Abstract
In recent years, the men who have sex with men (MSM) population has seen the fastest growing prevalence of HIV transmission in China. In addition, coinfection through sex and intravenous drug use is a major problem in HIV prevention and control. Recent studies have also revealed that three major viral strains (CRF07_BC, CRF01_AE, and subtype B) have been cocirculating among MSM in Sichuan, suggesting a high probability of generating new recombinants. This study reports a near full-length genome of a novel HIV-1 recombinant (MSM0720) between CRF07_BC and CRF01_AE. The analysis of MSM0720 shows that the genome is composed of at least 11 interlaced segments, including six CRF07_BC and five CRF01_AE segments, with CRF07_BC as the backbone; this is different from a previously identified CRF01_AE/07_BC recombinant strain in intravenous drug users from Jiangsu.
Since the first HIV/AIDS case was reported in China in 1989, an estimated 780,000 individuals have contracted HIV by the end of 2011.1 In recent years, one of the most at-risk populations for HIV infection in China are men who have sex with men (MSM), among whom the prevalence of HIV-1 has increased precipitously from 1.5% in 2006 to 10.8% in 2010 according to the National AIDS Case Reporting System.2 Multiple genotypes are involved in the HIV-1 epidemic (including A, B′, B, C, CRF01_AE, CRF02_AG, CRF07_BC, CRF08_BC, and other unique recombinant forms).3
Recombination between HIV-1 subtypes is a significant mechanism that contributes to the genetic complexity of HIV-1, and new strains are frequently found in regions and populations where multiple subtypes are circulating. One such region is Sichuan province in southwestern China. Sichuan has one of the highest HIV prevalence rates in the country, with CRF07_BC and CRF01_AE as the two major circulating strains among its large men who have sex with men (MSM) population.4 The coexistence of these two CRFs among MSM may facilitate the generation of new recombination forms. Here we identified and characterized a novel recombinant derived from CRF07_BC and CRF01_AE, which is highly different from a previously reported CRF01_AE/CRF07_BC recombinant from Jiangsu province.5
Sample MSM0720 was collected in 2007 from a male patient residing in Sichuan province who was diagnosed as HIV-1 positive in 2006 and died in 2008. Epidemiological survey data show that the patient was married, but had been infected with HIV through homosexual behavior. For the near full-length genome (NFLG) amplification and sequencing, RNA was extracted from the patient's blood plasma sample using a QIAamp Viral Mini Kit (QIAGEN, Germany). RNA was then transcribed into cDNA using a Superscript III First-strand synthesis system (Invitrogen, USA). With the near-endpoint diluted cDNA template, the NFLGs were amplified with TaKaRa LA Taq (TaKaRa, Dalian, China) using the same nested polymerase chain reaction (PCR) amplification conditions in both of the two rounds, including an initial denaturation at 94°C for 2 min followed by 10 cycles of 94°C for 10 s and 68°C for 8 min 30 s, and then 20 cycles of 94°C for 10 s and 68°C for 8 min 30 s with the cumulative addition of 20 s at 68°C with each successive cycle, followed by a final extension at 68°C for 20 min.6 The positive PCR products were purified using a QIAquick Gel Extraction Kit (QIAGEN, Germany) and sequenced by an ABI 3730XL sequencer using BigDye terminators (Applied Biosystems, Foster City, CA). The chromatogram data were cleaned and assembled using Sequencher v4.9 (Gene Codes, Ann Arbor, MI). A BLAST search was performed for the sequences using all sequences obtained in this laboratory as background, to check consistency with previous results and to identify and exclude possible cross-contamination events.
