Abstract
While the Polycomb complex is known to regulate cell identity in ES cells, its role in controlling tissue-specific stem cells is not well understood. Here we show that removal of Ezh1 and Ezh2, key Polycomb subunits, from mouse skin results in a marked change in fate determination in epidermal progenitor cells, leading to an increase in the number of lineage-committed Merkel cells, a specialized subtype of skin cells involved in mechanotransduction. By dissecting the genetic mechanism, we showed that the Polycomb complex restricts differentiation of epidermal progenitor cells by repressing the transcription factor Sox2. Ablation of Sox2 results in a dramatic loss of Merkel cells, indicating that Sox2 is a critical regulator of Merkel cell specification. We show that Sox2 directly activates Atoh1, the obligate regulator of Merkel cell differentiation. Concordantly, ablation of Sox2 attenuated the Ezh1/2-null phenotype, confirming the importance of Polycomb-mediated repression of Sox2 in maintaining the epidermal progenitor cell state. Together, these findings define a novel regulatory network by which the Polycomb complex maintains the progenitor cell state and governs differentiation in vivo.
Keywords: Merkel cells, Polycomb, skin, Sox2, stem cells
Introduction
There is increasing evidence that chromatin structure and epigenetic regulation of gene expression play a fundamental role in the control of stem cells, development, and tissue homeostasis. One of the earliest described chromatin regulators is the Polycomb repressive complex (PRC), which establishes and maintains gene repression. PRC activity is divided between two multi-subunit complexes that work sequentially: first, PRC2 is recruited to chromatin, where it catalyses trimethylation of lysine 27 on histone H3 (H3K27me3) through the activity of Ezh1 and Ezh2 histone methyltransferases (Cao et al, 2002; Shen et al, 2008). This histone mark then allows the second complex, PRC1, to be recruited, where it assists in chromatin compaction and gene silencing (Min et al, 2003; Zhang et al, 2012). Misregulations of Polycomb function have been associated with diverse human pathologies, including cancer (Sauvageau and Sauvageau, 2010; Delgado-Olguin et al, 2012).
Most mammalian studies of the role of the Polycomb complex in control of stem cells and development to date have been done in vitro with either embryonic stem cells or progenitor cells, but the roles of Polycomb in regulating tissue-specific stem cells and governing organogenesis remain poorly understood (Caretti et al, 2004; Benoit et al, 2012; Sher et al, 2012). Importantly, profiling of the association of Polycomb with genomic regions in many stem cell systems identified its presence at a large set of differentiation genes (Boyer et al, 2006; Lee et al, 2006), suggesting a model wherein this complex represses differentiation. Published functional studies, however, have so far failed to support this model. Indeed, in many systems, Polycomb-null phenotypes were linked to activation of the Ink4a/Ink4b/Arf locus (Bracken et al, 2007) leading to loss of cell proliferation rather than aberrant differentiation (Molofsky et al, 2003; Park et al, 2003; Martinez and Cavalli, 2006; Chen et al, 2009). In skin, loss of Ezh1/2 also results in an upregulation of the Ink4a/Ink4b/Arf locus, leading to loss of hair follicle stem cell proliferation and ultimately degeneration of the hair follicles (Ezhkova et al, 2011). Thus, the importance of Polycomb-mediated repression and the gene regulatory networks involved in controlling stem cell differentiation in vivo need to be investigated.
Skin has proven to be an excellent model system to study the mechanisms controlling stem cell self-renewal and differentiation (Zhang et al, 2012). During embryonic development, a single layer of multipotent embryonic epidermal stem cells that reside in the basal layer produce multiple lineages, including the epidermis that provides barrier function, hair follicles that provide thermal protection, and Merkel cells that are involved in mechanotransduction (Blanpain and Fuchs, 2009; Mascre et al, 2012). While the mechanisms controlling hair follicle and epidermal development are well studied (Blanpain and Fuchs, 2009), the mechanisms controlling Merkel cell specification are largely unknown.
Merkel cells were described over a century ago (Merkel, 1875) as clusters of cells located in touch-sensitive areas of the skin, where they transduce mechanical stimuli via sensory neurons to aid in the perception of curvature, texture, and shape of objects (Haeberle and Lumpkin, 2008). Consistent with this function, Merkel cells express voltage-gated ion channels, neuropeptides, components of the presynaptic machinery such as Rab3c, and are innervated by sensory neurons; this is surprising, however, considering the epithelial origin of these cells (Maricich et al, 2009; Morrison et al, 2009; Van Keymeulen et al, 2009; Woo et al, 2010). The intermediate filament cytokeratins 18 and 20 (K18 and K20) are often used as a tool for the analysis and diagnosis of Merkel cell carcinoma due to their highly specific expression in Merkel cells (Houben et al, 2010; Donepudi et al, 2012). Furthermore, a variety of transcription factors involved in neuronal differentiation, such as Sox2 and Isl1 (Haeberle et al, 2004), are also found in Merkel cells, though how these factors control Merkel cell lineage specification is unknown. It has been shown that in mice, Merkel cell lineage development depends on the basic helix–loop–helix transcription factor Atoh1 (Maricich et al, 2009), but despite the importance of these cells, and the previous determination of the Merkel cell signature (Haeberle et al, 2004), little is known about the mechanism orchestrating their development.
In this report, we provide evidence that Ezh1 and Ezh2 repress Merkel cell lineage differentiation in epidermal stem cells. We show that conditional ablation of Ezh1 and Ezh2 in mouse skin results in an increase in the number of Merkel cells due to increased differentiation of progenitor cells. We delineate the molecular pathway through which the Polycomb complex controls Merkel cell specification, and show that the PRC-dependent H3K27me3 histone mark directly targets and represses Sox2, which we posit as a novel regulator of Merkel cell lineage specification. Finally, we show that ablation of Sox2 in Ezh1/2 2KO skin attenuates the Polycomb loss-of-function phenotype, confirming the critical role of the Ezh1/2 repression of Sox2 in maintaining the epidermal stem cell state. Through these experiments, we have not only elucidated the molecular pathway that controls Merkel cell differentiation, but have shown that the Polycomb complex can act as a specific lineage regulator through repression of transcription factor networks in a mammalian stem cell system.
Results
Loss of Ezh1/2 leads to expansion of Merkel cells
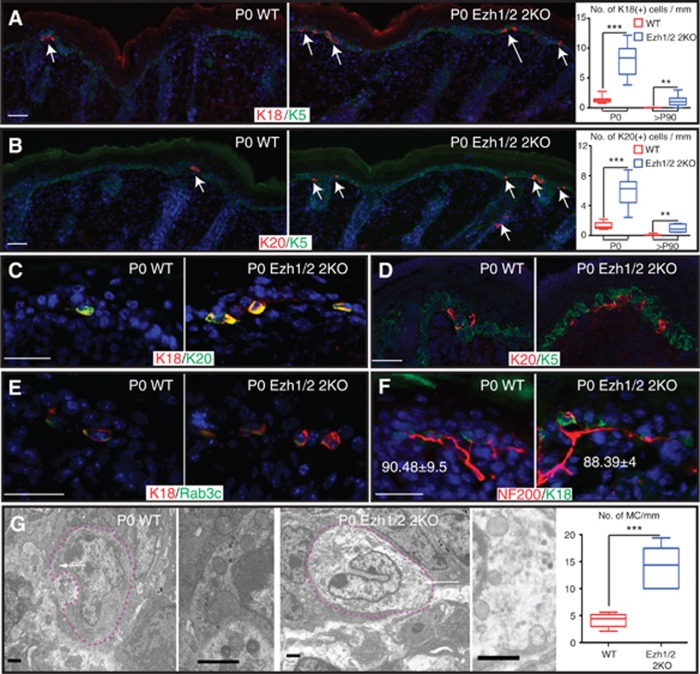
Analysis of transcriptional profiling data of genes expressed in Merkel cells revealed that several components of the Polycomb complex (Ezh2, Eed, Pcgf2) are expressed in skin epithelium but are downregulated in Merkel cells (Haeberle et al, 2004). To explore the role of the Polycomb complex in controlling the Merkel cell lineage, we analysed the Merkel cell population in Ezh1/2 2KO mice (Ezhkova et al, 2011), where key subunits of the Polycomb complex, histone methyltransferases Ezh1 and Ezh2, were conditionally ablated in the skin epithelium. Immunofluorescent (IF) studies with antibodies against the well-characterized Merkel cell markers, cytokeratin 18 and 20 (K18 and K20), revealed an increase in the number of K18(+) and K20(+) cells in newborn (P0) and adult (P90) Ezh1/2 2KO back skin compared to WT (Figure 1A and B and Supplementary Figure 1A). Increase in the number of K18(+) or K20(+) cells was also found in the skin of the ventral, paw, and whisker interfollicular epidermis (IFE), tissues that normally contain Merkel cells (Supplementary Figure 1B–D, quantified in F). In whisker follicles, Merkel cells are about 10-fold more abundant in comparison to the other analysed zones, and interestingly, increase in Merkel cell number was not observed in Ezh1/2 2KO mice in the whiskers area (Supplementary Figure 1E and F). We speculate that the whisker follicle region has reached a maximum density in WT animals, and thus loss of Ezh1/2 has little consequence there. As expected, at P0, all Merkel cells co-express K18 and K20 (Figure 1C) and in accordance with previous reports (Maricich et al, 2009; Morrison et al, 2009; Van Keymeulen et al, 2009; Woo et al, 2010), they lose expression of epidermal progenitor marker K5 (Figure 1D). Merkel cells have been suggested to form near guard hair follicles, but histological analysis indicated no difference in the number of guard hair follicles present in Ezh1/2 2KO skin compared to WT (Supplementary Figure 1G), discounting the possibility that the observed phenotype was due to alterations in guard hair number.
