteinPilot™ software version 4.5 using Paragon™ Algorithm 4.5.0.0; a search was made on the C. violaceum Uniprot complete proteome database entry version 19-Oct-2011, and a reversed database to deduce the global false discovery rate. Sample type was iTRAQ 8plex (peptide labeled) with MMTS cysteine alkylation and an ID Focus on biological modifications. The cut-off value used for protein regulation was 1.3 for up-regulation and 0.7 for down-regulation39.
The bars in Figures 2 to 6 represent the standard deviation for the various points in the plots for the various experiments.
Author Contributions
W.S.Y. and Y.P.T. conceived the project. T.S.B.T. constructed the engineered pBAD and pTAC strains. G.N. carried out the bioleaching studies and the metal and cyanide analyses. M.C.M.C. designed the comparative proteomics studies, and M.N.A.R. and H.T.T. carried out the comparative proteome analyses. All authors contributed to the preparation of the manuscript.
Supplementary Material
Supplementary Information
Acknowledgments
We would like to thank Professor Herbert P. Schweizer for the kind gifts of the mini-Tn7 delivery plasmids. We also thank Mr. Lim Teck Kwang for his technical assistance in mass spectrometry. This work was supported by grants from the Academic Research Fund of the Ministry of Education, Singapore.
References
- Tsydenova O. & Bengtsson M. Chemical hazards associated with treatment of waste electrical and electronic equipment. Waste Management 31, 45–58 (2011). [DOI] [PubMed] [Google Scholar]
- Cui J. & Zhang L. Metallurgical recovery of metals from electronic waste: a review. Journal of hazardous materials 158, 228–256 (2008). [DOI] [PubMed] [Google Scholar]
- Pham V. & Ting Y. P. Gold bioleaching of electronic waste by cyanogenic bacteria and its enhancement with bio-oxidation. Advanced Materials Research 71, 661–664 (2009). [Google Scholar]
- Korte F., Spiteller M. & Coulston F. The cyanide leaching gold recovery process is a nonsustainable technology with unacceptable impacts on ecosystems and humans: the disaster in Romania. Ecotoxicology and environmental safety 46, 241–245 (2000). [DOI] [PubMed] [Google Scholar]
- Schluep M. et al. UNEP. Recycling - From E-Waste to Resources, in Sustainable Innovation and Technology Transfer Industrial Sector Studies, http://www.unep.org/PDF/PressReleases/E-Waste_publication_screen_FINALVERSION-sml.pdf (2009). [Google Scholar]
- Greenpeace International. The e-waste problem, http://www.greenpeace.org/international/en/campaigns/toxics/electronics/the-e-waste-problem/ (2005). [Google Scholar]
- Fields S. Tarnishing the earth: gold mining's dirty secret. Environ Health Perspect 109, A474–481 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs W., Brombacher C., Bosshard P. P., Bachofen R. & Brandl H. Microbial recovery of metals from solids. FEMS Microbiology Reviews 20, 605–617 (1997). [Google Scholar]
- Valenzuela L. et al. Genomics, metagenomics and proteomics in biomining microorganisms. Biotechnology Advances 24, 197–211 (2006). [DOI] [PubMed] [Google Scholar]
- Faramarzi M. A., Stagars M., Pensini E., Krebs W. & Brandl H. Metal solubilization from metal-containing solid materials by cyanogenic Chromobacterium violaceum. Journal of Biotechnology 113, 321–326 (2004). [DOI] [PubMed] [Google Scholar]
- Rawlings D. E. Heavy Metal Mining Using Microbes. Annual Review of Microbiology 56, 65–91 (2002). [DOI] [PubMed] [Google Scholar]
- Knowles C. J. & Bunch A. W. Microbial cyanide metabolism. Advances in microbial physiology 27, 73–111 (1986). [DOI] [PubMed] [Google Scholar]
- Laville J. et al. Characterization of the hcnABC gene cluster encoding hydrogen cyanide synthase and anaerobic regulation by ANR in the strictly aerobic biocontrol agent. Pseudomonas fluorescens CHA0. Journal of bacteriology 180, 3187–3196 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels R. & Corpe W. A. Cyanide Formation by Chromobacterium Violaceum. Journal of bacteriology 89, 106–112 (1965). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels R., Hankes L. V. & Corpe W. A. Cyanide formation from glycine by nonproliferating cells of Chromobacterium violaceum. Arch Biochem Biophys 111, 121–125 (1965). [DOI] [PubMed] [Google Scholar]
- Brazilian National Genome Project, C. The complete genome sequence of Chromobacterium violaceum reveals remarkable and exploitable bacterial adaptability. Proceedings of the National Academy of Sciences of the United States of America 100, 11660–11665 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motokawa Y. & Kikuchi G. Glycine metabolism in rat liver mitochondria: V. Intramitochondrial localization of the reversible glycine cleavage system and serine hydroxymethyltransferase. Archives of Biochemistry and Biophysics 146, 461–466 (1971). [DOI] [PubMed] [Google Scholar]
