Abstract
Polycomb group (PcG) proteins form essential epigenetic memory systems for controlling gene expression during development in plants and animals. However, the mechanism of plant PcG protein functions remains poorly understood. Here, we probed the composition and function of plant Polycomb repressive complex 2 (PRC2). This work established the fact that all known plant PRC2 complexes contain MSI1, a homologue of Drosophila p55. While p55 is not essential for the in vitro enzymatic activity of PRC2, plant MSI1 was required for the functions of the EMBRYONIC FLOWER and the VERNALIZATION PRC2 complexes including trimethylation of histone H3 Lys27 (H3K27) at the target chromatin, as well as gene repression and establishment of competence to flower. We found that MSI1 serves to link PRC2 to LIKE HETEROCHROMATIN PROTEIN 1 (LHP1), a protein that binds H3K27me3 in vitro and in vivo and is required for a functional plant PcG system. The LHP1–MSI1 interaction forms a positive feedback loop to recruit PRC2 to chromatin that carries H3K27me3. Consequently, this can provide a mechanism for the faithful inheritance of local epigenetic information through replication.
Keywords: arabidopsis , chromatin, epigenetics, MSI1, polycomb group proteins
Introduction
Most developmental decisions are based on tight regulation of transcription to establish and maintain specific gene expression patterns, and polycomb group (PcG) proteins are among the master regulators of different developmental programmes. PcG proteins were first identified in Drosophila as regulators of Hox gene expression (Lewis, 1978) and were subsequently found to represent an ancient and evolutionarily conserved mechanism of gene silencing (for reviews see Hennig and Derkacheva, 2009; Butenko and Ohad, 2011; Margueron and Reinberg, 2011). Animal and plant PcG proteins function by forming multi-subunit protein complexes such as Polycomb repressive complex 1 (PRC1) and PRC2. PRC2 is recruited to target genes and catalyses the trimethylation of lysine 27 of histone H3 (H3K27me3). Animal PRC1 binds to H3K27me3 and establishes monoubiquitylation of H2AK119. H3K27me3 is, however, not always required for PRC1 recruitment to target genes. Eventually, animal PcG proteins repress transcription by means of mechanisms that are not fully understood and that probably involve compaction of nucleosomes and interference with transcription elongation. In Drosophila and Arabidopsis, silencing by PcG proteins involves local restriction of DNA accessibility (Shu et al, 2012).
The PRC1 complex was originally characterized in Drosophila, where it consists of four main subunits: polycomb (Pc), polyhomeotic (PH), posterior sex combs (Psc) and RING (Francis et al, 2001; Mohd-Sarip et al, 2002). Pc binds to H3K27me3 (Fischle et al, 2003), and RING catalyses H2AK119 monoubiquitylation (Wang et al, 2004; de Napoles et al, 2004). Similar to animals, plant PcG function seems to involve RING proteins that can monoubiquitylate H2A (Sanchez-Pulido et al, 2008; Xu and Shen, 2008; Bratzel et al, 2010; Li et al, 2011). Although plants lack Pc homologues, LIKE HETEROCHROMATIN PROTEIN 1 (LHP1), also known as TERMINAL FLOWER 2, is considered to fulfil the role of Pc in plants based on its ability to bind to H3K27me3 in vitro and its genome-wide co-localization with H3K27me3 in vivo (Turck et al, 2007; Zhang et al, 2007). LHP1 binding to H3K27me3 is required for its function (Exner et al, 2009), and LHP1 is required for repression of several PcG protein targets such as FLOWERING LOCUS C (FLC), FLOWERING TIME (FT) and AGAMOUS (AG) (Kotake et al, 2003; Libault et al, 2005). However, it remains unknown whether LHP1 has additional functions independent of the plant PcG system.
In contrast to PRC1, homologues of all four core subunits of animal PRC2 exist in plants. The Arabidopsis genome encodes three homologues of the histone methyltransferase enhancer of zeste (E(z)): CURLY LEAF (CLF), SWINGER (SWN) and MEDEA (MEA); three homologues of the suppressor of zeste: EMBRYONIC FLOWER 2 (EMF2), FERTILIZATION INDEPENDENT SEED 2 (FIS2) and VERNALIZATION 2 (VRN2); a single extra sex comb homologue: FERTILIZATION INDEPENDENT ENDOSPERM (FIE); and five homologues of p55: MULTICOPY SUPRESSOR OF IRA 1–5 (MSI1–5). The diverse PRC2 subunit homologues in Arabidopsis probably form at least three different PRC2-like complexes with distinct functions. The VERNALIZATION (VRN) complex comprises VRN2, FIE, CLF or SWN and MSI1, and accelerates flowering in response to prolonged exposure to cold (Wood et al, 2006; De Lucia et al, 2008). The EMBRYONIC FLOWER (EMF) complex was proposed to control vegetative development and the transition to flowering and to comprise EMF2, FIE, CLF or SWN and one p55 homologue. An interaction of EMF2 with CLF was shown in vitro and in yeast two-hybrid assays (Chanvivattana et al, 2004), but the in vivo composition of the EMF complex awaits confirmation. Both EMF2 and VRN2 contribute to repression of the FLC (Gendall et al, 2001; Jiang et al, 2008). The FERTILIZATION INDEPENDENT SEED (FIS) complex has specific functions in the female gametophyte and the endosperm and comprises FIS2, FIE, MEA and MSI1 (Köhler et al, 2003; Spillane et al, 2000).
