Abstract
The G-protein coupled C-X-C chemokine receptor type 4 (CXCR4) is highly overexpressed in a range of cancers and is therefore an excellent biomarker for cancer imaging. To this end targeted iron oxide nanoparticles were developed and utilised for in vitro imaging of MDA-MB-231 breast cancer cells overexpressing the CXCR4 receptor. Nanoparticles comprising an iron oxide core, encapsulated in a stabilising epichlorohydrin crossed-linked dextran polymer, were conjugated to a cyclopentapeptide with affinity to the CXCR4 receptor. The particles were characterized for their size, surface charge and r2 relaxivity at 4.7 T. MR imaging of the CXCR4 receptor with targeted iron oxide nanoparticles revealed an approximately 3-fold increase in T2 signal enhancement of MDA-MB-231 cells compared to non-targeted controls. Prussian blue staining of labeled MDA-MB-231 cells revealed darker and more intense staining of the cellular membrane. This study demonstrates the potential of targeted iron oxide nanoparticles for the imaging of the CXCR4 receptor by magnetic resonance imaging (MRI).
Keywords: CXCR4, iron oxide nanoparticles, tumor MRI, targeted nanoparticles, T2 imaging, cyclopentapeptide
Introduction
Superparamagnetic iron oxide nanoparticles (IONPs) are used in MRI due to their biocompatible nature and strong effect on T2 relaxation. IONPs are commonly composed of a monocrystalline iron oxide core with a polysaccharide coating such as dextran [1-3]. Clinical IONPs are non-targeted and are generally used to image the blood pool, macrophage activity and sites of macrophage infiltration such as the liver, spleen and lymph nodes [4,5]. IONPs have been used for in vivo imaging, in particular for stem cell tracking, as excellent signal-to-noise ratios are obtainable [6,7]. Another attractive feature of IONPs is the relative ease of modifying their surface chemistry and thus the possibility of targeting specific biological targets by functionalization with targeting ligands [8-13]. In this study, we created a CXCR4 receptor targeted IONP by conjugating a cyclic pentapeptide moiety to epichlorohydrin cross-linked dextran coated IONPs and used these particles to detect MDA-MB-231 cells overexpressing the CXCR4 receptor.
The chemokine receptor CXCR4 [14] is a seven trans-membrane G-protein coupled receptor that is overexpressed in numerous types of cancers [15], including breast, brain [16], ovarian, pancreatic and prostate [17]. It has been shown to be involved in the metastasis of cancers such as breast, kidney, prostate, lung, pancreas, melanoma, neuroblastoma, non-Hodgkin’s lymphoma, multiple myeloma, ovarian cancer and malignant gliomas [15]. The interaction of the CXCR4 receptor with its natural ligand, stromal cell-derived factor 1α (SDF-1α) is critical for tumor development, growth and metastasis [18]. SDF-1α is the only major ligand of the CXCR4 chemokine receptor thus making this receptor-ligand pair an attractive target for imaging studies [15]. The receptor’s role in breast cancer and metastasis is particularly significant [19]. Metastases express high levels of CXCR4; it has therefore been suggested that CXCR4 levels could be predictive of metastatic potential, particularly in breast cancers [18,19]. A number of studies have explored CXCR4 as a biomarker target over the past decade. Various radionuclides have been used to label CXCR4’s natural ligand SDF-1α [20], as well as anti-CXCR4 antibodies [16], peptide-based inhibitors [21-25] and small molecule inhibitors of CXCR4 [26-28] for nuclear imaging by single photon emission computed tomography (SPECT) and positron emission tomography (PET). Fluorescence and bioluminescence imaging probes have been developed using SDF-1α [29,30], various peptides [31,32], as well as small molecule inhibitors [33,34]. Recent studies have experimented with more innovative imaging concepts, using metal nanoshells [35] and bimodal (fluorescence and SPECT) imaging agents for the detection of CXCR4 [36]. The advances in the field are summarized in two recent reviews by Knight et al [37] and van Leeuwen et al [38].
Since the discovery of the CXCR4 receptor and its role in HIV infection and cancer, several antagonists to the receptor have been identified and developed. One of the most potent compounds identified is cyclo-[Nal1-Gly2-(D-Tyr3)-Arg4-Arg5] with an IC50 of 4 nM, which is a cyclic peptide commonly known as FC131 (Figure 1A) [39]. Based on the structure of FC131, an ornithine substituted cyclic pentapeptide was developed [40], which possesses CXCR4 antagonist activity and provides an amine group for functionalization and subsequent bioconjugation with imaging probes.
Figure 1.
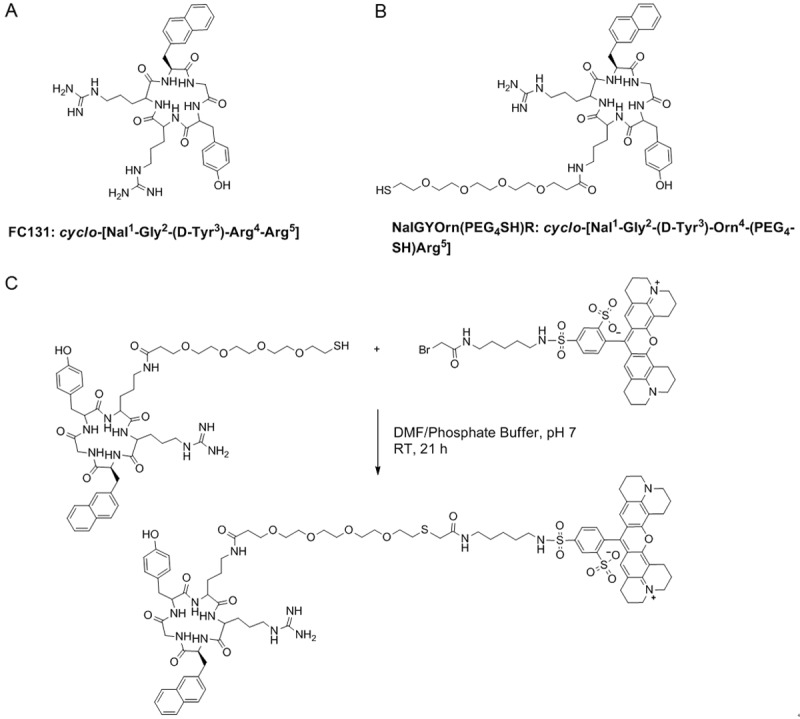
Structures of the original cyclopentapeptide CXCR4 antagonist FC131 (A), novel analogue with ornithine substitution and polyethylene glycol (PEG)-thiol linker (B) and synthesis of CXCR4 targeted fluorescent probe NalGYOrn(PEG4SH)R-TxR; (DMF/Phosphate buffer (pH 7), room temperature, 21 h) followed by semi-preparative HPLC purification (C).