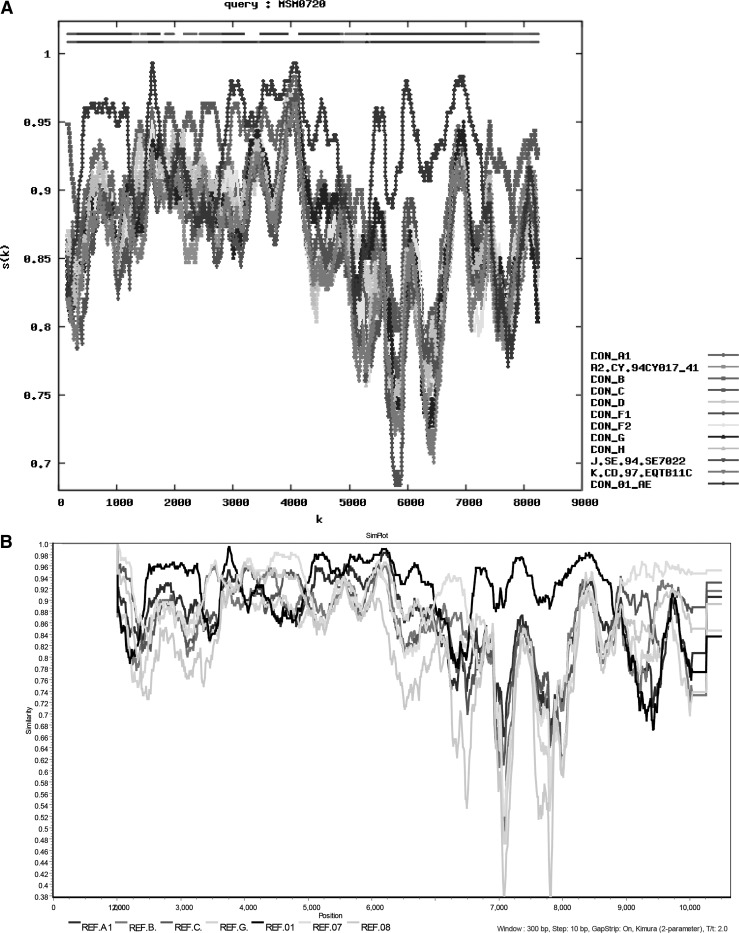
The standard subtype reference alignment file, which includes subtype A1, A2, B, C, D, F, G, H, J, P, CRF01_AE, CRF07_BC, and CRF08_BC, was downloaded from the Los Alamos National Laboratory HIV Database (www.hiv.lanl.gov/content/sequence/NEWALIGN/align.html#ref). Nucleotide sequences were first aligned with the HXB2 standard reference strain using Clustal W, merged into the reference file, and adjusted manually using BioEdit (version 7.1).7 To analyze the recombination form of the MSM0720 strain, the sequence was submitted to RIP (www.hiv.lanl.gov/content/sequence/RIP/RIP.html) using all default settings except the window size of 300. The result revealed a recombination form consisting mostly of subtype A1, B, C, and CRF01_AE (Fig. 1A). Similarity plot analysis was performed with SimPlot (version 3.5.1)8 using the reference file mentioned above. The result shows that the genome was most likely comprised of CRF07_BC and CRF01_AE (Fig. 1B). Bootscan analysis was subsequently carried out using the same software to position the recombination breakpoint. The query sequence MSM0720 was bootscanned with reference sequences (CRF01_AE and CRF07_BC as parents, subtype G as outgroup). The parameters were a window size of 300, a step size of 10, and the neighbor-joining method using the Kimura two-parameter model with replicates of 100. Phylogenetic trees were constructed with MEGA 5.0 using the neighbor-joining method with 1,000 bootstrap replications to confirm the subtype of the mosaic fragments.9
FIG. 1.
Recombinant Identification Program (RIP) and similarity plots analysis were performed for MSM0720 to identify parental subtypes. (A) Similarity distance analysis was performed using RIP from the Los Alamos National Laboratory HIV Database with default setting except for the window size of 300. (B) Similarity plots for MSM0720 using SimPlot software, with subtype reference strains A1, B, C, D, 01_AE, 07_BC, and 08_BC. These reference strains were verified above by RIP, which may contain the components of MSM0720.
