Abstract
The spatial localization and molecular distribution of metabolites and neurotransmitters within biological organisms is of tremendous interest to neuroscientists. In comparison to conventional imaging techniques such as immunohistochemistry, matrix-assisted laser desorption/ionization (MALDI) mass spectrometric imaging (MSI) has demonstrated its unique advantage by directly localizing the distribution of a wide range of biomolecules simultaneously from a tissue specimen. Although MALDI-MSI of metabolites and neurotransmitters is hindered by numerous matrix-derived peaks, high-resolution and high-accuracy mass spectrometers (HRMS) allow differentiation of endogenous analytes from matrix peaks, unambiguously obtaining biomolecular distributions. In this study, we present MSI of metabolites and neurotransmitters in rodent and crustacean central nervous systems acquired on HRMS. Results were compared with those obtained from a medium-resolution mass spectrometer (MRMS), tandem time-of-flight instrument, to demonstrate the power and unique advantages of HRMSI and reveal how this new tool would benefit molecular imaging applications in neuroscience.
Keywords: Mass spectrometric imaging, high spectral resolution, metabolites, neurotransmitters, central nervous system, matrix-assisted laser desorption/ionization
Metabolites are the end products of regulatory processes in cells and could directly reveal the physiological states of cells. Therefore, large scale analyses of metabolites have been used to discriminate cell populations of different physiological conditions, such as diseased vs healthy,1 young vs old,2 and normal vs stressed,3 to potentially identify specific biomarkers.4 Currently, changes in metabolic profiles have been clinically utilized to characterize ∼200 inherited enzymatic disorders.4 Neurotransmitters (NTs) are a class of endogenous small molecules that transmit intercellular signals from a neuron to a target cell at chemical synapses.5 Neurotransmitters include amino acids such as serine and glycine, monoamines such as dopamine, and other classical NTs such as acetylcholine (ACh).6 Abnormal neurotransmitter expression levels have also been associated with various diseases. For example, decreased synthesis of ACh has been proposed as the cause of Alzheimer’s disease (AD),7 and the loss of dopaminergic neurons is implicated in Parkinson’s disease.8 Studies of metabolites and NTs could not only deepen our understanding of related pathogenic mechanisms but could also lead to potential therapeutic treatments.
Examination of metabolites and NTs from cells and tissues of homogenized extracts can be performed via well-established gas chromatography (GC) or liquid chromatography (LC) mass spectrometry (MS) methods.9 Unfortunately, these techniques do not account for the spatial distributions of endogenous metabolites and NTs. Therefore, a complementary approach that directly characterizes and localizes metabolites and NTs from tissues is needed.
Previously, metabolite and NT imaging was typically achieved by imaging antibodies against the targets’ conjugates or against the enzymes involved in synthesis or degradation of the low molecular weight (MW) targets. One example is immunohistochemical (IHC) imaging of ACh by localizing the ACh-synthesizing enzyme choline acetyltransferase.10 Although high-resolution images of low MW targets can be provided via IHC, several drawbacks exist: sample preparation is tedious, a limited number of targets can be imaged simultaneously, highly specific antibodies are difficult to obtain, and discovery of unknown molecules is impossible because molecule-specific antibodies are used.11 In contrast, mass spectrometric imaging (MSI) examines the biodistribution of multiple molecules without prior knowledge of their identities through direct detection from tissue samples.11,12 Its unique capability enables mapping of low-MW species with minimal sample preparation and high specificity, revolutionizing our understanding of biological processes by visualizing the dynamics of metabolites and NTs.13−17
MALDI is a sensitive, soft ionization technique capable of ionizing a wide range of molecules,11,18,19 and its recent application to drug and metabolite imaging has gained enormous attention.19−22 However, MALDI-MSI of metabolites and NTs present at physiological concentrations in tissues is often complicated by numerous high-abundance matrix-derived peaks. High selectivity is therefore essential to successfully implement MSI in the low-MW region. One effective approach to increase selectivity is to perform tandem MS on selected low-MW metabolites and NTs.23 Instead of monitoring intact molecules in MS mode, the distribution of target molecules is measured in tandem MS mode by generating ion density maps of fragment ions exclusively produced by the targeted precursor ions.23 However, this method requires one tandem MSI analysis for each target, meaning N times more sample material and acquisition time to map N ions, whereas HRMSI solves this problem without compromising throughput and sensitivity.17,24 The high resolution, accurate mass measurements provided by HRMS discriminate low-MW chemical noise from endogenous metabolites and NTs, thereby verifying MS assignments with high confidence and increasing the information obtained from a biologically complex specimen.16,17,24,25
In this study, we developed HRMSI applications to measure the localizations of metabolites and NTs in rodent and crustacean central nervous systems (CNS) using a MALDI LTQ Orbitrap XL mass spectrometer. A comparison between results from complex CNS specimens obtained on an HRMS LTQ Orbitrap XL and an MRMS, tandem time-of-flight (TOF/TOF), was also made. Although the MALDI-TOF/TOF instrument (error < 10 ppm and resolution of up to 20 000 for the peptide range) is the most widely used instrument for MSI applications, the high mass-accuracy (<3 ppm) and mass-resolution (of up to 130 000 @ m/z 400) of HRMS enables unambiguous, confident identification of a number of metabolites and neurotransmitters directly from biologically complex tissue samples.