In the first instance we conjugated the peptide cyclo-[Nal1-Gly2-(D-Tyr3)-Orn4-(PEG4-SH)-Arg5], (NalGYOrn(PEG4SH)R, Figure 1B) to Texas Red and assessed the binding of the fluorescent probe to MDA-MB-231 cells. MDA-MB-231 cells have been shown to overexpress CXCR4 [41,42]. Subsequently the NalGYOrn(PEG4SH)R peptide was conjugated to IONPs for MR imaging of the CXCR4 receptor in vitro.
He et al have recently reported the development of anti-CXCR4 monoclonal antibody-labeled ultrasmall superparamagnetic iron oxide (USPIO) nanoparticles [43]. To the best of our knowledge, our research herein represents the first attempt to label an iron oxide-based MRI contrast agent with a small molecule CXCR4 specific targeting ligand.
Materials and methods
General methods
The NalGYOrn(PEG4SH)R peptide was obtained from Cambridge Research Chemicals (UK), Texas red bromoacetamide was purchased from Invitrogen (UK) and all other chemicals and reagents were purchased from Sigma-Aldrich (UK). High performance liquid chromatography (HPLC) studies were conducted on a Waters system with the following modules: Waters 600 Controller module, a Waters 2487 Dual λ Absorbance Detector and a Waters Fraction Collector using Empower Software for analysis. MRI experiments were conducted on a 4.7 T Magnex magnet (Oxford, UK) Varian Unity Inova console (Palo Alto, CA).
Synthesis of NalGYOrn(PEG4SH)R-TxR peptide probe
NalGYOrn(PEG4SH)R (0.6 mg, 0.616 μmol) was dissolved in phosphate buffer (500 μL, K2HPO4 10 mM, pH 7.0), to which was added Texas red bromoacetamide in dimethylformamide (DMF) (800 μL). The reaction was stirred for 21 h at room temperature. The mixture was then purified by semi-preparative HPLC on a Waters system with absorbance detection at 230 and 320 nm. A Waters Bondapak C18 column (7.8 × 300 mm) was used; tR: 19 min, gradient mixture: 0-15 min; 10/90 MeOH/H2O, 15-30 min; 75/25 MeOH/H2O, 30-45 min; 82/18 MeOH/H2O. HRMS: (TOF-MS ES+) calcd for C84H107N13O18S3, 1681.70 found 1683.7 (M+H)+.
In vitro incubation assay with NalGYOrn(PEG4SH)R-TxR
MDA-MB-231 (ATCC, USA) cells were cultured in L-15 growth medium (ATCC, USA) supplemented with 10% Fetal Calf Serum (FCS), 2% Penicillin/Streptomycin (Sigma) solution and 0.2% fungicide and incubated at 37°C in a 5% CO2/95% air atmosphere. NalGYOrn(PEG4SH)R-TxR was dissolved in dimethyl sulfoxide (DMSO) to give a 2 mg/mL stock solution. Stock solution was diluted to give desired concentrations in L-15 growth medium. Cells were seeded (1.2 × 103 cells/well) 24 hours prior to experiment in 100 μL growth medium. The peptide conjugate was then added to each well at the appropriate dose. Cells were incubated for 60 min in a 5% CO2/95% air atmosphere. The media was removed and the cells washed twice using ice-cold water. Phosphate-buffered saline (PBS) (150 μl) was added to each well and a fluorescent signal detected using a fluorescent plate reader (Victor) fitted with a Texas Red filter.
In vitro time course incubation assay with NalGYOrn(PEG4SH)R-TxR
MDA-MB-231 cells were cultured as described above, the cells were trypsinized (1 mM ethylenediaminetetraacetic acid (EDTA)), washed and centrifuged (2000 rpm, 3 min); cells were re-suspended in PBS, and centrifuged again (× 2). Cells were counted using Trypan blue staining and 2 × 105 cells were added to each Eppendorf containing 1000 nM of NalGYOrn(PEG4SH)R-TxR in PBS. Eppendorf tubes were incubated for 5, 30 or 60 min in a 5% CO2/95% air atmosphere at 37°C. The reaction was stopped by adding 500 μL ice-cold PBS and keeping the Eppendorf tubes on ice. Samples were then spun down (13000 rpm, 4°C, 5 min) and supernatant removed using a fine glass needle under vacuum. Cell pellets were then washed using 500 μL ice-cold PBS and samples further spun down (6,500 rpm, 4°C, 5 min). Supernatant was removed and cells re-suspended in 150 μL PBS. Cell suspensions were transferred to 96 well plates and fluorescence detected using a Victor fluorescence plate reader fitted with a Texas Red filter.
Fluorescence-activated cell sorting (FACS) analysis of MDA-MD-231 cells labeled with NalGYOrn(PEG4SH)R-TxR
NalGYOrn(PEG4SH)R-TxR was dissolved in DMSO to give 2 mg/mL stock solution. Stock solution was diluted to give 100 nM and 1000 nM peptide conjugate in PBS. Eppendorf tubes were prepared with peptide conjugate diluted to give a final volume of 500 μL/tube. MDA-MB-231 cells were cultured as mentioned previously, washed, trypsanized (1 mM EDTA), centrifuged (2000 rpm, 3 min), and washed with PBS (× 2). The cells were then re-suspended in PBS and counted using Trypan blue staining. 100 μL of cell suspension at the desired concentration (optimized at 2 × 105 cells/Eppendorf tube) was added to each Eppendorf tube. Eppendorf tubes were incubated for 60 min in a 5% CO2/95% air atmosphere at 37°C. The reaction was stopped by adding 500 μL ice-cold PBS and keeping the Eppendorf tubes on ice. Samples were then spun down (13,000 rpm, 4°C, 5 min) and supernatant removed using a fine glass needle under vacuum. Cell pellets were washed using 500 μL ice-cold PBS and samples spun down (6,500 rpm, 4°C, 5 min). Supernatant was removed and cells re-suspended in PBS (1 mL) and transferred to FACS tubes. Unlabeled cells were used as a standard. Samples were kept on ice post incubation and analyzed using a BD LSR2 FACS analysis machine. Live cells were gated using front and side scatter parameters. Fluorescence was measured using a 610/620 YG-A filter. Results were analyzed using FlowJo 8.8.6 software.
Fluorescence microscopy of MDA-MB-231 cells labeled with NalGYOrn(PEG4SH)R-TxR
MDA-MB-231 cells were seeded on glass chambers (6 × 104 cells/plate) in 500 μL L-15 growth medium (ATCC) supplemented with 10% Fetal Calf Serum (FCS), 2% Penicillin/Streptomycin solution and 0.2% fungicide (Sigma), and incubated 24 h prior to experiment at 37°C in a 5% CO2/95% air atmosphere. Cells were washed with media and PBS (250 μL) and incubated with NalGYOrn(PEG4SH)R-TxR at 10000 nM diluted growth medium for 60 min at 37°C in a 5% CO2/95% air atmosphere. Cells were washed (L-15 media, 1 mL × 2 and PBS, 1 mL × 2) and incubated with 4% paraformaldehyde for 20 min at 37°C. Cells were then washed (PBS, 1 mL × 2) and treated with 4’,6-diamidino-2-phenylindole (DAPI) containing fixing media. Images were obtained using an Olympus fluorescence microscope fitted with charge coupled device (CCD). Pictures were taken using Texas red and DAPI filters. Texas red and DAPI images were merged using ImageMerger software.