- Treumann A. & Thiede B. Isobaric protein and peptide quantification: perspectives and issues. Expert Review of Proteomics 7, 647–653 (2010). [DOI] [PubMed] [Google Scholar]
- The complete genome sequence of Chromobacterium violaceum reveals remarkable and exploitable bacterial adaptability. Proceedings of the National Academy of Sciences of the United States of America 100, 11660–11665 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K. H. et al. A Tn7-based broad-range bacterial cloning and expression system. Nat Methods 2, 443–448 (2005). [DOI] [PubMed] [Google Scholar]
- Choi K. H. & Schweizer H. P. mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat Protoc 1, 153–161 (2006). [DOI] [PubMed] [Google Scholar]
- Brandl H., Lehmann S., Faramarzi M. A. & Martinelli D. Biomobilization of silver, gold, and platinum from solid waste materials by HCN-forming microorganisms. Hydrometallurgy 94, 14–17 (2008). [Google Scholar]
- Campbell S. C., Olson G. J., Clark T. R. & McFeters G. Biogenic production of cyanide and its application to gold recovery. J Ind Microbiol Biotechnol 26, 134–139 (2001). [DOI] [PubMed] [Google Scholar]
- Kita Y., Nishikawa H. & Takemoto T. Effects of cyanide and dissolved oxygen concentration on biological Au recovery. J Biotechnol 124, 545–551 (2006). [DOI] [PubMed] [Google Scholar]
- Hiraga K. & Kikuchi G. The mitochondrial glycine cleavage system. Purification and properties of glycine decarboxylase from chicken liver mitochondria. J Biol Chem 255, 11664–11670 (1980). [PubMed] [Google Scholar]
- Yoshida T. & Kikuchi G. Comparative Study on Major Pathways of Glycine and Serine Catabolism in Vertebrate Livers. Journal of biochemistry 72, 1503–1516 (1972). [DOI] [PubMed] [Google Scholar]
- Miller D. A. & Simmonds S. The metabolism of L-threonine and glycine by. Escherichia coli. Proceedings of the National Academy of Sciences 43, 195–199 (1957). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carepo M. S. et al. Identification of Chromobacterium violaceum genes with potential biotechnological application in environmental detoxification. Genetics and molecular research : GMR 3, 181–194 (2004). [PubMed] [Google Scholar]
- Agarwal K. C., Agarwal R. P., Stoeckler J. D. & Parks R. E. Purine nucleoside phosphorylase. VI. Microheterogeneity and comparison of kinetic behavior of the enzyme from several tissues and species. Biochemistry 14, 79–84 (1975). [DOI] [PubMed] [Google Scholar]
- Ashley G. W. & Stubbe J. Current ideas on the chemical mechanism of ribonucleotide reductases. Pharmacology & Therapeutics 30, 301–329 (1985). [DOI] [PubMed] [Google Scholar]
- Camhi S. L., Lee P. & Choi A. M. The oxidative stress response. New horizons 3, 170–182 (1995). [PubMed] [Google Scholar]
- Mead J. F. Lipid Metabolism. Annual review of biochemistry 32, 241–268 (1963). [DOI] [PubMed] [Google Scholar]
- Zlosnik J. E. A. et al. Investigation of the physiological relationship between the cyanide-insensitive oxidase and cyanide production in Pseudomonas aeruginosa. Microbiology (Reading, England) 152, 1407–1415 (2006). [DOI] [PubMed] [Google Scholar]
- Ilyas S., Anwar M. A., Niazi S. B. & Afzal Ghauri M. Bioleaching of metals from electronic scrap by moderately thermophilic acidophilic bacteria. Hydrometallurgy 88, 180–188 (2007). [Google Scholar]
- Liang G., Mo Y. & Zhou Q. Novel strategies of bioleaching metals from printed circuit boards (PCBs) in mixed cultivation of two acidophiles. Enzyme and Microbial Technology 47, 322–326 (2010). [Google Scholar]
- Xiang Y. et al. Bioleaching of copper from waste printed circuit boards by bacterial consortium enriched from acid mine drainage. Journal of hazardous materials 184, 812–818 (2010). [DOI] [PubMed] [Google Scholar]
- Hagelüken C. Recycling of electronic scrap at Umicore's integrated metals smelter and refinery. World of Metallurgy – ERZMETALL 59, 152–161 (2006). [Google Scholar]
- Sadowski P. G. et al. Quantitative proteomic approach to study subcellular localization of membrane proteins. Nat Protoc 1, 1778–1789 (2006). [DOI] [PubMed] [Google Scholar]
- Tan H. T. et al. Quantitative and temporal proteome analysis of butyrate-treated colorectal cancer cells. Molecular & cellular proteomics : MCP 7, 1174–1185 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information