Results and Discussion
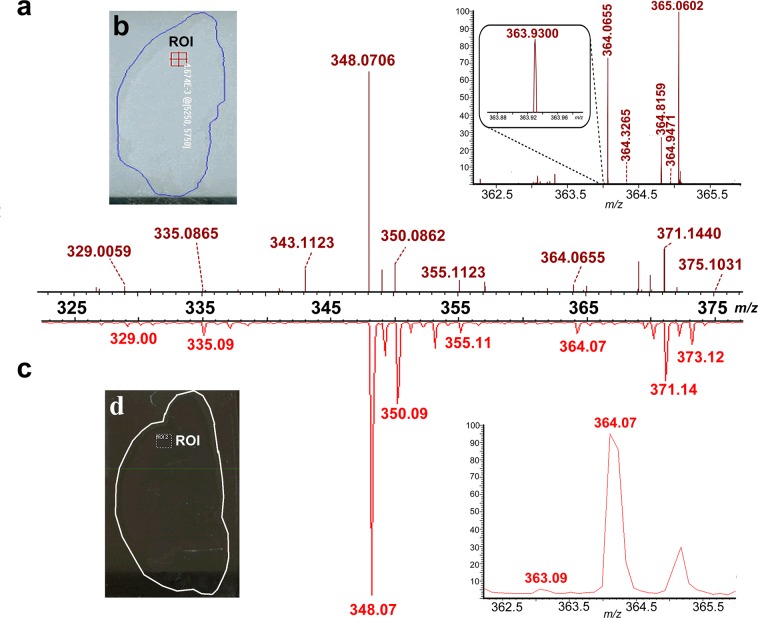
The difference in performance between HRMS and MRMS instrumentation is exhibited in Figure 1. In Figure 1a, the averaged mass spectrum acquired on the LTQ Orbitrap corresponds to the imaged region-of-interest (ROI) in rat brain cortex designated in Figure 1b. Figure 1c displays the averaged TOF/TOF spectrum of the ROI labeled in Figure 1d. Both ROIs are located in approximately the same regions of two adjacent sections, and the analytes desorbed/ionized from the two areas are assumed to produce similar mass spectra. The peaks present in the two representative spectra, Figure 1a and c, exhibited similar masses and relative intensities. However, the spectral resolution showed a clear distinction between the data generated by HRMS and MRMS. The insets display the explicit difference between the HRMS- and MRMS-produced spectra by enlarging the peak at m/z 364.07. The relatively low-abundance m/z 364.07 peak (at ∼3% of base peak height) had a high signal-to-noise ratio (S/N) in both zoomed-in spectra, indicating the high sensitivity of both instruments. Nevertheless, the ion obtained from the LTQ Orbitrap was detected at m/z 364.0655, whereas the ion detected by the TOF/TOF could only be assigned to m/z of 364.07 due to its limited resolution. The accurate, well-resolved peak in the LTQ Orbitrap spectrum contrasted the wide, asymmetrical peak at m/z 364.07 in the TOF/TOF spectrum and was confidently identified as guanosine monophosphate (GMP) with a mass error of 0.5 ppm. Additionally, peaks of small m/z differences at m/z 363.9300 or of low abundance compared to the base peak (Figure 1a inset at m/z 364.3265) were detected and well-resolved in the zoomed-in LTQ Orbitrap spectrum. Conversely, those peaks were merged in the shoulder of the m/z 364.07 peak in the enlarged TOF/TOF spectrum. This comparison demonstrates that HRMS can resolve nearly isobaric species and identify metabolites unequivocally from rat CNS specimen.
Figure 1.
Comparison between HRMS- and MRMS-produced spectra acquired from serial rat CNS sections. (a) HRMS-generated spectrum corresponding to the region of interest (ROI) localized in the cortex region of a rat brain section as highlighted in (b). The inset shows an enlarged m/z range of 362–366 obtained on a MALDI LTQ Orbitrap XL. (c) MRMS-generated mass spectrum corresponding to an identical ROI from the cortex region of a serial rat brain section as shown in (d). The inset shows an enlarged m/z range of 362–366 obtained on a MALDI-TOF/TOF.