Ferrozine assay
Iron content of IONPs was determined using a previously published protocol [44]. The ferrozine reagent was made by dissolving ferrozine (51.4 mg) and hydroxylamine hydrochloride (1 g) in a minimal amount of water, followed by the addition of concentrated (37%) hydrochloric acid (5 mL) and diluting the solution to a final volume of 10 mL using distilled water. An ammonium buffer was made by dissolving ammonium acetate (10 g) in ammonium hydroxide (8.75 mL) and diluting with distilled water to give a final volume of 25 mL. Hydrochloric acid was used to titrate the buffer to pH 5.5. Ferrozine (100 μL) reagent was added to iron oxide nanoparticle sample (50 μL) in an Eppendorf tube. The tube was heated using an oil bath at 60°C for 1 h. Samples were then treated with ammonium buffer (200 μL) and left for full color development (30 min). Samples were diluted to a final volume of 2 mL using distilled water for UV-Vis spectrometry (Perkin-Elmer Lambda 2 UV-Vis spectrometer). Endorem® (Guerbet, France) was used as a standard to produce a calibration curve for comparison.
Synthesis of iodo-functionalized IONPs
Succinimide iodoacetate (8.49 mg, 0.03 mmol) was dissolved in DMSO (30 μL) and added to amine-functionalized iron oxide nanoparticles (total volume 3 mL, 1.5 mg/mL) in HEPES buffer (0.01 M pH 8.5) and the reaction mixture stirred at room temperature for 1 h. The sample was dialyzed using a spectropore dialysis membrane (MWCO 3000, SpectrumLabs; Rancho Dominguez, CA, USA) in HEPES buffer (0.01 M pH 7.0) overnight to remove unbound iodoacetate.
Conjugation of NalGYOrn(PEG4SH)R to iodo-IONPs
NalGYOrn(PEG4SH) (31.40 mg, 0.033 mmol) was dissolved in DMSO (70 μL) and added to succinimide iodoacetate-functionalized iron oxide nanoparticles (IONP) (4.5 mg) in 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) buffer (7 mL, 0.01 M pH 7.0). The reaction mixture was stirred gently for 21 h and thereafter purified by dialysis using a spectropore dialysis membrane (MWCO 3000, SpectrumLabs; Rancho Dominguez, CA, USA) in HEPES buffer (0.01 M pH 7.0) overnight in order to remove unbound material.
Prussian blue staining of MDA-MB-231’s incubated with NalGYOrn(PEG4SH)R-IONPs
MDA-MB-231 cells were seeded on glass plates 24 h prior to experiment and cultured in L-15 growth medium (ATCC) supplemented with 10% Fetal Calf Serum (FCS), 2% Penicillin/Streptomycin solution and 0.2% fungicide (Sigma) and incubated at 37°C in a 5% CO2/95% air atmosphere. NalGYOrn(PEG4SH)R-IONPs, in addition to non-targeted IONPs (NH2-IONPs) and Endorem® were diluted with growth media (250 μl) to obtain a final [Fe] of 0.1 mg/mL. The cells were then incubated for 60 min at 37°C in a 5% CO2/95% air atmosphere and washed with media (1 mL, × 2) and PBS (1 mL, × 2) and fixed with 4% paraformaldehyde (20 min at 37°C). The slides were then washed with PBS (1 mL, × 2) and fixed with mounting media containing DAPI reagent (ProLong antifade reagent with DAPI, Invitrogen, UK). Microscopy images were obtained using an Olympus fluorescence microscope fitted with CCD and analysis software.
MRI relaxivity measurements
NalGYOrn(PEG4SH)R-IONPs and Endorem® were diluted in HEPES (0.01 M, pH 7.0) to obtain 4 samples with varying amounts of Fe content ([Fe]: 0.24 mg/mL 0.12 mg/mL; 0.06 mg/mL; 0.03 mg/mL). Samples were then added to Eppendorf tubes (200 μL) and placed in a quadrature 1H coil and measurements taken at room temperature. MRI experiments were carried out using a 4.7 T Magnex magnet (Oxford, UK) Varian Unity Inova console (Palo Alto, CA, USA). T2 values were obtained using saturation recovery experiments performed with a standard spin-echo sequence and a 9 mm single slice acquisition (TR = 3000 ms, TE values = 11.18, 15, 30, 60 and 120 ms), slice thickness: 9 mm, number of signal averages: 10, FOV: 100 × 50 mm2, collected into a matrix of 256 × 128. MR images were analyzed using ImageJ (National Institutes of Health, USA), with a region of interest (ROI) drawn around the sample area. The T2 values were calculated by plotting the mean signal intensity (Y) against the ROI taken at each TR and fitted to the equation: Y = M0*(exp(-x/T2)) using GraphPad Prism software (San Diego, USA). The r2 relaxivity was obtained from the linear fit of 1/T2 vs [Fe] mM [45].
MRI analysis of MDA-MB-231 cells incubated with NalGYOrn(PEG4SH)R-IONPs
MDA-MB-231 cells were seeded in T25 flasks (7.5 × 104 cells/flask) and incubated at 37°C for 24 hours in a 5% CO2/95% air atmosphere. NalGYOrn(PEG4SH)R-IONPs, non-targeted IONPs and Endorem® were diluted in HEPES buffer (0.01 M, pH 7.0) to give [Fe] = 0.2 mg/mL. Cells were then incubated with 500 μL nanoparticle solution and 4500 μL growth medium to give a final concentration of 0.02 mg/mL. Cells were incubated for 240 min at 37°C in a 5% CO2/95% air atmosphere. After incubation cells were washed with PBS (× 3) and detached using trypsin (500 μL). Trypsin was neutralized using 1 mL media and cells were centrifuged (13,000 rpm, 5 min). Media was removed and cells resuspended in 200 μL PBS. Samples were transferred to 200 μL Eppendorf tubes and cells were centrifuged again (2500 rpm, 5 min) in PBS (200 μL) and were placed in a quadrature 1H coil and measurements taken at room temperature. MRI experiments were carried out as described above.
Results
In vitro evaluation of NalGYOrn(PEG4SH)R-TxR
The cyclopentapeptide NalGYOrn(PEG4-SH)R bearing a short PEG linker and reactive thiol functionality was coupled to Texas red bromoacetamide and dissolved in DMF to obtain NalGYOrn(PEG4SH)R-TxR (Figure 1C). The coupling reaction was carried out in phosphate buffer and followed by HPLC purification. In order to assess the potential of the peptide conjugate to bind CXCR4, various concentrations of the synthesized fluorescent peptide conjugate NalGYOrn(PEG4SH)R-TxR were incubated with CXCR4 overexpressing MDA-MB-231 cells for 60 min at 37°C (Figure 2A). The cells were then washed to remove any unbound probe and their fluorescence detected. Fluorescence signal was shown to increase three-fold between probe concentrations of 100 nM and 1000 nM and increase another three-fold between probe concentrations of 1000 nM and 10000 nM, while no appreciable signal was detected at concentrations of 1 nM and 10 nM.