Figure 1.
Number of Merkel cells is increased in Ezh1/2 2KO epidermis. (A, B) Immunofluorescent (IF) staining of WT and Ezh1/2 2KO skin with antibodies against Merkel markers K18 (A) or K20 (B) shows increase in the number of K18(+) and K20(+) cells. Arrows indicate positively stained K18(+) and K20(+) cells. Skins counterstained with basal layer marker K5. Quantifications are provided at the right and show statistically significant increase in the number of Merkel cells (P0 WT/2KO K18 P<0.0001, n=127/150 mm, 310/1285 cells; P90 WT/2KO K18 P=0.0026, n=84/83 mm, 2/95 cells; P0 WT/2KO K20 P=0.0003, n=72/52 mm, 94/298 cells; P90 WT/2KO K20 P=0.0017, n=81/39mm, 5/33 cells). (C) IF studies with K18 and K20 antibodies show overlap of the two markers. (D) Confocal analysis of K20-labeled cells showing lack of overlap with anti-K5-labeled cells. (E, F) IF studies show overlap of K18 with Merkel cell protein Rab3c (E), and innervation of K18(+) cells by neurons, as labeled by NF200 (F). Numbers in (F) show similar levels of innervation in WT and Ezh1/2 2KO skin (WT/2KO P=0.8497, n=78/220 cells). (G) TEM imaging shows fully differentiated Merkel cells in the whisker area of P0 WT and Ezh1/2 2KO mice. Dotted lines outline Merkel cells. Arrows indicate characteristic dense core neuroendocrine granules, magnified at the right. TEM quantifications are shown at the right (WT/2KO P=0.0009, n=9/8.7 mm, 37/120 cells) and confirm increase in the number of Merkel cells in Ezh1/2 2KO skin. Scale bars for (A–F) are 50 μm. Scale bars for (G) are 1 μm.
To carry out their mechanosensory functions, Merkel cells are innervated by sensory neurons and express neuronal proteins. IF analysis showed the presence of Rab3c, a component of the presynaptic machinery (Fischer von Mollard et al, 1994; Haeberle et al, 2004), in Ezh1/2 2KO K18(+) cells (Figure 1E) and innervation by sensory neurons, as shown by IF staining with antibodies to the neurofilament protein NF200 (Figure 1F). Interestingly, the percentage of innervated K18(+) cells in WT and Ezh1/2 2KO skins was comparable (Figure 1F, percentages), indicating that the additional K18(+) cells formed in Ezh1/2 2KO skin become innervated successfully. Finally, transmission electron microscopy (TEM) confirmed the presence of additional Merkel cells in Ezh1/2 2KO skin, as detected by the presence of characteristic neuroendocrine granules (Tachibana et al, 1983); (Figure 1G) and corroborated by quantifications (Figure 1G, right). Overall, these data indicate that loss of Ezh1/2 leads to an increase in the number of fully differentiated Merkel cells.
Ezh1 and Ezh2 repress Merkel cell differentiation
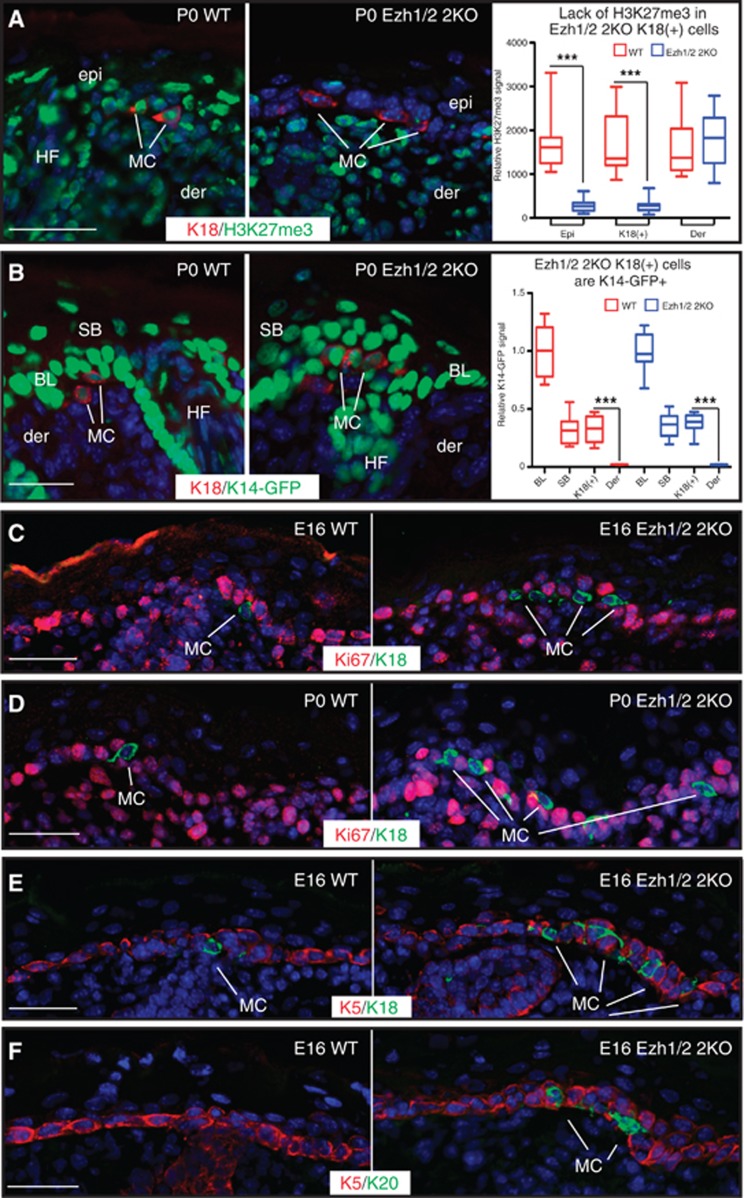
We next explored the origin of Merkel cells in Ezh1/2 2KO skin. We have previously reported that in K14-Cre Ezh1/2 2KO mice, epithelial cells are Ezh1/2 null and thus completely lack the H3K27me3 histone mark (Ezhkova et al, 2011), whereas cells of non-epithelial origins retain this mark. By examining the pattern of H3K27me3, we found that K18(+) cells in Ezh1/2 2KO skin also lacked H3K27me3 (Figure 2A), whereas mesenchymal dermal cells and neural crest-derived melanocytes remained H3K27me3 positive (Figure 2A and Supplementary Figure 2A). To further confirm the epithelial origin of K18(+) cells in Ezh1/2 2KO skin, we crossed WT and Ezh1/2 2KO mice with K14-GFP-H2B mice (Tumbar et al, 2004), where histone H2B is tagged with GFP and is expressed under the keratin 14 (K14) promoter that is active only in epidermal progenitor cells. IF analysis of K18(+) cells in WT and Ezh1/2 2KO mice showed that they were GFP positive (Figure 2B), whereas dermal or melanocyte cells were GFP negative (Figure 2B, and Supplementary Figure 2B and C). The GFP level in WT and Ezh1/2 2KO K18(+) cells was significantly lower than in the K14-expressing basal epidermal cells, but comparable to the level seen in K14-derived suprabasal cells (Figure 2B, right). Together, these data indicate that Ezh1/2 2KO Merkel cells are indeed K14 derived and therefore epithelial in origin, consistent with previous reports (Morrison et al, 2009; Van Keymeulen et al, 2009; Woo et al, 2010).