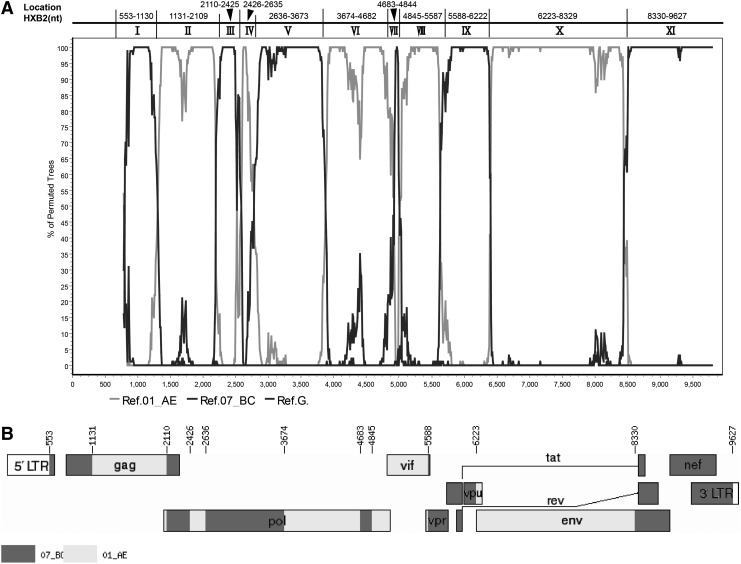
MSM0720 contained at least 11 interlaced mosaic segments corresponding to CRF01_AE (II, IV, VI, VIII, and X) and CRF07_BC (I, III, V, VII, IX, and XI) (Fig. 2A). Five mosaic CRF01_AE segments and four CRF07_BC segments (I, V, IX, and XI) were well confirmed by phylogenetic trees with bootstrap value >0.7 (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/aid). Segment III corresponds to a subtype B segment in CRF07_BC. Although the bootstrap value of Segment III is 0.68 with CRF07_BC, it is also found in one cluster containing both reference sequences subtype B and CRF07_BC with a high bootstrap value of 0.98. The phylogenetic tree for Segment VII is unstable; this may be due to the shortness of the fragment (16 2bp) as well as the very high degrees of conservation within the pol gene, which together cannot provide enough information to distinguish differences between subtypes. However, MSM0720 is inclined to cluster with CRF07_BC.
FIG. 2.
Bootscan plots and schematic representation of the MSM0720 near full-length genome's mosaic structure. (A) Bootscan plots of MSM0720 using CRF01_AE, CRF07_BC, and subtype G as subtype references. The parameters of SimPlot bootscan analysis were set as default except for a window size of 300 and step size of 10. The breakpoint positions refer to HXB2 coordinates located by the HIV Sequence Locator (www.hiv.lanl.gov/content/sequence/LOCATE/locate.html). (B) Genomic map of MSM0720. The mosaic map was generated using the Recombinant HIV-1 Drawing Tool (www.hiv.lanl.gov/content/sequence/DRAW_CRF/recom_mapper.html).
The breakpoints positions refer to the HXB2 coordinates, and were located by the HIV Sequence Locator (www.hiv.lanl.gov/content/sequence/LOCATE/locate.html). Therefore, the map of the new recombinant virus MSM0720 is as follows: 07_BC (553–1,130 nt), 01_AE (1,131–2,109 nt), 07_BC (2,110–2,425 nt), 01_AE (2,426–2,635 nt), 07_BC (2,636–3,673 nt), 01_AE(3,674–4,682 nt), 07_BC (4,683–4,844 nt), 01_AE (4,845–5,587 nt), 07_BC (5,588–6,222 nt), 01_AE (6,223–8,329 nt), and 07_BC (8,330–9,627 nt) (Fig. 2B).