More chemical information generated from HRMS is shown in Table 1, and the resulting images of the identified metabolites and NTs are shown in Figure 2. Since we measured analytes’ distribution as a proof-of-principle study to demonstrate the usefulness of HRMSI-based platform in imaging metabolites and NTs, we set a relatively large step-size to reduce the analysis time. Figure 2a shows an optical image of a rat brain section prepared for MSI experiments. Anatomical regions such as the olfactory bulb, cortex, medulla, and pons can be visualized clearly. The optical image was compared to MS images to interpret identified analytes’ localization. For example, Figure 2b displays the localization of choline (m/z at 104.1070) over almost the entire rat brain section. Choline’s ubiquitous distribution could be explained by its role as an essential nutrient reported in many plants and animal organs.26 It is also the precursor to a classic, synaptic neurotransmitter, ACh. Mapping ACh is of interest to numerous researchers due to its involvement in various normal and abnormal neuronal functions, association with learning, memory and diseases like AD.27 Conventional IHC methods studied the distributions of ACh-synthesizing enzyme choline acetyltransferase28 or ACh-metabolizing enzyme acetylcholinesterase29 to indirectly represent the localization of ACh. In this study, ACh was directly imaged from a rat brain section via single-stage MSI for the first time. As shown in Figure 2c, the relative abundance of the ACh peak was higher in the cortex and brainstem but lower in the olfactory bulb and cerebellum. The ACh localization image acquired via HRMSI correlated well with the distribution map of acetylcholinesterase from the Allen brain atlas (www.brain-map.org). Although this study attained low-spatial-resolution images due to the relatively large step-size (500 μm) during acquisition, high-spatial-resolution MS images of ACh can be obtained by implementing the HRMS-based platform with a highly focused laser beam and smaller step-size on tissue prepared with dryer matrix application method like sublimation. Previously, Sugiura et al. reported the application of tandem MSI to study the localization of ACh in mouse brain because of an unresolved, interfering matrix-related peak close to the m/z of ACh in MS using a MRMS MALDI-LTQ linear ion trap.23 The image of a signature ACh fragment ion instead of intact ACh was used to represent the distribution of ACh. Although their method successfully circumvented matrix interferences and specifically imaged ACh from tissue samples, its throughput was compromised because only one precursor ion could be selected for monitoring in a single experiment. Moreover, tandem MSI results are based on the assumption that the production of fragment ions can be correlated to the abundance of the precursor ion. However, this could be complicated due to factors like instrument instability and sample inhomogeneity. Additionally, the low intensity of the precursors might result in poor fragmentation, reducing the sensitivity of MSI for selected molecules. In contrast, the HRMSI-based approach is simple, relatively fast, and it eliminates limitations resulting from the reliance on tandem MS events.
Table 1. Metabolites and NTs Identified from the Rat CNS Specimen.
| name | formula | theoretical monoisotopic m/z | Orbitrap XL measurement m/z | Δm (ppm) |
|---|---|---|---|---|
| choline | C5H14NO | 104.1070 | 104.1070 | 0 |
| acetylcholine | C7H16NO2 | 146.1176 | 146.1175 | –0.7 |
| AMP | C10H15N5O7P | 348.0704 | 348.0706 | 0.6 |
| GMP | C10H15N5O8P | 364.0653 | 364.0655 | 0.5 |
| cholestrol | C27H45 | 369.3516 | 369.3519 | 0.8 |
| nicotinamide adenine dinucleotide | C21H28N7O14P2 | 664.1164 | 664.1177 | 2.0 |
| PC(32:0) + K | C40H80NO8PK | 772.5253 | 772.5258 | 0.6 |
Figure 2.
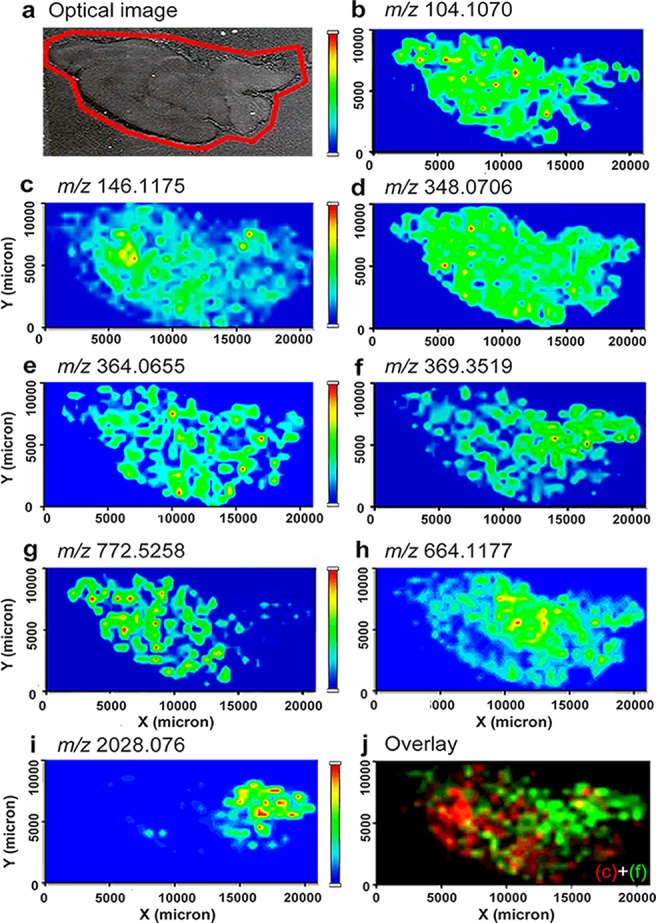
MS images of metabolites and NTs identified from rat CNS. (a) Optical image of a rat brain section subjected to MSI sample preparation. (b–g) MS image of metabolites and NTs, including (b) choline at m/z 104.1070, (c) ACh at m/z 146.1175, (d) AMP at m/z 348.0706, (e) GMP at m/z 364.0655, (f) cholesterol with neutral loss at m/z 369.3519, (g) potassiated PC(32:0) at m/z 772.5258, and (h) nicotinamide adenine dinucleotide at m/z 664.1177. Other than low MW molecules, (i) MS image of the acetylated peptide ASQKRPSQRHGSKYLATA at m/z 2028.076 was also shown. (j) Overlaid image of a NT, ACh, as in (c) and cholesterol-derived ion as in (f).