Figure 2.
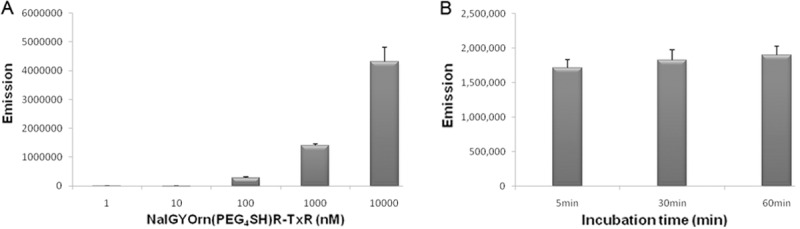
Incubation of MDA-MB-231 cells with NalGYOrn(PEG4SH)R-TxR at 1 nM, 10 nM, 100 nM, 1000 nM and 10000 nM at 37°C for 60 min, followed by analysis of fluorescence intensity using a plate reader (A), Incubation of MDA-MB-231 cells with 1000 nM of NalGYOrn(PEG4SH)R-TxR at 37°C, followed by analysis of fluorescence intensity at 5 min, 30 min and 60 min (B).
The fluorescence readings measured from the MDA-MB-231 cells post incubation with the NalGYOrn(PEG4SH)R-TxR probe were also analyzed at various time points leading up to 60 min (Figure 2B). Comparable cellular fluorescence was detected at just 5 min and also at 60 min incubation suggesting rapid saturation-of-binding post incubation with NalGYOrn(PEG4SH)R-TxR.
The binding properties of the fluorescent conjugated peptide were further assessed by flow cytometry experiments, where a concentration-dependent fluorescence shift from the MDA-MB-231 cells was observed following labeling with NalGYOrn(PEG4SH)R-TxR (Figure 3A).
Figure 3.
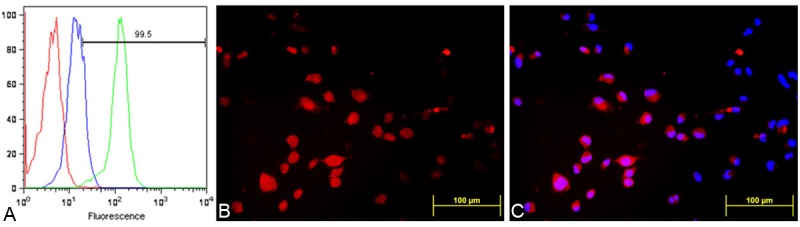
FACS analysis of MDA-MB-231 cells alone (red), incubated with a 100 nM (blue) and 1000 nM (green) dose of NalGYOrn(PEG4SH)R-TxR for 60 min at 37°C (A), fluorescence microscopy images of MDA-MB-231 cells after incubation with NalGYOrn(PEG4SH)R-TxR (1000 nM, 60 min, 37°C) (B), co-localization with DAPI (C).
Cellular localization of the conjugate MDA-MB-231 cells was investigated by fluorescence microscopy imaging; in these experiments, cells were incubated with 1000 nM of NalGYOrn(PEG4SH)R-TxR for 60 min at 37°C and then fixed and imaged. The fluorescence microscopy studies confirmed NalGYOrn(PEG4SH)R-TxR retention by MDA-MB-231 cells (Figure 3B and 3C) with a ubiquitous level of fluorescence observed throughout the cells indicating internalization of the peptide conjugate.
To establish MRI capability of the cyclic pentapeptide, NalGYOrn(PEG4SH)R (Figure 1B) was additionally conjugated to the surface of IONPs for MR imaging of the CXCR4 receptor.
NalGYOrn(PEG4SH)R conjugation to IONPs
IONPs with terminal amines (NH2-IONPs) were prepared using the method described by Palmicci et al (72). The iron and amine content of the nanoparticles was determined using ferrozine and fluorescamine assays respectively [44,47]. Conjugation of the peptide to the surface of the IONPs was carried out as described previously [48]. Briefly, terminal amines on the surface of the IONPs were chemically modified to produce iodo-iron oxide nanoparticles (I-IONPs) using succinimide iodoacetate under basic conditions. The NalGYOrn(PEG4SH)R peptide was subsequently coupled to I-IONPs at neutral pH in (Figure 4). HEPES buffer yielding NalGYOrn(PEG4SH)R-IONPs. The IONPs were purified using Spectropore dialysis membranes (MWCO 3000) to remove unbound peptide. IONPs were analyzed for their size and zetapotential following each modification procedure (Table 1) with NalGYOrn(PEG4SH)R-IONPs diameter measuring ~55 nm. The size and zetapotential of the NalGYOrn(PEG4SH)R-IONPs increased due to aggregation effects induced by the peptide conjugation and the presence of the guanidinium group of arginine respectively. The guanidinium group (pKa 12.48) is positively charged under neutral conditions making the zetapotential of the IONPs more basic.
Figure 4.
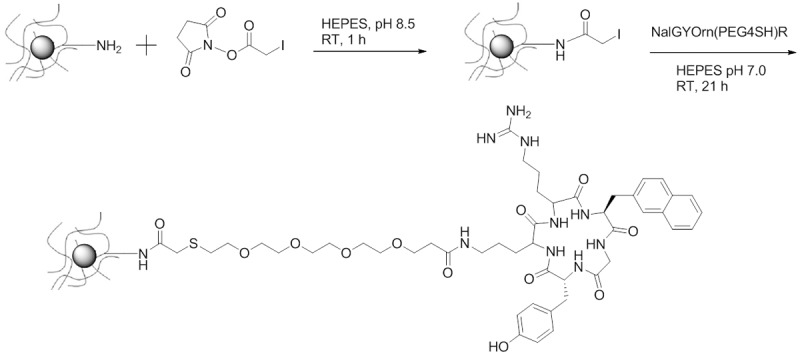
Scheme of NalGYOrn(PEG4SH)R peptide conjugation to IONPs; Step 1: succinimide iodoacetate, amine functionalized IONPs, HEPES buffer (pH 8.5), room temperature, 1 hour; Step 2: NalGYOrn(PEG4SH)R, SIA-functionalized IONPs, HEPES buffer (pH 7), room temperature, 21 hours; Purification by dialysis.
Table 1.
Iron oxide nanoparticles (IONPs) and physical characteristics
| Nanoparticle | Buffer | Size (nm) | Zeta Potential (mV) |
|---|---|---|---|
| NH2-IONPs | HEPES, 0.01 M, pH 8.5 | 39.64 | -0.75 |
| I-IONPs | HEPES, 0.01 M, pH 7.0 | 21.45 | 0.29 |
| NalGYOrn(PEG4SH)R-IONPs | HEPES, 0.01 M, pH 7.0 | 55.49 | 8.06 |
The concentration of peptide bound to the surface of the particles was determined using UV-Vis spectroscopy and the concentration of peptides was determined to be 6.0 mM, corresponding to approximately 90% peptide conjugation to the IONPs.