CRF01_AE and CRF07_BC are two major HIV-1 circulating strains in Sichuan. Among MSM, the proportion of CRF01_AE and CRF07_BC is 54.5% and 31.2%, respectively.4 In China, CRF01_AE is historically mainly associated with sexual transmission, while CRF07_BC accounted for a larger proportion among intravenous drug users (IDUs). The emergence of the 07_BC/01_AE recombinant strain in MSM indicates that there is cross-transmission between IDUs and those mainly at risk through sexual contact. Regarding this study, the patient who provided this sample has been married to a female partner for several decades. In China, due to a high degree of stigma and social pressure, a large number of MSM do not disclose their sexual orientation and have multiple sex partners, including female marital partners. Such situations facilitate the transmission of HIV-1 and increase the difficulty of HIV/AIDS prevention and control work. In addition, demographic data from the National AIDS Direct Reporting System show that HIV-1 infection in MSM is mainly found in young adults, approximately 22.5% of whom are college students.2 Among these, 11.9% are detected through prescreening during voluntary blood donations.2 As young students are currently the main donors of blood in China, this may pose challenges to blood safety.
The first CRF01_AE/CRF07_BC recombinant form in China was isolated from IDUs from Jiangsu province.5 Here we characterize a new unique NFLG sequence isolated from MSM in Sichuan province, consisting of different recombinant breakpoints between the CRF07_BC and CRF01_AE strains. The emergence of such recombinants demonstrates increasing cross-transmission between previously segregated risk populations, and could further complicate the HIV-1 epidemic in the MSM population in Sichuan.
Sequence Data
The nucleotide sequence of MSM0720 has been submitted to GenBank with the accession number KC833436.
Supplementary Material
Acknowledgments
This work was supported by National Major Projects for Infectious Diseases Control and Prevention (grant 2012ZX10001008, 2012ZX10001002), the National Natural Science Foundation of China (grant 81020108030), and the State Key Laboratory for Infectious Disease Development (grant 2012SKLID103).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Ministry of Health of China. UNAIDS, WHO: The estimation of HIV/AIDS in China in 2011. Ministry of Health of China; Beijing: 2011. [Google Scholar]
- 2.Hei FX. Wang L. Qin QQ, et al. Epidemic characteristics of HIV/AIDS among men who have sex with men from 2006 to 2010 in China. Zhonghua Liu Xing Bing Xue Za Zhi. 2012;33(1):67–70. [PubMed] [Google Scholar]
- 3.Kijak GH. McCutchan FE. HIV diversity, molecular epidemiology, and the role of recombination. Curr Infect Dis Rep. 2005;7(6):480–488. doi: 10.1007/s11908-005-0051-8. [DOI] [PubMed] [Google Scholar]
- 4.Yuan D. Feng L. Xi J. Moleculo-epidemiological study of HIV-1/AIDS among 77 men having sex with men in Sichuan province. Chinese J STD/AIDS Prev Cont. 2011;17(4):410–413. [Google Scholar]
- 5.Guo H. Guo D. Wei JF, et al. First detection of a novel HIV Type 1 CRF01_AE/07_BC recombinant among an epidemiologically linked cohort of IDUs in Jiangsu, China. AIDS Res Hum Retroviruses. 2009;25(4):463–467. doi: 10.1089/aid.2008.0250. [DOI] [PubMed] [Google Scholar]
- 6.Rousseau CM. Birditt BA. McKay AR, et al. Large-scale amplification, cloning and sequencing of near full-length HIV-1 subtype C genomes. J Virol Methods. 2006;136(1–2):118–125. doi: 10.1016/j.jviromet.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 7.Hall TA. BioEdit: A user-friendly biological sequence alignment editor, analysis program for Windows 95/98/NT. Paper presented at the Nucleic Acids Symposium Series. 1999.
- 8.Lole KS. Bollinger RC. Paranjape RS, et al. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 1999;73(1):152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tamura K. Dudley J. Nei M. Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.