Figure 2d,e shows the distribution of two nucleotides, adenosine monophosphate (AMP) (m/z 348.0706) and GMP (m/z 364.0655), from the same rat brain section. AMP and GMP are both ubiquitously distributed on the rat brain with a higher abundance in the cortex. To further confirm the HRMSI results, we performed tandem MSI on the TOF/TOF by selecting m/z 364.07 as the precursor ion and monitoring the distribution of its major fragment ion at m/z 135.03. Supporting Information (SI) Figure 1a depicts the fragmentation pathway of this transition. The tandem MS image shown in SI Figure 1c confirms that the fragment ion is distributed throughout the brain region with a higher abundance in the cortex when correlated with the optical image in SI Figure 1b. SI Figure 1d corresponds to the image of an interfering ion produced simultaneously during fragmentation and is assigned as matrix-derived due to its distribution outside the tissue section. The difference in origin is shown in SI Figure 1e. Other than primary metabolites, lipid distributions are of interest to researchers because they have been reported to regulate a variety of biological functions in the CNS, including energy storage, membrane composition, and chemical signaling.25,30,31 In Figure 2f, the ion at m/z 369.3519 was identified as cholesterol with a neutral loss of water, agreeing with the previous observation of an m/z 369.3 peak by Jackson et al. using a MRMS MALDI-TOF.32 The abundance of the cholesterol peak was higher in brain stem regions such as the medulla, pons, and midbrain, as shown in Figure 2f. This result agrees with previous work showing that cholesterol is more concentrated in areas that are rich in myelinated axons.33 Figure 2g displays the localization of a potassiated lipid phosphotidylcholine (PC) (32:0) at m/z 772.5258. Although both of the molecules are lipids, they belong to different lipid families and exhibited distinct distributions, indicating their different biological roles involved in rat CNS functioning. Figure 2h shows the distribution of a coenzyme extensively used in cells’ redox reactions, nicotinamide adenine dinucleotide, at m/z 664.1177. This coenzyme is distributed differently than other metabolites shown in Figure 2 by its elevated abundance in the diencephalon and decreased expression levels in the forebrain. HRMSI also enables direct mapping of peptide neurotransmitters in the CNS and makes it possible to investigate the colocalization of low-MW and peptide NTs. Figure 2i displays the MS image of a truncated peptide of myelin basic protein detected at m/z 2028.076, which shares a similar distribution pattern with cholesterol in myelin-rich regions. Figure 2j is an overlay of Figure 2c and f, showing the distinct distribution of these two biomolecules (ACh and cholesterol-derived ions) from a single rat brain section. These representative images demonstrate the ability of HRMSI to map metabolites and NTs, and investigate their biological interactions through spatial correlation. It is worth mentioning that the HRMS LTQ Orbitrap is also capable of tandem MSI compared to TOF/TOF, where it outperforms TOF/TOF by providing better precursor ion isolation and enabling isolation of multiple precursor ions in a single experiment. A multiplexing imaging approach has been developed to obtain untargeted and targeted imaging data simultaneously.17,24 By incorporating both full MS scan and multiple tandem MS scans as one experiment, one could maximize the information gained from precious sample material with minimal acquisition time.17,24
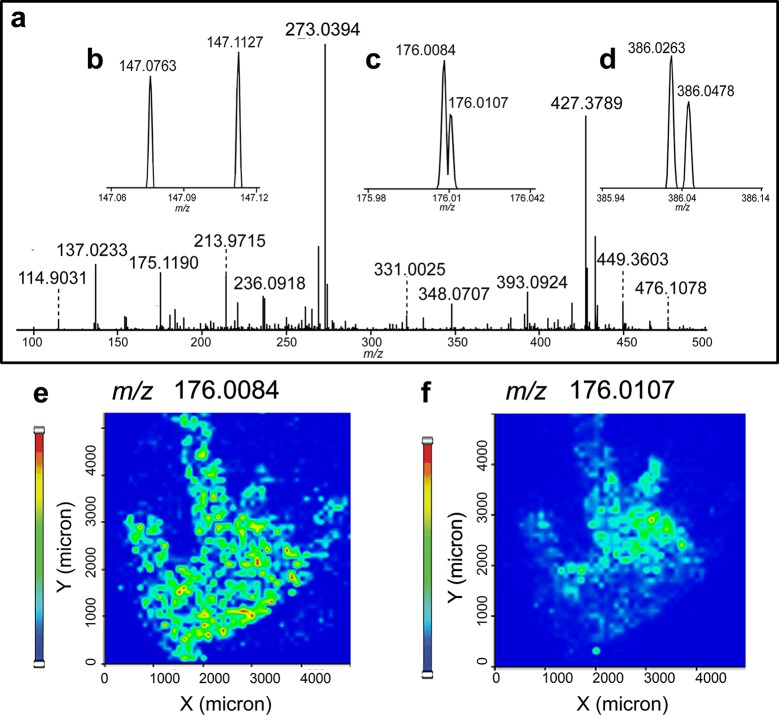
In order to further validate the HRMSI platform, we used it to map metabolites from another CNS model, blue crab C. sapidus brain, for the first time. Figure 3 shows the HRMS spectra of metabolites recorded from the crab brain. Figure 3a shows the overall HRMS spectrum averaged across the whole crab brain section, and Figure 3b–d shows three enlarged mass spectra, exemplifying the importance of high-resolution and high-accuracy in metabolite identification. Specifically, Figure 3b examines the m/z range of 147.06–147.12. Two peaks at m/z 147.0763 and 147.1127 were well-resolved (Figure 3b) and identified as glutamine and lysine, respectively. Figure 3c demonstrates that our HRMSI approach provides superior mass resolution by separating two peaks differing by only 0.23 mDa at m/z 176.0084 and 176.0107. The corresponding ion density maps are shown in Figure 3e and f, where the potassiated aminobenzoic acid (m/z 176.0107) was distributed distinctly from another ion (m/z 176.0084), illustrating the necessity of acquiring accurate m/z values to resolve low-MW compounds in MSI experiments. Otherwise, the two peaks would have been observed as a solitary peak in MRMS data, leading to a shift in observed m/z and incorrect MS images of the two molecules as one. Figure 3d displays the zoomed-in spectrum encompassing m/z 385.94–386.14, in which two peaks with close m/z values (m/z 386.0263 and 386.0478) were identified as potassiated AMP and sodiated GMP.