In vitro MRI of CXCR4 with targeted IONPs
To assess cellular binding of the NalGYOrn(PEG4SH)R peptide conjugated IONPs (NalGYOrn(PEG4SH)R-IONPs), MDA-MB-231 cells were incubated with nanoparticles for 60 min at 37°C. To compare specificity of binding, CXCR4 overexpressing MDA-MB-231 cells were incubated with NalGYOrn(PEG4SH)R-IONPs, non-peptide conjugated particles (IONPs) or the clinically used iron oxide nanoparticle Endorem® (total [Fe]: 0.1 mg/mL). To visualize the nanoparticles, cells were then incubated with a Prussian blue reagent and cell nuclei were counterstained with DAPI (Figure 5). Darker staining was observed from cells stained with NalGYOrn(PEG4SH)R-IONPs, which revealed dark blue rings around the cellular membrane, confirming NalGYOrn(PEG4SH)R-IONPs targeting to MDA-MB-231 cells. Minimal staining was observed for the non-targeted IONPs and Endorem controls (Figure 5B and 5D). Cell surface binding of the targeted IONPs to the cells and lack of intracellular uptake of the particles suggest/confirm that the peptide acts as an antagonist of the CXCR4 receptor. Therefore, the internalization of nanoparticles does not occur to any appreciable extent within the 60 min incubation period.
Figure 5.
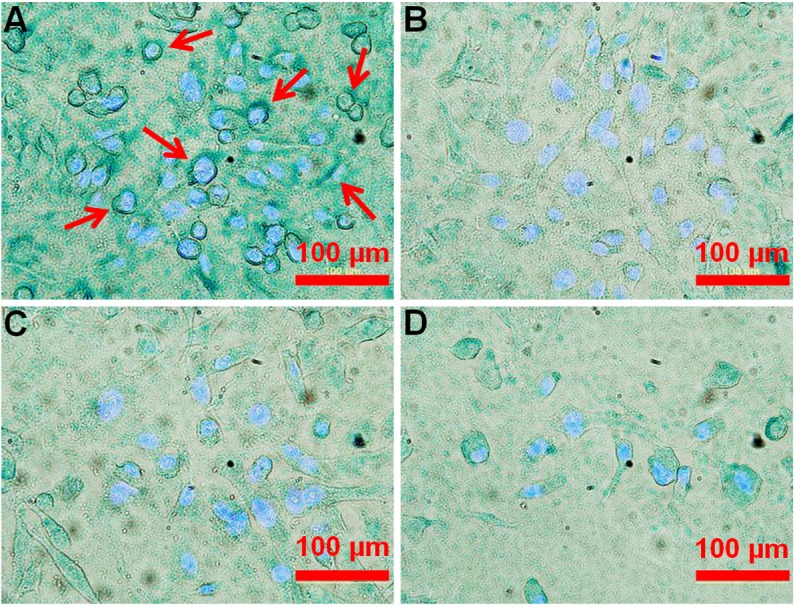
Bright field images of MDA-MB-231 cells incubated with NalGYOrn(PEG4SH)R-IONPs (A), IONPs (B), Endorem® (C) ([Fe]=0.1 mg/ml, 60 min, 37°C) and cells alone (D); Iron was stained using Prussian Blue, cell nuclei were stained using DAPI, white arrows point to cell membrane stained areas.
The MRI efficacy of the synthesized NalGYOrn(PEG4SH)R-IONPs was investigated by measuring r2 relaxivity at 4.7 T and ambient temperature. Both NalGYOrn(PEG4SH)R-IONPs and non-targeted IONPs were incubated with CXCR4 overexpressing MDA-MB-231 cells in order to assess their MR signal enhancing potential. The particles were incubated with the cells for 4 h at 37°C at a total [Fe] 0.01 mg/mL. Following this incubation period, the cells were washed, trypsinized, pelleted in PBS and imaged at 4.7 T. The r2 relaxivity of NalGYOrn(PEG4SH)R-IONPs was measured to be 3.66 mM-1s-1 which is comparable to other commercially available T2 contrast agents.
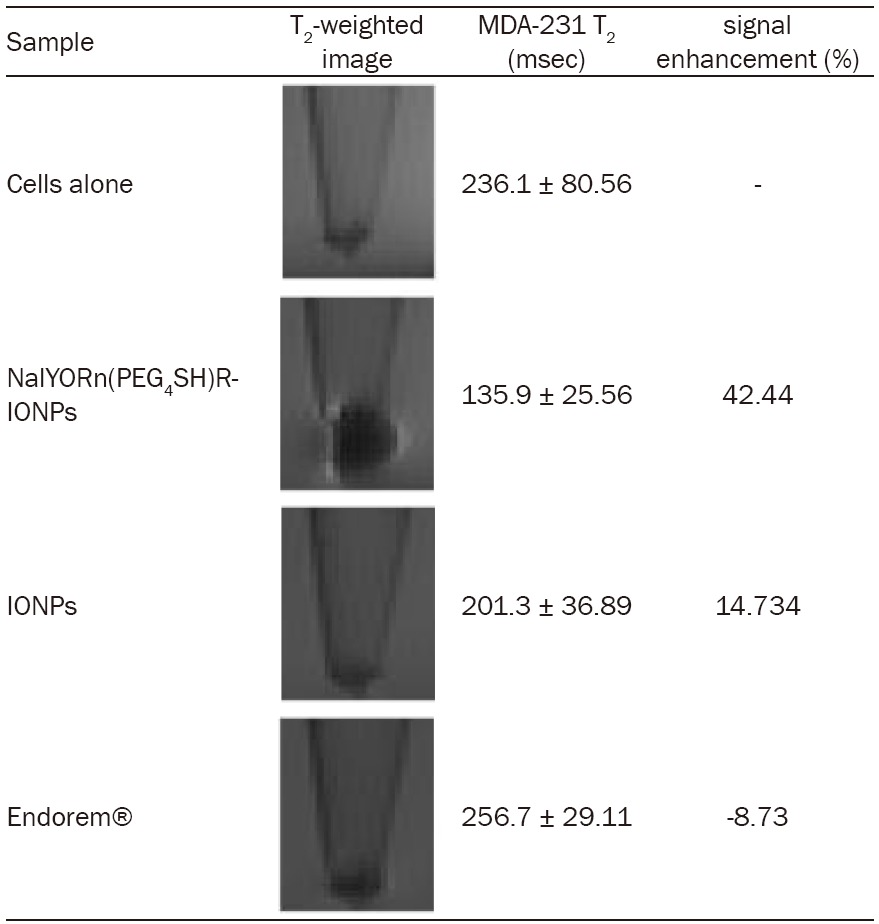
MRI relaxation data obtained using region of interest quantification showed approximately 3-fold increase in T2 signal from MDA-MB-231 cells labeled with NalGYOrn(PEG4SH)R-IONPs in comparison to control non-targeted IONPs (Table 2). Additionally, Endorem® did not reduce T2 signal post incubation with MDA-MB-231 cells suggesting no binding or uptake of this non-targeted agent with these cells.
Table 2.