Figure 2.
Loss of Ezh1/2 leads to accelerated differentiation of epidermal progenitors to Merkel cells. (A) IF studies of P0 WT and Ezh1/2 2KO skin using antibodies against K18 and H3K27me3 show that K18(+) Merkel cells lack H3K27me3 signal. Fluorescence intensity quantification of H3K27me3 signal (arbitrary units) is shown at the right (WT versus 2KO epi P<0.0001, n=20/20 cells; WT versus 2KO K18 P<0.0001, n=13/14 cells). (B) IF studies of P0 WT and Ezh1/2 2KO mice expressing K14-GFP-H2B show that K18(+) cells are GFP(+). Relative fluorescence intensity quantification of K14-GFP-H2B signal is shown at the right and normalized to the average basal layer signal (WT K18 versus der P<0.0001, n=7; 2KO K18 versus der P<0.0001, n=6). (C, D) IF analysis of E16 (C) or P0 (D) WT and Ezh1/2 2KO skin using antibodies against K18 and the proliferation marker Ki67 show that Merkel cells are postmitotic. (E, F) IF studies of E16 WT and Ezh1/2 2KO skin using antibodies against K5 and either K18 (E) or K20 (F) showing the premature formation of Merkel cells in Ezh1/2 2KO skin. In all panels, epi: epidermis; der: dermis; HF: hair follicle; BL: basal layer; MC: Merkel cell. All scale bars are 50 μm.
It has been shown that Merkel cells are postmitotic under normal conditions (Woo et al, 2010). To test whether increase in the number of Ezh1/2-null Merkel cells is due to their aberrant proliferation, we analysed the proliferation marker Ki67 (S+M-phase cells) and observed that, as in WT, Ezh1/2 2KO K18(+) cells were Ki67(−) both at E16 and at P0 (Figure 2C and D, respectively, and Supplementary Figure 2H). Furthermore, 4-h pulses of 5-bromo-2′-deoxyuridine (BrdU) to WT and Ezh1/2 2KO mice at E16, P0, and P14 showed that Merkel cells were BrdU negative (Supplementary Figure 2D–G), further indicating that these cells are non-proliferative. These results thus show that loss of Ezh1 and Ezh2 does not affect the proliferative status of Merkel cells and that they remain postmitotic.
Since Merkel cells showed no proliferation defects, we next analysed if differentiation of Ezh1/2-null progenitors to Merkel cells might be enhanced. Remarkably, K18(+) and K20(+) Merkel cells were precociously acquired in Ezh1/2 2KO skin at E16 as compared with their WT littermates, where appearance of fully differentiated Merkel cells was observed only by E17 (Figures 2E, F and 3E) in accordance with previous reports (Van Keymeulen et al, 2009). This suggests that the commitment of epidermal stem cells to the Merkel cell lineage is accelerated in the Ezh1/2 2KO. In hairy portions of mouse skin, Merkel cells appear in clusters referred to as touch domes (Woo et al, 2010), which are complexes of Merkel cells and neurons located around the guard hairs. However, in Ezh1/2 2KO skin, Merkel cell clusters form around different hair types, and can be seen in the IFE (Figure 1A and B). Our analysis of the size of these clusters, by number of Merkel cells per cluster, did not reveal significant differences between WT and Ezh1/2 2KO skin, but strikingly, we observed an increase in the number of distinct clusters in Ezh1/2-null skin compared to WT (Supplementary Figure 2I). Overall, these data suggest that loss of Ezh1/2 leads to increased commitment of stem cells to the Merkel lineage, leading to formation of new Merkel cell clusters.
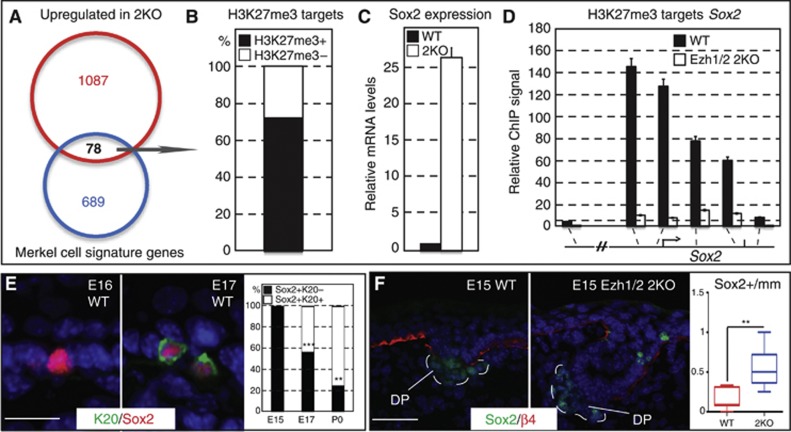
Figure 3.
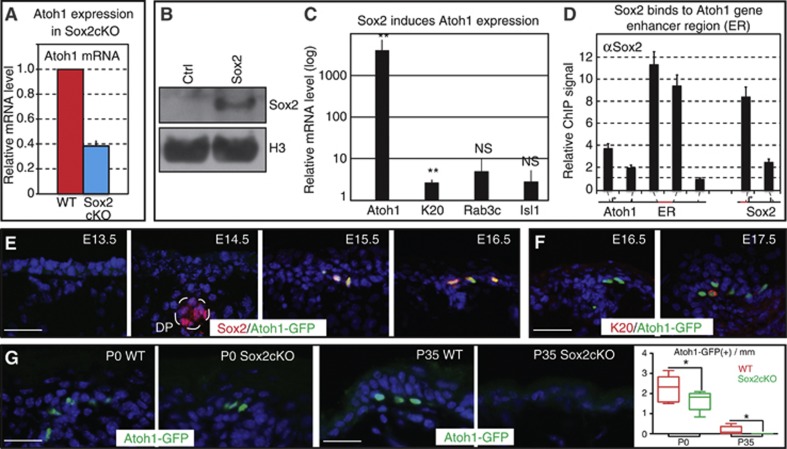
The Merkel signature gene Sox2 is directly regulated by Ezh1/2-mediated repression in epidermal stem cells. (A) Venn diagram of Merkel cell signature genes with genes that are upregulated in Ezh1/2 2KO epidermal progenitors shows overlap of 78 genes. (B) Of those 78 genes, 55 (70.5%) are direct targets of H3K27me3 repression, and Sox2 is among the most upregulated. (C) qPCR confirmation of microarray data shows that Sox2 is 26-fold upregulated in Ezh1/2 2KO progenitors. (D) In vivo ChIP-qPCR confirmation of ChIP-seq data showing that Sox2 is directly targeted by the Ezh1/2-dependent H3K27me3 histone mark, and that the H3K27me3 ChIP signal is drastically decreased in total skin cells isolated from P0 Ezh1/2 2KO mice. Please note that total skin contained both epithelial and a small fraction of non-epithelial cells (melanocytes, immune cells, etc.) that were not targeted by the K14-Cre-mediated ablation of Ezh1/2. Thus, a small residual level of H3K27me3 at Sox2 is observed in 2KO cells. (E) IF studies of E16 and E17 WT skin using antibodies against K20 and Sox2 reveal that Sox2 is expressed prior to other Merkel cell markers. Quantifications are provided at the right and show statistically significant decrease in the number of Sox2+K20− cells throughout development (E15 versus E17 P=0.0002; E17 versus P0 P=0.0036; E15 n=109mm/0 Sox2(+)K20(+) cells/19 Sox2+K20− cells; E17 n=97/99/151; P0 n=68/74/26). (F) IF analysis of E15 WT and Ezh1/2 2KO skin using antibodies against Sox2 and ß4-integrin shows that Sox2 is expressed prematurely in Ezh1/2 2KO skin. Quantifications are provided at the right and show statistically significant increase in the number of Sox2(+) cells in 2KO versus WT (WT versus 2KO P=0.0013, n=90/83mm, 14/45 cells). All scale bars are 50 μm.