Figure 3.
HRMSI spectra averaged from a blue crab brain section acquired on a MALDI LTQ Orbitrap XL. (a) Averaged HRMS spectra displaying the m/z range of 90–500. (b–d) Zoomed-in spectra shown in (a) over the range of (b) m/z 147.06–147.12, (c) m/z 175.98–176.04, and (d) m/z 385.94–386.14. Panels (e) and (f) display different distributions of two ions present in (c) from blue crab brain.
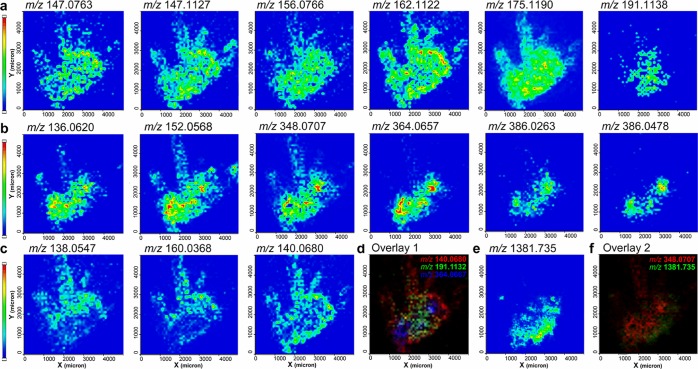
SI Figure 2a is an illustration of a crab brain with the main anatomical regions annotated.34SI Figure 2b shows the MS image of a major matrix-related ion at m/z 137.0233 that was identified as [M – H2O + H]+ and was detected over the entire imaged region. Conversely, the MS image in SI Figure 2c shows that the endogenous metabolites were localized exclusively in the brain section, facilitating the differentiation between matrix peaks and endogenous molecules. Various classes of metabolites’ MS images were collected by the HRMSI-based technique and shown in Figure 4. Specifically, amino acids such as glutamine (m/z 147.0763), lysine (m/z 147.1127), histidine (m/z 156.0766), arginine (m/z 175.1190), and carnitine (m/z 162.1122) were found to be distributed over the entire crab brain (Figure 4a). In comparison, an m/z 191.1138 ion corresponding to hydroxyarginine was localized to different regions of the brain, including the protocerebrum and median antenna I neuropil (MAN) from the deutocerebrum. Moreover, HRMS images of other amino acids such as proline, leucine, phenylalanine, and tyrosine were also obtained. Nucleobases were also imaged, including adenine (m/z 136.0620) and guanine (m/z 152.0568), as shown in Figure 4b. Interestingly, adenine and guanine were both localized to the large, spherical olfactory lobes (ON) in the deutocerebrum, whereas AMP and GMP ions (m/z 348.0707 and 364.0657, respectively) displayed nearly identical distributions. Based on HRMS measurements, the well-separated peaks in Figure 3d were identified as potassiated AMP and sodiated GMP, and their distributions agreed with the protonated forms of AMP and GMP accordingly. Organic acids and their adducts were also detected by HRMSI, as shown in Figure 4c. For example, the protonated, sodiated, and potassiated forms of aminobenzoic acid, m/z 138.0547, 160.0368 (Figure 4c), and 176.0107 (Figure 3f), were shown to colocalize in the outer, whitish sheath of the crab brain section, which validated our HRMS-based assignment. Another example is aminopentanoic acid. Its sodiated peak, m/z 140.0680, was also concentrated in the outer, fatty layer outside the neuronal structure. Figure 4d overlays three metabolites that exhibited drastically different localizations on the same crab brain section, aminopentanoic acid (red), hydroxyarginine (green) and GMP (blue). Their different localizations suggest each metabolite has a different function in the crustacean CNS. Using crustacean CNS as a model, HRMSI’s role in MSI of metabolites and NTs is once again validated by assigning molecular identities (detailed in Table 2 and SI Table 1) and quickly delivering accurate spatial images of multiple ions from biological samples simultaneously. Notably, unambiguous identification has to be verified via structural elucidation by MS/MS. Besides the presence of isobaric ions, in-source decay (ISD) fragments could further complicate the molecular assignment if solely relying on accurate mass measurement using HRMS.35
Figure 4.