Measured MRI data and images obtained at 4.7 T; MDA-MB-231 cells incubated with NalGYOrn(PEG4SH)R-IONPs, IONPs and Endorem® ([Fe]=0.02 mg/ml, 240 min, 37°C)
|
Discussion
CXCR4 expression has been found to be an important biomarker for several cancers [49]; and CXCR4 up-regulation has been implicated in metastases and tumor invasion [19]. Therefore CXCR4 targeted imaging agents can be valuable for both the detection of primary tumors and secondary tumors arising as a result of metastasis.
In order to establish the targeting capability of the selected NalGYOrn(PEG4SH)R for binding to the CXCR4 receptor, the peptide was conjugated to Texas Red to produce a NalGYOrn(PEG4SH)R-TxR probe for fluorescence imaging. MDA-MB-231 cell associated fluorescence was measured post incubation with the fluorescent probe in a concentration dependent manner (Figure 2A). Interestingly, the time course incubation assay did not reveal a significant increase in the level of uptake of the probe from 5 min to 60 min suggesting rapid saturation of binding sites (Figure 2B). To assess binding of the peptide conjugate to CXCR4 cells, flow cytometry experiments were carried out and a concentration-dependent increase in fluorescence signal was observed (Figure 3A). Fluorescence microscopy studies of MDA-MB-231 cells labeled with the peptide conjugate revealed a uniform level of fluorescence visible in the cell cytoplasm, indicating the likely internalization of the probe into cells. It was postulated that intracellular fluorescence should not be observable as the targeting peptide is an antagonist of the CXCR4 receptor. It is likely, however, that the observed internalization is due to the lipophilic nature of the Texas Red conjugate. Although fluorescent probe conjugation to active targeting ligands is a relatively cheap and facile method of establishing specificity, it is important to note that cellular internalization due to the lipophilicity of aromatic structures in fluorophores via diffusion or endocytosis is a limiting factor. It is possible that active transport mechanisms lead to the internalization of the peptide conjugate as other fluorescent CXCR4 receptor antagonists bearing a similar fluorophore have also been shown to become internalized into cells as a result of active transport mechanisms [33].
Incubation with targeted nanoparticles, on the other hand, showed distinct cell surface association of the IONPs on MDA-MB-231 cells. For these studies, amino-coated IONPs were synthesized and characterized for their charge and size. Incubation of MDA-MB-231 cells with NalGYOrn(PEG4SH)R-IONPs led to the appearance of intense blue rings around the Prussian blue stained cells (Figure 5). This intense staining was not visible in cells incubated with non-targeted IONPs or Endorem®. These images confirm the cell surface binding of the targeted nanoparticles. Non-targeted nanoparticles below 60 nm in diameter have been shown to enter cells through receptor-mediated endocytosis [50], with ~50 nm being an optimal nanoparticle size for cellular entry [51], however, we do not observe this behavior with NalGYOrn(PEG4SH)R-IONPs suggesting “antagonistic” binding to the cell surface membrane as a dominant binding mechanism.
MRI of MDA-MB-231 cells incubated with NalGYOrn(PEG4SH)R-IONPs, control non-targeted-IONPs, and Endorem® was carried out and T2 measurements obtained at 4.7 T (Table 2). A hypointense signal was observed from the cell pellet region post incubation with NalGYOrn(PEG4SH)R-IONPs, where the susceptibility artifact of the superparamagnetic IONPs was also apparent (Table 2). The measured T2 % signal enhancement was up to 3-fold higher from MDA-MB-231 cells labeled with the targeted nanoparticles than with non-targeted-IONPs or Endorem®. The measured T2 signal from non-targeted IONPs was lower than from Endorem®. This is likely to be due to the fact that the control IONPs, although non-targeted, have a surface decorated with amino groups which may become positively charged in cell culture media leading to attractive forces with the negatively charged phosphate anions of the cell surface membrane. The T2 signal intensity of cells labeled with Endorem® did not decrease because the surface properties of particles in this formulation are neutral, deterring interactions with the cell membrane.
Further work to establish the cytotoxicity of the targeted-IONPs on MDA-MB-231 and other cell lines overexpressing CXCR4 is currently underway, furthermore we aim to establish the utility of NalGYOrn(PEG4SH)R-IONPs for MR imaging of CXCR4 overexpressing tumors in vivo.
Conclusion
In summary, we have synthesized a novel MRI contrast agent for the imaging of the CXCR4 receptor. Using an in vitro model based on the MDA-MB-231 cell line, we have demonstrated the feasibility of using targeted-fluorescent or -IONPs for this purpose. The majority of commonly used superparamagnetic IONPs in the clinic are non-targeted agents. Targeted imaging of specific receptors on cancer cells allows for more effective detection of cancer.
Acknowledgements
The authors thank Dr. William Jones for helpful discussions. We acknowledge support by staff at the Biological Imaging Centre at Imperial College London. Funding for this project was provided from CRUK, EPSRC, MRC and the Department of Health Centre grant (C2536/A10337). NJL would like to thank the Leverhulme Trust for a Research Fellowship.
References
- 1.Mahmoudi M, Sant S, Wang B, Laurent S, Sen T. Superparamagnetic iron oxide nanoparticles (SPIONs): Development, surface modification and applications in chemotherapy. Adv Drug Deliv Rev. 2011;63:24–46. doi: 10.1016/j.addr.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 2.Dias A, Hussain A, Marcos A, Roque A. A biotechnological perspective on the application of iron oxide magnetic colloids modified with polysaccharides. Biotechnol Adv. 2011;29:142–155. doi: 10.1016/j.biotechadv.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Ling D, Hyeon T. Chemical Design of Biocompatible Iron Oxide Nanoparticles for Medical Applications. Small. 2013;9:1450–1466. doi: 10.1002/smll.201202111. [DOI] [PubMed] [Google Scholar]