Polycomb represses the Merkel cell gene Sox2 in epidermal stem cells
To gain insight into how loss of Ezh1/2 might lead to the observed phenotype, we analysed genes that were (1) activated in Ezh1/2-null epidermal stem cells compared to WT (Ezhkova et al, 2011) and (2) expressed in Merkel cells (Haeberle et al, 2004). The analysis revealed that 78 Merkel cell signature genes (∼10% of total) (Haeberle et al, 2004) became activated in Ezh1/2 2KO cells, while they were silenced in WT cells (Figure 3A and Supplementary Table 1). Interestingly, expression of many bona fide Merkel cell genes, such as keratin 20 and chromogranin B, were unchanged in 2KO basal cells compared to WT, indicating that only a specific subset of Merkel signature genes becomes activated in Ezh1/2 2KO epidermal stem cells (Supplementary Figure 3A). About two-thirds of these 78 genes were direct targets of the H3K27me3 repressive mark in WT epidermal stem cells (Figure 3B) (Ezhkova et al, 2011; Lien et al, 2011), indicating the direct role of the Polycomb-mediated repression in their control. Remarkably, the transcription factor Sox2 was one of the most highly upregulated genes (26-fold) of this subset, which we confirmed by semi-quantitative real-time PCR (RT-PCR) analysis (Figure 3C) and by performing IF studies where Sox2 was detectable in Ezh1/2 2KO skin in both K18(+) Merkel cells and K18(−) epidermal stem cells (Supplementary Figure 3C). We also confirmed that Sox2 is a direct target of Polycomb repression by performing chromatin immunoprecipitation (ChIP) and showed the presence of Polycomb-dependent H3K27me3 histone marks as well as Ezh1 and Ezh2 at the Sox2 gene (Figure 3D and Supplementary Figure 3B). Notably, IF studies indicated that Sox2 is expressed in the WT skin at an early developmental stage, just prior to expression of the first Merkel cell markers (Figure 3E), as well as precociously expressed in Ezh1/2 2KO epidermis (Figure 3F, quantified at right), suggesting that Sox2 is an early marker of Merkel cell differentiation. Importantly, the percentage of Sox2(+)K20(−) cells decreases consistently between E15 and P0, suggesting that these Sox2(+) cells eventually become fully differentiated Sox2(+)K20(+) Merkel cells (Figure 3E, graph).
Sox2 controls Merkel cell development
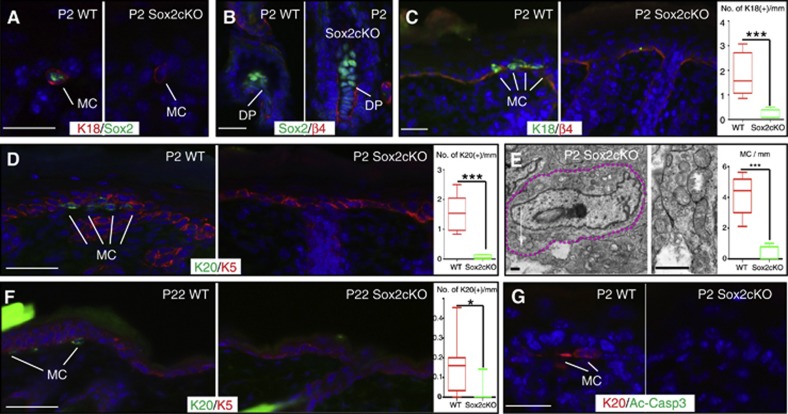
We next hypothesized that the loss of Polycomb repression led to activation of Sox2 expression in epidermal stem cells and resulted in increased differentiation of cells to the Merkel lineage. To test this, we first analysed the role of Sox2 in control of the Merkel cell lineage through conditional ablation of Sox2 in skin epithelium by crossing Sox2 fl/fl mice with K14-Cre mice (Sox2cKO). IF studies confirmed that Sox2 expression was abolished in Sox2cKO epithelium (Figure 4A), but not in dermal papilla cells (Figure 4B) that are not targeted by the K14-Cre ablation strategy (Vasioukhin et al, 1999). A dramatic decrease in the number of fully differentiated Merkel cells was observed in Sox2cKO skin, as shown by IF analysis of the Merkel cell markers K18 and K20, as well as by EM (Figure 4C–E). This loss of Merkel cells was observed in all body regions analysed, including paws, whisker IFE, and whisker follicles (Supplementary Figure 4A–D, quantified in E). The observed phenotype persisted into adulthood (Figure 4F), indicating that the observed phenotype is not due to a delay in the development of Merkel cells. Besides the alteration in the Merkel cell number, mice deficient for Sox2 in the skin were born alive at a Mendelian ratio, bred normally, and had no gross pathological abnormalities (Supplementary Figure 4F). Furthermore, histological and IF analysis of Sox2cKO epidermis showed no apparent defects in epidermal differentiation, hair formation, or skin innervation by NF200(+) neurons (Supplementary Figure 4G–J). Importantly, no increase in apoptosis of Merkel cells was observed in Sox2cKO back skin, paws, or whiskers either prior to Merkel cell specification at E16 or postnatally at P0 and P22, as shown by IF with antibodies to activated-caspase 3 (Figure 4G and Supplementary Figure 4K–S). Overall, the requirement of Sox2 for Merkel cell development positions this gene as a new critical regulator of Merkel cell lineage specification.
Figure 4.
Sox2 is required for proper Merkel cell development. (A, B) IF analysis using antibodies against Sox2 and either K18 (A) or ß4-integrin (B) shows loss of Sox2 from Merkel cells but not from the dermal papilla. (C, D) IF using antibodies against either K18 (C) or K20 (D) shows dramatic reduction of Merkel cells from Sox2cKO skin. Quantifications are shown at right (WT versus Sox2cKO K20 P<0.0001, n=82/80mm, 156/16 cells; K18 P=0.0002, n=73/80mm, 112/5 cells). (E) TEM imaging (left) shows that few Merkel cells present are fully developed. Dotted lines outline Merkel cells. Arrows indicate characteristic dense core neuroendocrine granules, magnified at right. TEM quantifications are shown at right and confirm drastic decrease in the number of fully differentiated Merkel cells present in the whisker area of P0 Sox2cKO mice (WT versus Sox2cKO P=0.0003, n=9/9.5mm, 37/3 cells). (F) Decrease in the number of fully differentiated Merkel cells in Sox2cKO persists into adulthood as shown by IF analysis of K20. Quantifications are provided at right (WT versus Sox2cKO P=0.0163, n=99/95mm 15/2 cells). (G) Analysis of apoptosis using antibodies against Act-Casp3 and K20 shows no difference between WT and Sox2cKO skin. All scale bars are 50 μm.