MS images of metabolites identified from crustacean CNS. (a) MS images of amino acids, including (from left to right) glutamine, lysine, histidine, carnitine, arginine, and hydroxyarginine. (b) MS images of nucleobase-derived metabolites, including (from left to right) adenine, guanine, AMP, GMP, potassiated AMP, and sodiated GMP. (c) MS images of organic acids, including the protonated and sodiated aminobenzoic acid and the sodiated aminopentanoic acid. (d) Overlaid image of the sodiated aminopentoic acid in red, the hydroxyarginine in green, and GMP in blue. Other than small molecules, (e) MS image of the neuropeptide GYRKPPFNGSIFa displayed a distinct distribution from all the metabolites shown above. (f) Overlaid image of AMP coded in red and the neuropeptide SIFamide shown in green.
Table 2. Metabolites Identified from the Crab Brain Specimen Based on HRMSI.
| name | formula | theoretical monoisotopic m/z | Orbitrap XL measurement m/z | Δm (ppm) |
|---|---|---|---|---|
| glutamine | C5H11N2O3 | 147.0764 | 147.0763 | –0.7 |
| lysine | C6H15N2O2 | 147.1128 | 147.1127 | –0.7 |
| histidine | C6H10N3O2 | 156.0768 | 156.0766 | –1.3 |
| carnitine | C7H16NO3 | 162.1125 | 162.1122 | –1.9 |
| arginine | C6H15N4O2 | 175.1190 | 175.1190 | 0 |
| hydroxyarginine | C6H15N4O3 | 191.1139 | 191.1138 | –0.5 |
| adenine | C5H6N5 | 136.0618 | 136.0620 | 1.5 |
| guanine | C5H6N5O | 152.0567 | 152.0568 | 0.7 |
| AMP | C10H15N5O7P | 348.0704 | 348.0707 | 0.9 |
| GMP | C10H15N5O8P | 364.0653 | 364.0657 | 1.1 |
| AMP + K | C10H14N5O7PK | 386.0262 | 386.0263 | 0.3 |
| GMP + Na | C10H14N5O8PNa | 386.0472 | 386.0478 | 1.6 |
| aminobenzoic acid | C7H8NO2 | 138.0550 | 138.0547 | –2.2 |
| aminobenzoic acid + Na | C7H7NO2Na | 160.0369 | 160.0368 | –0.6 |
| aminopentanoic acid + Na | C5H11NO2Na | 140.0682 | 140.0680 | –1.4 |
HRMSI has also become integral in neuropeptide imaging studies of neural tissue.11 Figure 4f is an overlaid MS image of the metabolite AMP (red) and a protocerebrum-concentrated neuropeptide SIFamide at m/z 1381.735 (Figure 4e) (green). Small molecule and peptide distribution data may assist investigations that study the dynamic interaction and synergetic relationship between neurotransmitters and neuropeptides.
In conclusion, our study presents a novel HRMSI-based platform to investigate the distribution of metabolites and NTs in a biologically complex and important specimen. The high spectral resolution and mass accuracy provided by HRMS instrumentation employed in this study was advantageous when identifying analytes of interest in comparison to a MRMS instrument. We successfully generated MS images of multiple metabolites and NTs in rat and crustacean CNS, demonstrating the potential of HRMSI in biomedical applications. We anticipate that the novel knowledge of metabolite and NT distributions gained via HRMSI in this study will provide insight into related neuroscience research and present a powerful approach for future studies.
Methods
Animal Dissection
Animal experiments were performed according to institutional guidelines (UW-Madison IACUC) and can be found in detail in the Supporting Information (SI).
Sample Preparation
The CNS tissue was placed in gelatin solution, flash-frozen, and sectioned on a cryostat. The resulting sections were then dehydrated and applied with matrix DHB. The details can be found in the SI.
MALDI LTQ Orbitrap XL
The MALDI LTQ Orbitrap XL instrument (Thermo Scientific, Bremen, Germany) equipped with a commercial 60 Hz N2 laser at 337 nm (Lasertechnik Berlin GmbH, Berlin, Germany) was used for MSI.36 Automatic gain control (AGC) was on. MSI experiments were acquired using a step-size of 50 μm for crab brain and 500 μm for rat brain. The details can be found in the SI.
MALDI-TOF/TOF
An Autoflex III MALDI-TOF/TOF mass spectrometer (Bruker Daltonics, Billerica, MA) equipped with a 200 Hz smartbeam was employed for MALDI-MSI and tandem MSI. The mass spectra data were acquired over a mass range of m/z 40–1000. Each spectrum consists of 200 laser shots, and a step-size of 500 μm was used. Positive LIFT mode was employed for tandem MSI acquisition. The details can be found in the SI.
Data Analysis
The regional MS spectra and MS images obtained on the MALDI LTQ Orbitrap XL were processed using ImageQuest (Thermo Scientific). Briefly, the overall spectra of the rat or crab brain sections were generated by selecting the whole section as an ROI. The representative spectrum of the rat brain section was acquired by randomly selecting a region in the cortex as an ROI. A list of peaks with accurate mass information was obtained and verified by comparing the peaks from the off-the-tissue region, which is only covered with matrix, to ascertain the origin of the ions. The peak lists were then searched against the hmdb (http://www.hmdb.ca) and Metlin (http://metlin.scripps.edu/) databases with a mass accuracy window of 3 ppm. The identified metabolites were then searched in MetaCyc (http://metacyc.org/) and KEGG (http://www.genome.jp/kegg/kegg2.html) to ensure their biological roles in the CNS. For identified metabolites, distribution maps of selected metabolites were generated as spatially resolved color-coded extracted ion chromatograms with narrow extraction windows. The smallest window set in ImageQuest software had a width of 2 mDa (± 1 mDa). This equaled to a ±3 ppm window width at m/z 330. Overlaying metabolites’ distribution was accomplished via the “combo” feature in ImageQuest by selecting up to three metabolites’ MS images extracted from a single data set.