MSI1–5 proteins belong to a subfamily of WD-40 repeat proteins, which are subunits of several chromatin-remodelling complexes in animals, plants and yeast. They do not have enzymatic activity but can bind to histones and serve as protein scaffolds (for a review, see Hennig et al, 2005). Although MSI1-like proteins were usually found among the core subunits of animal PRC2, they are not required for enzymatic activity in vitro (Cao and Zhang, 2004; Ketel et al, 2005; Schmitges et al, 2011). Similarly, the role of plant MSI1-like proteins in PcG gene silencing has been under debate. Arabidopsis MSI1 was shown to be part of the FIS complex and is essential for gametophyte and seed development (Köhler et al, 2003; Guitton et al, 2004; Guitton and Berger, 2005; Leroy et al, 2007). MSI1 co-purified with VRN2 (De Lucia et al, 2008), but it is not known whether MSI1 is required for VRN complex function and the vernalization response. Finally, which of the five MSI1-like proteins function in the EMF complex has not been established yet. Deficiency of MSI1 affects shoot apical meristems and floral meristems and primordia, suggesting a role in vegetative plant development and transition to flowering (Hennig et al, 2003; Bouveret et al, 2006; Schönrock et al, 2006), possibly as part of the EMF complex. Similar to MSI1, MSI4 and MSI5 regulate the transition to flowering (Kim et al, 2004; Ausin et al, 2004; Gu et al, 2011). Recently, co-immunoprecipitation of MSI4 with CLF was shown, suggesting that MSI4 instead of MSI1 could be part of the EMF complex (Pazhouhandeh et al, 2011).
In this study, we have analysed the function of MSI1 in sporophytic PRC2 complexes in Arabidopsis. Purification of the EMF complex established MSI1 but not MSI4 as a core subunit. Similarly, MSI1 but not MSI4 interacts with EMF2. MSI1 is recruited to the chromatin of EMF target genes, where it is required for transcriptional silencing. Further, we find that MSI1 is recruited to the FLC locus where it is required for stable repression by cold and for a normal vernalization response. Our data indicate that MSI1 is an indispensable subunit of all PRC2 complexes in Arabidopsis. MSI1 was found to interact with LHP1, a major protein for PRC1-like functions in plants. We suggest that a physical link between plant PRC2-like and PRC1-like complexes contributes to the inheritance of H3K27me3 during DNA replication and to the maintenance of H3K27me3 levels during interphase.
Results
MSI1 is a core subunit of the EMF complex
EMF2 is essential for vegetative plant development (Yang et al, 1995; Yoshida et al, 2001), but the proposed EMF complex has not been isolated yet. To uncover the composition of the EMF complex in vivo, we expressed a FLAG-tagged EMF2 in Arabidopsis and immunoaffinity-purified the FLAG–EMF2 complex from inflorescences. Wild-type plants served as controls. The purified fractions from four independent experiments were analysed by mass spectrometry. Measured spectra were searched with Mascot against the Arabidopsis TAIR9 protein database using a concatenated decoy database and imported into Scaffold. Cutoffs of 90% minimal confidence for protein identification and of 95% minimal confidence for peptide identification were applied. These criteria resulted in a spectrum false-discovery rate below 1%. Only proteins identified with at least two peptides in at least two replicates but not in control samples were taken into account. Three plant PcG proteins were found to co-purify with EMF2: FIE, SWN and MSI1 (Table I and Supplementary Table S1). This is the first demonstration of the composition of the plant EMF complex in vivo, showing that the core EMF complex consists of the four main subunits EMF2, MSI1, FIE and SWN. MSI2, 3, 4 and 5 were not found in any experiment, suggesting that these MSI1 homologues are not part of the core EMF complex in inflorescences.
Table 1. EMF2 co-purifies with PcG proteins.