- 4.Heredia V, Altun E, Bilaj F, Ramalho M, Hyslop B, Semelka R. Gadolinium- and superparamagnetic-iron-oxide-enhanced MR findings of intrapancreatic accessory spleen in five patients. Magn Reson Imaging. 2008;26:1273–1278. doi: 10.1016/j.mri.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 5.Te Boekhorst BC, van Tilborg GA, Strijkers GJ, Nicolay K. Molecular MRI of inflammation in atherosclerosis. Curr Cardiovasc Imaging Rep. 2012;5:60–68. doi: 10.1007/s12410-011-9114-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chapon C, Jackson J, Aboagye E, Herlihy A, Jones W, Bhakoo K. An In Vivo Multimodal Imaging Study Using MRI and PET of Stem Cell Transplantation after Myocardial Infarction in Rats. Mol Imaging Biol. 2009;11:31–38. doi: 10.1007/s11307-008-0174-z. [DOI] [PubMed] [Google Scholar]
- 7.Bulte JW, Duncan ID, Frank JA. In vivo magnetic resonance tracking of magnetically labeled cells after transplantation. J Cereb Blood Flow Metab. 2002;22:899–907. doi: 10.1097/00004647-200208000-00001. [DOI] [PubMed] [Google Scholar]
- 8.Maeng J, Lee D, Jung K, Bae Y, Park I, Jeong S, Jeon Y, Shim C, Kim W, Kim J. Multifunctional doxorubicin loaded superparamagnetic iron oxide nanoparticles for chemotherapy and magnetic resonance imaging in liver cancer. Biomaterials. 2010;31:4995–5006. doi: 10.1016/j.biomaterials.2010.02.068. [DOI] [PubMed] [Google Scholar]
- 9.Rerat V, Laurent S, Burtea C, Driesschaert B, Pourcelle V, Vander L, Muller R, Marchand-Brynaert J. Ultrasmall particle of iron oxide-RGD peptidomimetic conjugate: synthesis and characterisation. Bioorg Med Chem Lett. 2010;20:1861–1865. doi: 10.1016/j.bmcl.2010.01.150. [DOI] [PubMed] [Google Scholar]
- 10.Chen B, Wu W, Wang X. Magnetic iron oxide nanoparticles for tumor-targeted therapy. Curr Cancer Drug Targets. 2011;11:184–9. doi: 10.2174/156800911794328475. [DOI] [PubMed] [Google Scholar]
- 11.Tassa C, Shaw S, Weissleder R. Dextran-Coated Iron Oxide Nanoparticles: A Versatile Platform for Targeted Molecular Imaging, Molecular Diagnostics, and Therapy. Acc Chem Res. 2011;44:842–852. doi: 10.1021/ar200084x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mout R, Moyano D, Rana S, Rotello V. Surface functionalization of nanoparticles for nanomedicine. Chem Soc Rev. 2012;41:2539–2544. doi: 10.1039/c2cs15294k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghosh D, Lee Y, Thomas S, Kohli A, Yun D, Belcher A, Kelly K. M13-templated magnetic nanoparticles for targeted in vivo imaging of prostate cancer. Nat Nanotechnol. 2012;7:677–682. doi: 10.1038/nnano.2012.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu B, Chien EY, Mol CD, Fenalti G, Liu W, Katritch V, Abagyan R, Brooun A, Wells P, Bi FC, Hamel DJ, Kuhn P, Handel TM, Cherezov V, Stevens RC. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science. 2010;330:1066–1071. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsutsumi H, Tanaka T, Ohashi N, Masuno H, Tamamura H, Hiramatsu K, Araki T, Ueda S, Oishi S, Fujii N. Therapeutic potential of the chemokine receptor CXCR4 antagonists as multifunctional agents. Biopolymers. 2007;88:279–289. doi: 10.1002/bip.20653. [DOI] [PubMed] [Google Scholar]
- 16.Nimmagadda S, Pullambhatla M, Pomper MG. Immunoimaging of CXCR4 Expression in Brain Tumor Xenografts Using SPECT/CT. J Nucl Med. 2009;50:1124–30. doi: 10.2967/jnumed.108.061325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh S, Singh U, Grizzle W, Lillard J. CXCL12-CXCR4 interactions modulate prostate cancer cell migration, metalloproteinase expression and invasion. Lab Invest. 2004;84:1666–1676. doi: 10.1038/labinvest.3700181. [DOI] [PubMed] [Google Scholar]
- 18.Tamamura H, Hori A, Kanzaki N, Hiramatsu K, Mizumoto M, Nakashima H, Yamamoto N, Otaka A, Fujii N. CXCR4 antagonists identified as anti-cancer-metastatic agents. Pept Sci. 2004;2003:65–68. doi: 10.1016/s0014-5793(03)00824-x. [DOI] [PubMed] [Google Scholar]
- 19.Smith M, Luker K, Garbow J, Prior J, Jackson E, Piwnica-Worms D, Luker G. CXCR4 regulates growth of both primary and metastatic breast cancer. Cancer Res. 2004;64:8604–8612. doi: 10.1158/0008-5472.CAN-04-1844. [DOI] [PubMed] [Google Scholar]
- 20.Misra P, Lebeche D, Ly H, Schwarzkopf M, Diaz G, Hajjar R, Schecter A, Frangioni J. Quantitation of CXCR4 expression in myocardial infarction using 99mTc-labeled SDF-1α. J Nucl Med. 2008;49:963–969. doi: 10.2967/jnumed.107.050054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirofumi H, Mukai T, Tamamura H, Mori T, Ishino S, Ogawa K, Iida Y, Doi R, Fujii N, Saji H. Development of a 111In-labeled peptide derivative targeting a chemokine receptor, CXCR4, for imaging tumors. Nucl Med Biol. 2006;33:489–494. doi: 10.1016/j.nucmedbio.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 22.Jacobson O, Weiss I, Kiesewetter D, Farber J, Chen X. PET of tumor CXCR4 expression with 4-18F-T140. J Nucl Med. 2010;51:1796–1804. doi: 10.2967/jnumed.110.079418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gourni E, Demmer O, Schottelius M, D’Alessandria C, Schulz S, Dijkgraaf I, Schumacher U, Schwaiger M, Kessler H, Wester HJ. PET of CXCR4 expression by a 68Ga-labeled highly specific targeted contrast agent. J Nucl Med. 2011;52:1803–1810. doi: 10.2967/jnumed.111.098798. [DOI] [PubMed] [Google Scholar]
- 24.Demmer O, Dijkgraaf I, Schumacher U, Marinelli L, Cosconati S, Gourni E, Wester HJ, Kessler H. Design, synthesis, and functionalization of dimeric peptides targeting chemokine receptor CXCR4. J Med Chem. 2011;54:7648–7662. doi: 10.1021/jm2009716. [DOI] [PubMed] [Google Scholar]
- 25.Åberg O, Pisaneschi F, Smith G, Nguyen QD, Stevens E, Aboagye EO. 18F-Labelling of a Cyclic Pentapeptide Inhibitor of the Chemokine Receptor CXCR4. J Fluorine Chem. 2012;135:200–206. [Google Scholar]