Sox2 regulates Atoh1 and Isl1 gene expression
To further characterize the mechanism of how Sox2 controls Merkel cells, we analysed the expression of the Atoh1 gene, which has been shown to be an obligate transcription factor for Merkel cell differentiation (Maricich et al, 2009). We observed a >3-fold decrease in Atoh1 mRNA levels in Sox2cKO skin (Figure 5A). Genome scanning identified a Sox2 binding site at the enhancer region of the Atoh1 gene, and a direct role for Sox2 in controlling the Atoh1 gene transcription has been reported in other developmental systems (Yoon et al, 2011; Neves et al, 2012). To determine if a similar mechanism is operating in Merkel cells, we performed in vitro studies on cultured epidermal progenitor cells overexpressing Sox2 (Figure 5B) and observed an ∼5000-fold upregulation of Atoh1 mRNA, and a small (∼5-fold) increase in K20 mRNA, compared to Control (Figure 5C). To analyse if Sox2 directly binds the Atoh1 gene, we performed ChIP assay with Sox2 antibodies. Indeed, we found that Sox2 targets the Atoh1 enhancer region, and that the signal was significantly higher than at the 3′ end of Atoh1, which does not contain Sox2-binding sites (Figure 5D). We also found an association of Sox2 with its own promoter region (Figure 5D), consistent with reports in embryonic stem cells that show Sox2 autoregulation is important for stemness control (Heng et al, 2010). Together these data indicate that Sox2 directly activates transcription of the Atoh1 gene.
Figure 5.
Sox2 regulates expression of Atoh1, a critical regulator of Merkel cell specification. (A) Expression of the obligate Merkel cell transcription factor Atoh1 is reduced in total skin isolated from P0 Sox2cKO mice as measured by RT-qPCR. (B) Overexpression of Sox2 in WT epidermal stem cells was confirmed by western blot. Histone H3 serves as a loading control. (C) Sox2-overexpressing cells had a dramatic increase in the level of Atoh1 mRNA and a slight upregulation of K20 mRNA, while expression of other Merkel cells markers remained unchanged. (D) ChIP studies on Sox2-overexpressing cells showed Sox2 binding at the Atoh1 enhancer region and the known Sox2 autoregulatory site. ChIP-qPCR signal is normalized to experimental control cells infected with an empty vector. (E) Concomitant expression of Sox2 and Atoh1 is observed throughout Markel cell development, as shown by IF studies of Sox2 on Atoh1-GFP back skin samples. (F) As with Sox2, Atoh1 is an early marker of Merkel cell differentiation as shown by lack of co-staining between Atoh1-GFP and K20 at E16. (G) IF studies show that the presence of Atoh1-GFP(+) cells is dependent on Sox2, as shown by decrease in the number of Atoh1-GFP+ cells at P0 and their absence at P35. Quantifications are provided at right (P0 WT versus Sox2cKO P=0.0283, n=103/86mm 226/144 cells. P35 WT versus Sox2cKO n=103/92mm 16/0 cells). All scale bars are 50 μm.
Source data for this figure is available on the online supplementary information page.
To characterize the onset of expression of Sox2 and Atoh1, we performed a detailed temporal analysis of the appearance of Sox2(+) cells in the skin of Atoh1-GFP mice, where EGFP is fused to the 3′ end of the Atoh1 gene. In these mice, Merkel cells are the only skin cells with GFP signal (Haeberle et al, 2004). In accordance with the published studies (Driskell et al, 2009; Clavel et al, 2012), Sox2 was found in the dermal papilla at E14, and those cells did not express Atoh1. However, neither Atoh1-GFP nor Sox2 is expressed in the epidermis before E15, at which point they are consistently co-expressed (Figure 5E). These Atoh1-GFP cells then begin to express K20 starting at E17 (Figure 5F). These data suggest that Sox2 and Atoh1-GFP are concomitantly expressed during Merkel cell specification and are both early markers of Merkel cell development.
To confirm the involvement of Sox2 in Atoh1 expression and Merkel cell differentiation in vivo, we crossed Sox2cKO mice with Atoh1-GFP mice. In these animals, Atoh1-GFP(+) cells were found in significantly reduced numbers at P0 (Figure 5G, left and graph) and completely lacked K20 staining (Supplementary Figure 5A). Fluorescence-activated cell sorting (FACS) analysis of P0 epidermis confirmed the reduction of Atoh1 expression, as indicated by decrease in the Atoh1-GFP signal intensity (Supplementary Figure 5B, right) and showed a 3-fold decrease in the number of Atoh1-GFP(+) cells in Sox2cKO compared to WT (Supplementary Figure 5B). Importantly, by P35, no Atoh1-GFP(+) cells were observed in Sox2cKO skin, whereas they were still present in WT mice (Figure 5G, right and graph). These data indicate that Sox2 is required to sustain Atoh1 expression and to promote Merkel cell specification.
RT–PCR analysis of the Merkel cell marker Isl1 also showed a >5-fold decrease for Isl1 mRNA in Sox2cKO skin compared to WT (Supplementary Figure 5D, left) and both the number of Isl1+ cells and the IF fluorescence intensity for Isl1 in Sox2cKO K18(+) Merkel cells were reduced (Supplementary Figure 5D, right and E). Together, these data demonstrate that Sox2 is required to properly specify Merkel cells during development and suggest that Sox2 modulates transcription of Atoh1, as well as Isl1. Since Atoh1 is absolutely required for Merkel cell differentiation and Sox2 can induce Atoh1 transcription (Maricich et al, 2009; Van Keymeulen et al, 2009), we propose that Sox2 controls Merkel cell development by activating Atoh1. Importantly, ChIP analysis of the Atoh1 gene revealed the presence of the Polycomb-dependent H3K27me3 histone mark (Supplementary Figure 5C), and analysis of ChIP-sequencing data (Ezhkova et al, 2011; Lien et al, 2011) at the promoters of Isl1 and Atoh1 also revealed the presence of the H3K27me3 repressive mark. These data thus reveal a complex regulation of the Merkel cell lineage by Polycomb in epidermal stem cells, with tight control of both the activator Sox2 and its effector genes, Atoh1 and Isl1.
Loss of Ezh1/2 promotes Merkel cell differentiation by derepression of Sox2
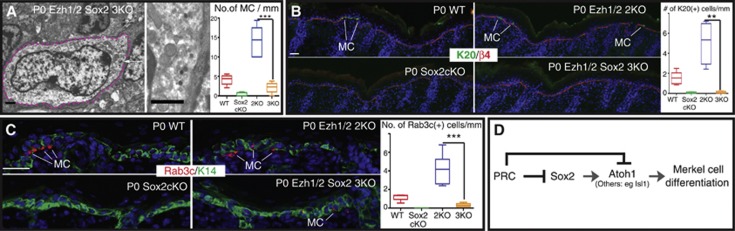
Finally, to determine the extent to which the increase in the number of Merkel cells in Ezh1/2-null skin was directly attributable to the activation of Sox2, we generated Ezh1/2 Sox2 3KO mice. IF analysis of P0 Ezh1/2 Sox2 3KO skin confirmed a complete loss of H3K27me3 and Sox2 specifically in the knockout epithelium, but not in the K14-Cre-negative dermal cells (Supplementary Figure 6A–C). No apparent defects in epidermal differentiation, skin innervation by NF200-neurons, or increase in apoptosis were observed in 3KO mice compared to WT (Supplementary Figure 6D–I). TEM analysis of the Ezh1/2 Sox2 3KO skin showed a drastic decrease of the number of fully differentiated Merkel cells when compared to Ezh1/2 2KO skin (Figure 6A) and was corroborated by IF analysis of K20, Rab3c, and K18 Merkel cell markers (Figure 6B and C, and Supplementary Figure 6J). The decrease was also observed throughout all body regions of the mice, including paws, whisker IFE, and whisker follicles (Supplementary Figure 6K–O). As shown in Supplementary Figure 6J, the number of K18(+) cells in Ezh1/2 Sox2 3KO mice was similar to that seen in WT mice. This is consistent with data showing that the K18 gene is directly targeted by Polycomb, and leads us to speculate that loss of Polycomb allows partial activation of K18 in the absence of Sox2, while addition of Sox2 greatly amplifies K18 expression. The collected evidence in this work suggests a mechanistic model for Polycomb repression of the Merkel cell lineage in epidermal stem cells (Figure 6D). We propose that Polycomb complexes are responsible for the direct repression of Sox2, a novel critical regulator of Merkel cell lineage specification, and Sox2-regulated Merkel cell genes (Atoh1, Isl1). Loss of Polycomb repression leads to activation of Sox2, which in turn promotes transcription of Atoh1, leading to Merkel cell differentiation.