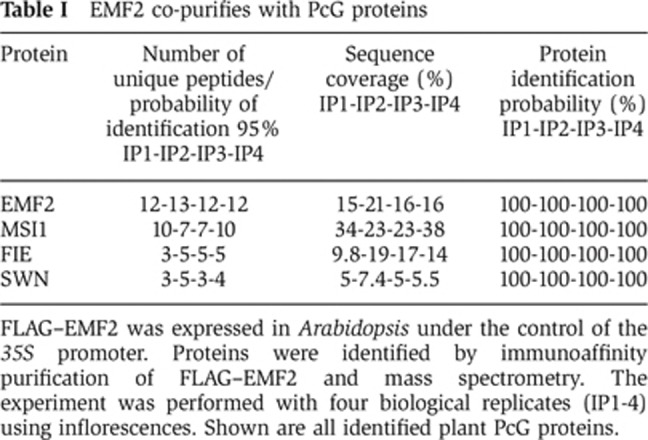
FLAG–EMF2 was expressed in Arabidopsis under the control of the 35S promoter. Proteins were identified by immunoaffinity purification of FLAG–EMF2 and mass spectrometry. The experiment was performed with four biological replicates (IP1-4) using inflorescences. Shown are all identified plant PcG proteins.
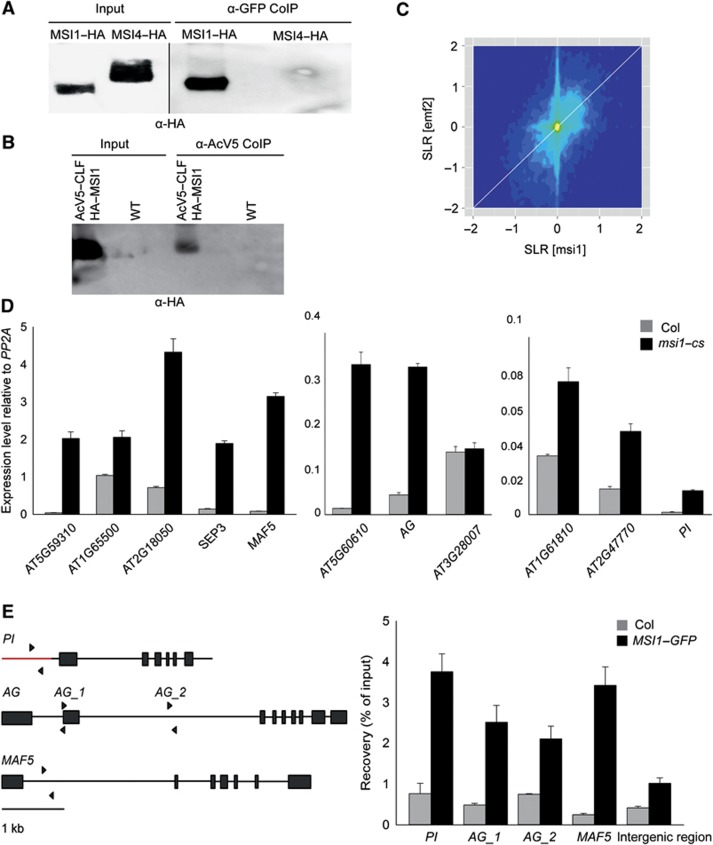
To verify the presence of MSI1 in the EMF complex, we tested the interaction of MSI1 and EMF2 in vivo. YFP-tagged EMF2 (YFP–EMF2) and HA-tagged MSI1 (HA–MSI1) or MSI4 (HA–MSI4) were transiently co-expressed in Nicotiana benthamiana leaves. YFP–EMF2 was immunoaffinity-purified, and the presence of the co-precipitating proteins was analysed on protein immunoblots. HA–MSI1 but not HA–MSI4 was co-precipitated with YFP–EMF2 (Figure 1A). This result confirms that MSI1 and EMF2 associate into a common complex in vivo. MSI4 did not interact with EMF2 in vivo in this assay. This finding not only establishes the specificity of the assay but also strengthens the notion that MSI1 but not MSI4 is a core EMF complex subunit in vivo.
Figure 1.
MSI1 is a key subunit of the EMF complex in vivo. (A) MSI1 co-purifies with EMF2. HA–MSI1 and YFP–EMF2 or HA–MSI4 and YFP–EMF2 were expressed in N. benthamiana leaves under the control of 35 S promoter. YFP–EMF2 was immunoprecipitated, and precipitates were analysed by immunoblotting using anti-HA antibodies. (B) MSI1 and CLF are present in the same complex in vivo. AcV5–CLF and HA–MSI1 were expressed in N. benthamiana leaves under the control of 35 S promoter. AcV5–CLF was immunoprecipitated, and the precipitates were analysed by immunoblotting using anti-HA antibodies. Wild-type N. benthamiana leaves were used as a control. (C) Lack of MSI1 and lack of EMF2 cause similar changes in the transcriptome. Transcript signal log ratios (SLR) for an MSI1 co-suppression line (msi1–cs) and an emf2 mutant were plotted. The colour gradient (dark blue to yellow) represents local data point density. The white diagonal line represents identical changes in msi1–cs and emf2. (D) MSI1 is needed for repression of EMF target genes. Quantitative RT–PCR was performed on cDNA from rosette leaves of 6-week-old plants. Relative expression values are shown as mean ±s.e. (n=3). Values were normalized to a PP2A gene (At1g13320). (E) MSI1 is recruited to the chromatin of the EMF target genes. Left: Genomic structure of PI, AG and MAF5. Black lines, introns; red line, promoter region; wide bars, exons. Arrows represent the position of primers used for qPCR. The intergenic control region is on chromosome 1 from 8383019 to 8383083 between At1G23700 and At1G23710. Values are recovery as percent of input; shown are mean ±s.d. (n=3).