The overall mass spectra and MS images of the rat brain sections obtained on the Autoflex III TOF/TOF were processed with flexImaging (Bruker Daltonics). A representative spectrum was generated by selecting an ROI at an approximately identical region on a serial section of the rat brain. For tandem MSI of the putative metabolite guanosine monophosphate (GMP), the image file was also processed with flexImaging by monitoring the selected fragment ion within a mass window of 0.5 Da.
Supporting Information Available
Additional information as noted in the text. This material is available free of charge via the Internet at http://pubs.acs.org.
This work is supported in part by the National Institutes of Health grants (1R01DK071801, 1R56DK071801, and 1S10RR029531) and the National Science Foundation grant (CHE-0957784). L.L. acknowledges a H.I. Romnes Faculty Research Fellowship.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Orešič M.; Hyötyläinen T.; Herukka S. K.; Sysi-Aho M.; Mattila I.; Seppänan-Laakso T.; Julkunen V.; Gopalacharyulu P. V.; Hallikainen M.; Koikkalainen J.; Kivipelto M.; Helisalmi S.; Lötjönen J.; Soininen H. (2011) Metabolome in progression to Alzheimer’s disease. Transl. Psychiatry 1, e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper R. H.; Argmann C.; Houten S. M.; Canto C.; Jeninga E. H.; Andreux P. A.; Thomas C.; Doenlen R.; Schoonjans K.; Auwerx J. (2011) The metabolic footprint of aging in mice. Sci. Rep. 1, 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasensky J.; Jonak C. (2012) Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J. Exp. Bot. 63, 1593–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Want E. J.; Cravatt B. F.; Siuzdak G. (2005) The expanding role of mass spectrometry in metabolite profiling and characterization. ChemBioChem 6, 1941–51. [DOI] [PubMed] [Google Scholar]
- Marc D. T.; Ailts J. W.; Campeau D. C. A.; Bull M. J.; Olson K. L. (2011) Neurotransmitters excreted in the urine as biomarkers of nervous system activity: validity and clinical applicability. Neurosci. Biobehav. Rev. 35, 635–644. [DOI] [PubMed] [Google Scholar]
- Westerink B. H. C.; Hofsteede H. M.; Damsma G.; de Vries J. B. (1988) The significance of extracellular calcium for the release of dopamine, acetylcholine and amino-acids in conscious rats, evaluated by brain microdialysis. Naunyn-Schmiedeberg’s Arch. Pharmacol. 337, 373–378. [DOI] [PubMed] [Google Scholar]
- Watanabe T.; Yamagata N.; Takasaki K.; Sano K.; Hayakawa K.; Katsurabayashi S.; Egashira N.; Mishima K.; Iwasaki K.; Fujiwara M. (2009) Decreased acetylcholine release is correlated to memory impairment in the Tg2576 transgenic mouse model of Alzheimer’s disease. Brain Res. 1249, 222–228. [DOI] [PubMed] [Google Scholar]
- Braak H.; Del Tredici K.; Rüb U.; de Vos R. A.; Jansen Steur E. N.; Braak E. (2003) Staging of brain pathology related to sporadic Parkinson’s disease. Neurobiol. Aging 24, 197–211. [DOI] [PubMed] [Google Scholar]
- Lei Z.; Huhman D. V.; Sumner L. W. (2011) Mass spectrometry strategies in metabolomics. J. Biol. Chem. 286, 25435–25442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B. E.; Beaudet A. (1987) Distribution of acetylcholine and catecholamine neurons in the cat brain stem: a choline acetyltransferase and tyrosine hydroxylase immunohistochemical study. J. Comp. Neurol. 261, 15–32. [DOI] [PubMed] [Google Scholar]
- Ye H.; Greer T.; Li L. (2012) Probing neuropeptide signaling at the organ and cellular domains via imaging mass spectrometry. J. Proteomics 75, 5014–5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnell L. A.; Heeren R. M. A.; Andren P. E.; Stoeckli M.; Corthals G. L. (2012) Going forward: increasing the accessibility of imaging mass spectrometry. J. Proteomics 75, 5113–5121. [DOI] [PubMed] [Google Scholar]
- Ifa D. R.; Wu C.; Ouyang Z.; Cooks R. G. (2010) Desorption electrospray ionization and other ambient ionization methods: current progress and preview. Analyst 135, 669–681. [DOI] [PubMed] [Google Scholar]
- Liu Q.; Xiao Y. S.; Pagan-Miranda C.; Chiu Y. M.; He L. (2009) Metabolite Imaging using matrix-enhanced surface-assisted laser desorption/ionization mass spectrometry (ME-SALDI-MS). J. Am. Soc. Mass Spectrom. 20, 80–88. [DOI] [PubMed] [Google Scholar]
- Liu J. J.; Cooks R. G.; Ouyang Z. (2011) Biological tissue diagnostics using needle biopsy and spray ionization mass spectrometry. Anal. Chem. 83, 9221–9225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun J. H.; Song Z.; Liu Z.; Nikolau B. J.; Yeung E. S.; Lee Y. J. (2010) High-spatial and high-mass resolution imaging of surface metabolites of Arabidopsis thaliana by laser desorption-ionization mass spectrometry using colloidal silver. Anal. Chem. 82, 3255–3265. [DOI] [PubMed] [Google Scholar]