Figure 6.
Loss of Sox2 attenuates the Ezh1/2 2KO Merkel cell phenotype. (A) TEM imaging showing fully developed Merkel cells in Ezh1/2 Sox2 3KO skin. Dotted lines outline Merkel cells. Arrow indicates dense core neuroendocrine granules shown in the magnified region at right. TEM quantifications shown at the right indicate the reduction in Merkel cells in Ezh1/2 Sox2 3KO skin relative to Ezh1/2 2KO (WT/3KO P=0.0003, 3KO n=9/5.8mm 37/13 cells). (B, C) IF analysis of Ezh1/2 Sox2 3KO skin using antibodies against either K20 (B) or Rab3c (C) confirms TEM data showing the reduction in Merkel cell number relative to Ezh1/2 2KO skin. Quantifications are shown at the right (K20 2KO versus 3KO P=0.0033, n=24/53mm 119/5 cells; Rab3c 2KO versus 3KO P=0.0002, n=56/64mm 128/10 cells). (D) Model. The Polycomb complex is responsible for the direct repression of Sox2 and Sox2-regulated Merkel cell genes (Atoh1, Isl1, others) to repress the Merkel cell differentiation program in epidermal stem cells. Scale bars for (A) are 1 μm, all others are 50 μm. WT: wild type; 2KO: Ezh1/2 2KO; 3KO: Ezh1/2 Sox2 3KO.
Discussion
Mechanisms involving Polycomb proteins are attracting a lot of attention in the stem cell field due to the importance of this complex in controlling in vitro cultured ES cells. Here, we uncovered the relevance of the Polycomb complex in control of the stem cell state in vivo, and showed that Polycomb is essential to repress the lineage commitment and differentiation of epidermal progenitor cells to Merkel cells. Importantly, we dissected the molecular mechanisms of this control by showing that the downstream target of Polycomb repression, Sox2, is a critical transcriptional regulator of Merkel cell differentiation and that its repression is required to maintain the stem cell state.
The role of the Polycomb complex in controlling epidermal progenitor cells is somewhat reminiscent of its function in ES cells, where PRC has also been shown to repress key transcriptional regulators of differentiation (Boyer et al, 2006; Lee et al, 2006). Interestingly, here we show that both Sox2 and its downstream target gene Atoh1, which is essential for Merkel cell differentiation, are under the control of the Polycomb. We therefore propose that Polycomb, Sox2, and Atoh1 form a regulatory network to control Merkel cell lineage specification in embryonic epidermal progenitors. We speculate that the Polycomb repression is first lost from Atoh1 and Sox2 genes, allowing their transcription to be initiated. Sox2 activity is then required to further promote Atoh1 expression and induce full differentiation of Merkel cells. This data thus reveal a tight regulatory network in place to maintain the epidermal stem cell state.
Despite the similarity to ES cells, our studies also uncovered clear differences between Polycomb regulation of ES and epidermal stem cells. In ES cells, the Polycomb complex has been shown to be required for proper execution of the differentiation program, and in vitro studies showed that Polycomb-null ES cells failed to differentiate the neuronal lineage due to failure to silence stemness genes (Pasini et al, 2007, 2010). Contrary to ES studies, we showed that Ezh1/2-null epidermal stem cells are capable of executing the terminal differentiation program and downregulating stemness genes. Taking into consideration our previous work on hair follicle stem cells (Ezhkova et al, 2011), we propose that in skin stem cells, the Polycomb complex is critical to maintain the stem cell state rather than to promote differentiation.
Decades of work dissecting the molecular mechanisms controlling hair follicle and epidermal suprabasal lineages identified several key regulators of these pathways (Fuchs and Raghavan, 2002; Blanpain and Fuchs, 2009). The processes regulating the Merkel cell differentiation program, however, are completely unknown despite the discovery of Merkel cells more than 100 years ago. Here we uncovered the critical role for the Sox2 transcription factor in promoting the Merkel cell lineage specification. Interestingly, in ES cells and several mammalian tissues (Masui et al, 2007; Arnold et al, 2011), Sox2 is expressed in stem cells and is required for stem cell maintenance. Our studies clearly show that in skin, Sox2 is absent from epidermal stem cells, is expressed in early committed Merkel cells, and is required for the Merkel lineage specification.
Together, our studies clearly show the different roles for both the Polycomb complex and Sox2 in different stem cell systems. As there is an increasing interest in the role of these proteins in controlling tumorigenesis, the clear understanding of their roles in regulation of stemness and differentiation in a specific tissue context will be essential to determine their functional involvement in cancer stem cells and, further, for the future design of proper cancer therapies. Our studies dissected that in at least one tissue, the skin, there is a clear role for the Polycomb-mediated repression of Sox2 in maintaining the stem cell state.
Materials and methods
Mice
All mice were housed and cared for according to MSSM and IACUC-approved protocols. At least two animals from independent litters were used for each analysis. Ezh1/2 2KO mice were previously reported (Ezhkova et al, 2011). As described, mice null for Ezh1 and Ezh2 die shortly after birth, and all analysis of these mice after P0 was performed on grafted skin obtained using the described grafting protocol (Ezhkova et al, 2011). K14-GFP-H2B mice were previously reported (Tumbar et al, 2004). K14-Cre and Atoh1-EGFP mice were obtained from The Jackson Laboratories. The Atoh1 and Ezh2 genes are located on chromosome 6, which prevents crossing Atoh1-GFP mice and Ezh2-flox mice to generate Ezh2 cKO Atoh1-GFP. Mice were genotyped by PCR using DNA extracted from tail skin. BrdU was administered as previously reported (Ezhkova et al, 2009). Briefly, BrdU was administered (50 μg BrdU per 1 g mouse weight) to mice or pregnant females 4 h before sacrificing. The back skin, whisker regions, and paws were embedded in OCT immediately following sacrifice.
Transmission electron microscopy
Tissues were fixed in glutaraldehyde-parformaldehyde in 0.1% sodium cacodylade (Karnovsky’s), post fixed in 1% osmium tetroxide and 1.5% potassium ferrocyanide in distilled water and embedded in epoxy-Araldite resin. Thin sections counterstained with lead citrate were imaged with the Hitachi H7000 TEM. Electron microscopic quantification was performed on five serial sections at 30 μm intervals for each animal on 2 × 1 mm butvar-coated slot grids (Tachibana et al, 1983). Both nucleated and non-nucleated Merkel cells were counted, as the 30-μm interval between sections ensured that cells were only counted once (the diameter of Merkel cells is less than 15 μm).
Immunofluorescence
Tissues were collected from mice and embedded fresh into OCT or PFA, fixed for 2 h, and equilibrated in Sucrose before embedding, and subsequently cut into 10-μm sections using a Leica Cryostat. Slides were then fixed for 10′ in 4% PFA and blocked for 1 h in phosphate-buffered saline (PBS)-Triton with BSA/NGS/NDS. Primary antibodies were diluted in blocking solution and incubations were carried out for 1 h at room temperature, followed by incubation in secondary antibody for 1 h at room temperature. Slides were then counterstained with Hoechst and mounted using antifade mounting media.
Antibodies for immunofluorescence
Antibodies were used as follows: Keratin 5 (1:500); H3K27Me3 (Millipore, 1:300); K18 (Abcam, 1:100); Isl1 (Abcam, 1:400); Rab3C (Abcam, 1:250); NF200 (Sigma, 1:1000); Ki67 (Novacostra, 1:250); Sox2 (Stemgent, 1:150); K20 (Dako, 1:70); AcCasp3 (R&D, 1:250); CD104 (B4, 1:100); GFP (Abcam, 1:1000); CD117 (eBioscience, 1:100); BrdU (Abcam, 1:250); K10 (Covance, 1:500); and Loricrin (Covance, 1:250).