- 26.Nimmagadda S, Pullambhatla M, Stone K, Green G, Bhujwalla Z, Pomper M. Molecular imaging of CXCR4 receptor expression in human cancer xenografts with [64Cu] AMD3100 positron emission tomography. Cancer Res. 2010;70:3935–3944. doi: 10.1158/0008-5472.CAN-09-4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weiss I, Jacobson O, Kiesewetter D, Jacobus J, Szajek L, Chen X, Farber J. Positron emission tomography imaging of tumors expressing the human chemokine receptor CXCR4 in mice with the use of 64Cu-AMD3100. Mol Imaging Biol. 2012;14:106–114. doi: 10.1007/s11307-010-0466-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Silva R, Peyre K, Pullambhatla M, Fox J, Pomper M, Nimmagadda S. Imaging CXCR4 expression in human cancer xenografts: evaluation of monocyclam 64Cu-AMD3465. J Nucl Med. 2011;52:986–993. doi: 10.2967/jnumed.110.085613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kollet O, Dar A, Shivtiel S, Kalinkovich A, Lapid K, Sztainberg Y, Tesio M, Samstein R, Goichberg P, Spiegel A, Elson A, Lapidot T. Osteoclasts degrade endosteal components and promote mobilization of hematopoietic progenitor cells. Nat Med. 2006;12:657–664. doi: 10.1038/nm1417. [DOI] [PubMed] [Google Scholar]
- 30.Meincke M, Tiwari S, Hattermann K, Kalthoff H, Mentlein R. Near-infrared molecular imaging of tumors via chemokine receptors CXCR4 and CXCR7. Clin Exp Metastasis. 2011;28:713–720. doi: 10.1007/s10585-011-9403-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oishi S, Masuda R, Evans B, Ueda S, Goto Y, Ohno H, Hirasawa A, Tsujimoto G, Wang Z, Peiper SC, Naito T, Kodama E, Matsuoka M, Fujii N. Synthesis and Application of Fluorescein-and Biotin labeled Molecular Probes for the Chemokine Receptor CXCR4. Chembiochem. 2008;9:1154–8. doi: 10.1002/cbic.200700761. [DOI] [PubMed] [Google Scholar]
- 32.Kuil J, Steunenberg P, Chin PT, Oldenburg J, Jalink K, Velders AH, van Leeuwen FW. Peptide Functionalized Luminescent Iridium Complexes for Lifetime Imaging of CXCR4 Expression. Chembiochem. 2011;12:1897–903. doi: 10.1002/cbic.201100271. [DOI] [PubMed] [Google Scholar]
- 33.Khan A, Silversides J, Madden L, Greenman J, Archibald S. Fluorescent CXCR4 chemokine receptor antagonists: metal activated binding. Chem Comm. 2007;4:416–418. doi: 10.1039/b614557d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knight J, Hallett A, Brancale A, Paisey SJ, Clarkson RW, Edwards PG. Evaluation of a Fluorescent Derivative of AMD3100 and its Interaction with the CXCR4 Chemokine Receptor. Chembiochem. 2011;12:2692–8. doi: 10.1002/cbic.201100441. [DOI] [PubMed] [Google Scholar]
- 35.Zhang J, Fu Y, Li G, Zhao R, Lakowicz J. Detection of CXCR4 receptors on cell surface using a fluorescent metal nanoshell. J Biomed Opt. 2011;16:016011. doi: 10.1117/1.3528623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuil J, Buckle T, Yuan H, van den Berg NS, Oishi S, Fujii N, Josephson L, van Leeuwen FW. Synthesis and evaluation of a bimodal CXCR4 antagonistic peptide. Bioconjug Chem. 2011;22:859–64. doi: 10.1021/bc2000947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knight J, Wuest F. Nuclear (PET/SPECT) and optical imaging probes targeting the CXCR4 chemokine receptor. Med Chem Comm. 2012;3:1039–1053. [Google Scholar]
- 38.Kuil J, Buckle T, van Leeuwen F. Imaging agents for the chemokine receptor 4 (CXCR4) Chem Soc Rev. 2012;41:5239–5261. doi: 10.1039/c2cs35085h. [DOI] [PubMed] [Google Scholar]
- 39.Fujii N, Oishi S, Hiramatsu K, Araki T, Ueda S, Tamamura H, Otaka A, Kusano S, Terakubo S, Nakashima H, Broach JA, Trent JO, Wang ZX, Peiper SC . Molecular-Size Reduction of a Potent CXCR4-Chemokine Antagonist Using Orthogonal Combination of Conformation- and Sequence-Based Libraries. Angew Chem Int Ed Engl. 2003;42:3251–3. doi: 10.1002/anie.200351024. [DOI] [PubMed] [Google Scholar]
- 40.Wester H, Koglin N, Schwaiger M, Kessle H, Laufer B, Demmer O, Anton M, inventors. Cancer imaging and treatment. WO 2007/096662 A2 . 2007
- 41.Huang E, Singh B, Cristofanilli M, Gelovani J, Wie C, Vincent L, Cook K, Lucci A. A CXCR4 antagonist CTCE-9908 inhibits primary tumor growth and metastasis of breast cancer. J Surg Res. 2009;155:231–236. doi: 10.1016/j.jss.2008.06.044. [DOI] [PubMed] [Google Scholar]
- 42.Hamaguchi T, Wakabayashi H, Matsumine A, Sudo A, Uchida A. TNF inhibitor suppresses bone metastasis in a breast cancer cell line. Biochem Biophys Res Comm. 2011;407:525–530. doi: 10.1016/j.bbrc.2011.03.051. [DOI] [PubMed] [Google Scholar]
- 43.He Y, Song W, Lei J, Li Z, Cao J, Huang S, Meng J, Xu H, Jin Z, Xue H. Anti-CXCR4 monoclonal antibody conjugated to ultrasmall superparamagnetic iron oxide nanoparticles in an application of MR molecular imaging of pancreatic cancer cell lines. Acta Radiol. 2012;53:1049–1058. doi: 10.1258/ar.2012.120055. [DOI] [PubMed] [Google Scholar]
- 44.Stookey LL. Ferrozine - a New Spectrophotometric Reagent for Iron. Anal Chem. 1970;42:779–781. [Google Scholar]
- 45.Bartolome R, Ferreiro S, Miquilena-Colina M, Martinez-Prats L, Soto-Montenegro M, Garcia-Bernal D, Vaquero J, Agami R, Delgado R, Desco M. The chemokine receptor CXCR4 and the metalloproteinase MT1-MMP are mutually required during melanoma metastasis to lungs. Am J Pathol. 2009;174:602–612. doi: 10.2353/ajpath.2009.080636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Palmacci S, inventor. Synthesis of polysaccharide covered superparamagnetic oxide colloids. 5,262,176 US Patent.
- 47.Bantan-Polak T, Kassai M, Grant K. A Comparison of Fluorescamine and Naphthalene-2,3-dicarboxaldehyde Fluorogenic Reagents for Microplate-Based Detection of Amino Acids. Anal Biochem. 2001;297:128–136. doi: 10.1006/abio.2001.5338. [DOI] [PubMed] [Google Scholar]
- 48.Pittet M, Swirski F, Reynolds F, Josephson L, Weissleder R. Labeling of immune cells for in vivo imaging using magnetofluorescent nano-particles. Nat Protoc. 2006;1:73–79. doi: 10.1038/nprot.2006.11. [DOI] [PubMed] [Google Scholar]
- 49.Darash-Yahana M, Pikarsky E, Abramovitch R, Zeira E, Pal B, Karplus R, Beider K, Avniel S, Kasem S, Galun E, Peled A. Role of high expression levels of CXCR4 in tumor growth, vascularization, and metastasis. FASEB J. 2004;18:1240–2. doi: 10.1096/fj.03-0935fje. [DOI] [PubMed] [Google Scholar]
- 50.Zhang S, Li J, Lykotrafitis G, Bao G, Suresh S. Size-Dependent Endocytosis of Nanoparticles. Adv Mater. 2009;21:419–424. doi: 10.1002/adma.200801393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang W, Kim B, Rutka J, Chan W. Nanoparticle-mediated cellular response is size-dependent. Nat Nanotechnol. 2008;3:145–150. doi: 10.1038/nnano.2008.30. [DOI] [PubMed] [Google Scholar]