- Yagnik G. B.; Korte A. R.; Lee Y. J. (2013) Multiplex mass spectrometry imaging for latent fingerprints. J. Mass Spectrom. 48, 100–104. [DOI] [PubMed] [Google Scholar]
- Ye H.; Greer T.; Li L. J. (2011) From pixel to voxel: a deeper view of biological tissue by 3D mass spectral imaging. Bioanalysis 3, 313–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. J.; Perdian D. C.; Song Z. H.; Yeung E. S.; Nikolau B. J. (2012) Use of mass spectrometry for imaging metabolites in plants. Plant J. 70, 81–95. [DOI] [PubMed] [Google Scholar]
- Pirman D. A.; Reich R. F.; Kiss A.; Heeren R. M.; Yost R. A. (2013) Quantitative MALDI tandem mass spectrometric imaging of cocaine from brain tissue with a deuterated internal standard. Anal. Chem. 85, 1081–1089. [DOI] [PubMed] [Google Scholar]
- Cornett D. S.; Frappier S. L.; Caprioli R. M. (2008) MALDI-FTICR imaging mass spectrometry of drugs and metabolites in tissue. Anal. Chem. 80, 5648–5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson A.; Forngren B.; Bjurstrom S.; Goodwin R. J.; Basmaci E.; Gustafsson I.; Annas A.; Hellgren D.; Svanhagen A.; Andren P. E.; Lindberg J. (2012) In situ mass spectrometry imaging and ex vivo characterization of renal crystalline deposits induced in multiple preclinical drug toxicology studies. PLoS One 7, e47353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura Y.; Zaima N.; Setou M.; Ito S.; Yao I. (2012) Visualization of acetylcholine distribution in central nervous system tissue sections by tandem imaging mass spectrometry. Anal. Bioanal. Chem. 403, 1851–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdian D. C.; Lee Y. J. (2010) Imaging MS methodology for more chemical information in less data acquisition time utilizing a hybrid linear ion trap-orbitrap mass spectrometer. Anal. Chem. 82, 9393–9400. [DOI] [PubMed] [Google Scholar]
- Manicke N. E.; Dill A. L.; Ifa D. R.; Cooks R. G. (2010) High-resolution tissue imaging on an orbitrap mass spectrometer by desorption electrospray ionization mass spectrometry. J. Mass Spectrom. 45, 223–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel S. H.; da Costa K. A. (2009) Choline: an essential nutrient for public health. Nutr. Rev. 67, 615–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderwolf C. H. (1988) Cerebral activity and behavior: control by central cholinergic and serotonergic systems. Int. Rev. Neurobiol. 30, 225–340. [DOI] [PubMed] [Google Scholar]
- Armstrong D. M.; Saper C. B.; Levey A. I.; Wainer B. H.; Terry R. D. (1983) Distribution of cholinergic neurons in rat brain: demonstrated by the immunocytochemical localization of choline acetyltransferase. J. Comp. Neurol. 216, 53–68. [DOI] [PubMed] [Google Scholar]
- Jacobowitz D. M.; Palkovits M. (1974) Topographic atlas of catecholamine and acetylcholinesterase-containing neurons in the rat brain. J. Comp. Neurol. 157, 13–28. [DOI] [PubMed] [Google Scholar]
- Johansson B. (2006) ToF-SIMS imaging of lipids in cell membranes. Surf. Interface Anal. 38, 1401–1412. [Google Scholar]
- Fernandis A. Z.; Wenk M. R. (2007) Membrane lipids as signaling molecules. Curr. Opin. Lipidol. 18, 121–128. [DOI] [PubMed] [Google Scholar]
- Jackson S. N.; Wang H. Y. J.; Woods A. S. (2005) Direct profiling of lipid distribution in brain tissue using MALDI-TOFMS. Anal. Chem. 77, 4523–4527. [DOI] [PubMed] [Google Scholar]
- Nemes P.; Woods A. S.; Vertes A. (2010) Simultaneous imaging of small metabolites and lipids in rat brain tissues at atmospheric pressure by laser ablation electrospray ionization mass spectrometry. Anal. Chem. 82, 982–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandeman D.; Sandeman R.; Derby C.; Schmidt M. (1992) Morphology of the brain of crayfish, crabs, and spiny lobsters: a common nomenclature for homologous structures. Biol. Bull. 183, 304–326. [DOI] [PubMed] [Google Scholar]
- Debois D.; Bertrand V.; Quinton L.; De Pauw-Gillet M. C.; De Pauw E. (2010) MALDI-in source decay applied to mass spectrometry imaging: a new tool for protein identification. Anal. Chem. 82, 4036–4045. [DOI] [PubMed] [Google Scholar]
- Strupat K.; Kovtoun V.; Bui H.; Viner R.; Stafford G.; Horning S. (2009) MALDI produced ions inspected with a linear ion trap-orbitrap hybrid mass analyzer. J. Am. Soc. Mass Spectrom. 20, 1451–1463. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.