Microscopy and quantification
Slides were imaged using either Leica DM6000 or Zeiss Axioplan 2IE inverted slide microscopes and either × 10 or × 20 objectives. Confocal microscopy was performed using a Leica SP5 DM and either × 20 or × 63 objectives. Fluorescence intensity was calculated from at least three raw, single-channel grayscale images per condition. Using either Leica LAS AF or NIH ImageJ software, the mean intensity was calculated for nuclei of K14-GFP or H3K27me3 cells from the lineages specified. Fluorescence intensity was normalized to non-nuclear background for Figure 2A. For Figure 2B, intensity was normalized to the average basal cell (BL) intensity to show reduction as relative to K14-GFP-expressing basal cells.
Merkel cell quantification
Merkel cell number quantifications were calculated using a fluorescence microscope. The length of each section was measured and the number of positively stained cells was counted. Typical section lengths were between 7–14 mm. Due to the highly variable number of Merkel cells between genotypes, we have included the total length of skin quantified. For instance, when comparing WT and Sox2cKO skins, we counted a large number of MCs in the WT condition (>100) and then counted the number of Merkel cells in a similar length of skin for the Sox2cKO condition. We defined a ‘MC cluster’ as a group of >3 K18+ Merkel cells in which no two cells are located more than 30 μm from each other. Please note that touch domes are defined as MC clusters located in and around guard hairs. In 2KO skin, however, Merkel cell clusters appear around different hair types as well as in the interfollicular epidermis. We thus called them Merkel cell clusters and not touch domes. The total length of skin analysed has been provided in each figure legend. Complete information on the total number of Merkel cells counted for each staining is in Supplementary Table 2.
Cell culture and overexpression
Primary epidermal progenitors were collected from WT CD1 mice and cultured in calcium-restricted media. Control (empty vector) or mSox2 overexpression lentiviruses were produced from transfected 293FT cells and used to transduce keratinocytes. After 3 days, infected cells were collected and analysed by ChIP and RT–qPCR.
Chromatin immunoprecipitation
ChIP assay was performed as described (Lien et al, 2011). Briefly, keratinocytes were obtained from in vitro culture or total epidermal isolation. (See the description in FACS analysis section. Cells from total epidermal isolation do contain small contaminating populations of melanocytes and immune cells, which accounts for the low level of H3K27me3 signal in Ezh1/2 2KO cells.) Cells were crosslinked by addition of 1/10th volume of fresh 11% formaldehyde solution for 10 min at room temperature and then rinsed twice with 1 × PBS prior to freezing in liquid nitrogen and storing at −80C. Before ChIP, cells were resuspended, lysed, and sonicated to solubilize and shear crosslinked DNAs. For sonication, lysates were treated with Triton X-100 to 1% and then subjected to a Bioruptor Sonicator (Diagenode, UCD-200) according to a 30 × regimen of 30 s sonication followed by 30 s rest. The resulting whole-cell extract was incubated overnight at 4°C with 10 ml of Dynal Protein G magnetic beads (Invitrogen), which had been preincubated with ∼2 ug of the Sox2 (Stemgent), H3K27me3 (Millipore), Ezh1 (Mousavi et al, 2012), Ezh2 (Active Motif), or pan-H3 (abcam) antibodies. After ChIP, samples were washed with low salt, high salt, LiCl, and Tris-EDTA buffers for 15 min at 4°C. Bound complexes were eluted and crosslinking was reversed by overnight (o/n) incubation at 65°C. Whole-cell extract DNA was also treated for crosslink reversal.
RNA analysis
Cells were lysed using Qiagen RLT buffer with ß-mercaptoethanol and RNA isolated using the Qiagen RNeasy Mini kit with DNaseI. Reverse transcription was performed using Invitrogen Superscript III and Oligo-dT primers. All qPCR was performed using Roche SYBR green reagents and a Lightcycler480 machine.
FACS analysis
For Atoh1-GFP analysis, whole back skin was collected from Atoh1-GFP or WT mice and placed in Dispase for 1 h at 37°C, after which the epidermis was peeled from the dermis and trypsinized to give a single-cell suspension. Cells were then stained with the viability marker DAPI and analysed using a BD LSRII and BD FACS Diva software.
FACS sorting of epidermal basal cells was performed as described in Ezhkova et al (2011). Briefly, cells were collected from WT and Ezh1/2 2KO mice as for Atoh1-GFP mice above. FACS gating was done on live CD140a−CD207−CD117−CD31−Sca1+α6-integrinhigh cells to collect basal epidermal cells to the exclusion of non-epithelial cell types.
Statistics
In all column bar graphs, mean value±1 standard deviation is presented. Box-and-whisker plots show first to third quartiles around the median, with whiskers showing maximum and minimum values. All quantifications were performed on multiple cell populations from different animals. To determine the significance between two groups (as indicated in the figures by a bracket), comparisons were made using Student’s t-test (GraphPad Prism 5). For all statistical tests, the 0.05 level of confidence was accepted for statistical significance and actual P-values (to four decimal places) are provided in the figure legends.
Supplementary Material
Acknowledgments
For help, advice, and critical suggestions on our work, we would like to thank Elaine Fuchs, H Amalia Passoli, Ross Cagan, Ihor Lemischka, Robert Krauss. The Ezh1 mutant mice were generated at the Research Institute of Molecular Pathology (IMP, Vienna) by Donal O’Carroll (laboratory of Thomas Jenuwein) with the help of Maria Sibilia (laboratory of Erwin Wagner). We would like to thank Alexander Tarakhovsky for previously providing us with Ezh2-floxed mice. We also thank Elaine Fuchs for sharing K14-GFP-H2B mice. We are grateful for the assistance provided by Michael Rendl and Carlos Clavel, Jianlong Wang and Junjun Ding, Helya Ghaffari, Chenleng Cai and Aya Kitabayashi, Vittorio Sartorelli, Cedric Blanpain and Alexandra Van Keymeulen, Pinxian Xu and Hans Snoeck. We would like to acknowledge Rumana Huq and Lauren O’Rourke at the MSSM Microscopy Core. JZ was supported by DOD fellowship GRANT11010440. CNP was supported by EMBO fellowship ALTF 552-2012. EE is a Basil O'Connor Scholar with the March of Dimes foundation and receives support from NIH/NIAMS grants (R00 AR057817) and (R01 AR063724).
Author contributions: ESB, VJV, JZ, and EE designed the study and performed the experiments. SN generated the Sox2-floxed mice. SAH performed the electron microscopy analysis. JMS analysed RNA microarray data. ESB, VJV, CP, and EE wrote the manuscript with input from all authors.
Footnotes
The authors declare that they have no conflict of interest.
References
- Arnold K, Sarkar A, Yram MA, Polo JM, Bronson R, Sengupta S, Seandel M, Geijsen N, Hochedlinger K (2011) Sox2(+) adult stem and progenitor cells are important for tissue regeneration and survival of mice. Cell Stem Cell 9: 317–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit YD, Lepage MB, Khalfaoui T, Tremblay E, Basora N, Carrier JC, Gudas LJ, Beaulieu JF (2012) Polycomb repressive complex 2 impedes intestinal cell terminal differentiation. J Cell Sci 125(Pt 14): 3454–3463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain C, Fuchs E (2009) Epidermal homeostasis: a balancing act of stem cells in the skin. Nat Rev Mol Cell Biol 10: 207–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, Bell GW, Otte AP, Vidal M, Gifford DK, Young RA, Jaenisch R (2006) Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349–353 [DOI] [PubMed] [Google Scholar]
- Bracken AP, Kleine-Kohlbrecher D, Dietrich N, Pasini D, Gargiulo G, Beekman C, Theilgaard-Monch K, Minucci S, Porse BT, Marine JC, Hansen KH, Helin K (2007) The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev 21: 525–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y (2002) Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298: 1039–1043 [DOI] [PubMed] [Google Scholar]
- Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V (2004) The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev 18: 2627–2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Gu X, Su IH, Bottino R, Contreras JL, Tarakhovsky A, Kim SK (2009) Polycomb protein Ezh2 regulates pancreatic beta-cell Ink4a/Arf expression and regeneration in diabetes mellitus. Genes Dev 23: 975–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavel C, Grisanti L, Zemla R, Rezza A, Barros R, Sennett R, Mazloom AR, Chung CY, Cai X, Cai CL, Pevny L, Nicolis S, Ma'ayan A, Rendl M (2012) Sox2 in the dermal papilla niche controls hair growth by fine-tuning BMP signaling in differentiating hair shaft progenitors. Dev Cell 23: 981–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado-Olguin P, Huang Y, Li X, Christodoulou D, Seidman CE, Seidman JG, Tarakhovsky A, Bruneau BG (2012) Epigenetic repression of cardiac progenitor gene expression by Ezh2 is required for postnatal cardiac homeostasis. Nat Genet 44: 343–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donepudi S, DeConti RC, Samlowski WE (2012) Recent advances in the understanding of the genetics, etiology, and treatment of Merkel cell carcinoma. Semin Oncol 39: 163–172 [DOI] [PubMed] [Google Scholar]
- Driskell RR, Giangreco A, Jensen KB, Mulder KW, Watt FM (2009) Sox2-positive dermal papilla cells specify hair follicle type in mammalian epidermis. Development 136: 2815–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhkova E, Lien WH, Stokes N, Pasolli HA, Silva JM, Fuchs E (2011) EZH1 and EZH2 cogovern histone H3K27 trimethylation and are essential for hair follicle homeostasis and wound repair. Genes Dev 25: 485–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhkova E, Pasolli HA, Parker JS, Stokes N, Su IH, Hannon G, Tarakhovsky A, Fuchs E (2009) Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell 136: 1122–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer von Mollard G, Stahl B, Khokhlatchev A, Sudhof TC, Jahn R (1994) Rab3C is a synaptic vesicle protein that dissociates from synaptic vesicles after stimulation of exocytosis. J Biol Chem 269: 10971–10974 [PubMed] [Google Scholar]
- Fuchs E, Raghavan S (2002) Getting under the skin of epidermal morphogenesis. Nat Rev Genet 3: 199–209 [DOI] [PubMed] [Google Scholar]
- Haeberle H, Fujiwara M, Chuang J, Medina MM, Panditrao MV, Bechstedt S, Howard J, Lumpkin EA (2004) Molecular profiling reveals synaptic release machinery in Merkel cells. Proc Natl Acad Sci USA 101: 14503–14508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeberle H, Lumpkin EA (2008) Merkel Cells in Somatosensation. Chemosens Percept 1: 110–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng JC, Orlov YL, Ng HH (2010) Transcription factors for the modulation of pluripotency and reprogramming. Cold Spring Harb Symp Quant Biol 75: 237–244 [DOI] [PubMed] [Google Scholar]
- Houben R, Shuda M, Weinkam R, Schrama D, Feng H, Chang Y, Moore PS, Becker JC (2010) Merkel cell polyomavirus-infected Merkel cell carcinoma cells require expression of viral T antigens. J Virol 84: 7064–7072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, Koseki H, Fuchikami T, Abe K, Murray HL, Zucker JP, Yuan B, Bell GW, Herbolsheimer E, Hannett NM, Sun K, Odom DT, Otte AP, Volkert TL, Bartel DP, Melton DA, Gifford DK, Jaenisch R, Young RA (2006) Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125: 301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien WH, Guo X, Polak L, Lawton LN, Young RA, Zheng D, Fuchs E (2011) Genome-wide Maps of Histone Modifications Unwind In Vivo Chromatin States of the Hair Follicle Lineage. Cell Stem Cell 9: 219–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maricich SM, Wellnitz SA, Nelson AM, Lesniak DR, Gerling GJ, Lumpkin EA, Zoghbi HY (2009) Merkel cells are essential for light-touch responses. Science 324: 1580–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez AM, Cavalli G (2006) The role of polycomb group proteins in cell cycle regulation during development. Cell Cycle 5: 1189–1197 [DOI] [PubMed] [Google Scholar]
- Mascre G, Dekoninck S, Drogat B, Youssef KK, Brohee S, Sotiropoulou PA, Simons BD, Blanpain C (2012) Distinct contribution of stem and progenitor cells to epidermal maintenance. Nature 489: 257–262 [DOI] [PubMed] [Google Scholar]
- Masui S, Nakatake Y, Toyooka Y, Shimosato D, Yagi R, Takahashi K, Okochi H, Okuda A, Matoba R, Sharov AA, Ko MS, Niwa H (2007) Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol 9: 625–635 [DOI] [PubMed] [Google Scholar]
- Merkel FS (1875) Tastzellen und tastkorperchen bei den haustieren und beim menschen. Arch Mikrosk Anat 11: 636–652 [Google Scholar]
- Min J, Zhang Y, Xu RM (2003) Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev 17: 1823–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AV, Pardal R, Iwashita T, Park IK, Clarke MF, Morrison SJ (2003) Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature 425: 962–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison KM, Miesegaes GR, Lumpkin EA, Maricich SM (2009) Mammalian Merkel cells are descended from the epidermal lineage. Dev Biol 336: 76–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousavi K, Zare H, Wang AH, Sartorelli V (2012) Polycomb protein Ezh1 promotes RNA polymerase II elongation. Mol Cell 45: 255–262 [DOI] [PubMed] [Google Scholar]
- Neves J, Uchikawa M, Bigas A, Giraldez F (2012) The prosensory function of Sox2 in the chicken inner ear relies on the direct regulation of Atoh1. PLoS One 7: e30871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IK, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, Morrison SJ, Clarke MF (2003) Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature 423: 302–305 [DOI] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K (2007) The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol 27: 3769–3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Cloos PA, Walfridsson J, Olsson L, Bukowski JP, Johansen JV, Bak M, Tommerup N, Rappsilber J, Helin K (2010) JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature 464: 306–310 [DOI] [PubMed] [Google Scholar]
- Sauvageau M, Sauvageau G (2010) Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell 7: 299–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, Yuan GC, Orkin SH (2008) EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell 32: 491–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sher F, Boddeke E, Olah M, Copray S (2012) Dynamic changes in Ezh2 gene occupancy underlie its involvement in neural stem cell self-renewal and differentiation towards oligodendrocytes. PLoS One 7: e40399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana T, Sakakura Y, Ishizeki K, Iida S, Nawa T (1983) An experimental study of the influence of sensory nerve fibers on Merkel cell differentiation in the labial mucosa of the rabbits. Arch Histol Jpn 46: 469–477 [DOI] [PubMed] [Google Scholar]
- Tumbar T, Guasch G, Greco V, Blanpain C, Lowry WE, Rendl M, Fuchs E (2004) Defining the epithelial stem cell niche in skin. Science 303: 359–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Keymeulen A, Mascre G, Youseff KK, Harel I, Michaux C, De Geest N, Szpalski C, Achouri Y, Bloch W, Hassan BA, Blanpain C (2009) Epidermal progenitors give rise to Merkel cells during embryonic development and adult homeostasis. J Cell Biol 187: 91–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasioukhin V, Degenstein L, Wise B, Fuchs E (1999) The magical touch: genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc Natl Acad Sci USA 96: 8551–8556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo SH, Stumpfova M, Jensen UB, Lumpkin EA, Owens DM (2010) Identification of epidermal progenitors for the Merkel cell lineage. Development 137: 3965–3971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon H, Lee DJ, Kim MH, Bok J (2011) Identification of genes concordantly expressed with Atoh1 during inner ear development. Anat Cell Biol 44: 69–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Bardot E, Ezhkova E (2012) Epigenetic regulation of skin: focus on the Polycomb complex. Cell Mol Life Sci 69: 2161–2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.