Source data for this figure is available on the online supplementary information page.
To provide independent confirmation for the presence of MSI1 in the EMF complex, we performed reciprocal immunoaffinity purification experiments using an Arabidopsis line expressing GFP-tagged MSI1 (MSI1–GFP) (Alexandre et al, 2009) and a GFP control line. Purified fractions were analysed by mass spectrometry in order to identify proteins co-precipitating with MSI1–GFP. Four independent experiments firmly established the presence of MSI1, EMF2, FIE and SWN in the complex (Table II). PcG proteins EMF2, FIE and SWN consistently co-purified with MSI1–GFP, confirming that MSI1 is a core subunit of the EMF complex in vivo. Consistent with earlier observations (De Lucia et al, 2008), the VRN2, VRN5 and VEL1 subunits of the VRN PRC2 complex were also found to associate with MSI1 in vivo (Table II). Several non-PcG proteins co-purified with MSI1, including homologues of yeast Rpd3 histone deacetylase complexes (Supplementary Table S2). To confirm these results, we performed additional immunoaffinity purification experiments using a modified protocol involving protein–protein cross-linking prior to protein extraction. These experiments confirmed the presence of the initially identified MSI1 interactors, except for VRN5, and revealed additional candidate interactions (Table III and Supplementary Table S2). Notably, the plant PcG protein LHP1 was found with high confidence in both additional experiments.
Table 2. MSI1 co-purifies with PcG proteins.
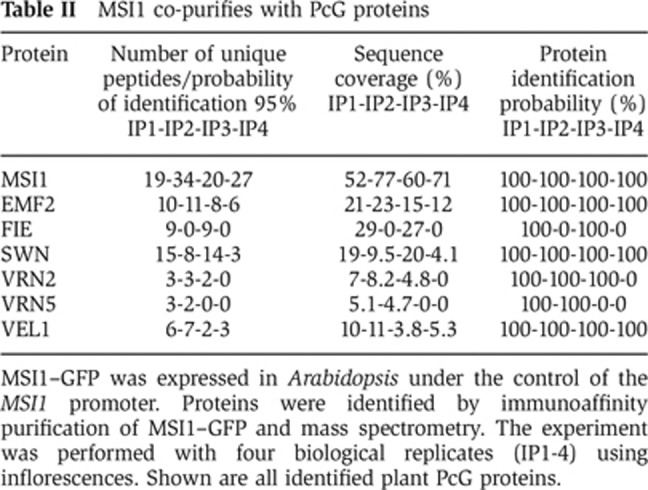
MSI1–GFP was expressed in Arabidopsis under the control of the MSI1 promoter. Proteins were identified by immunoaffinity purification of MSI1–GFP and mass spectrometry. The experiment was performed with four biological replicates (IP1-4) using inflorescences. Shown are all identified plant PcG proteins.
Table 3. Co-purification of MSI1 with PcG proteins from cross-linked protein extracts.
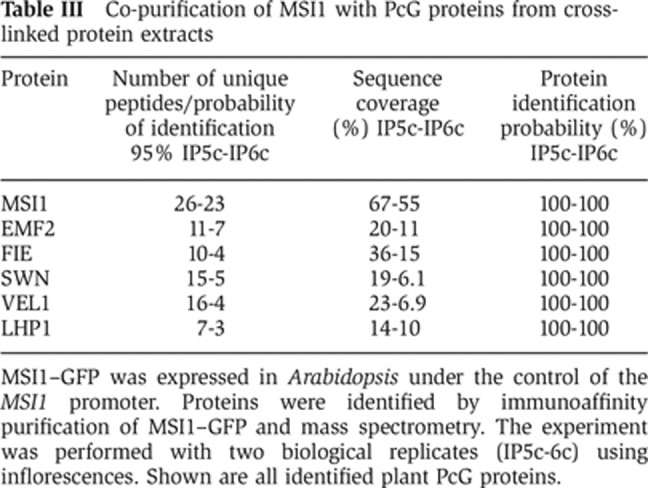
MSI1–GFP was expressed in Arabidopsis under the control of the MSI1 promoter. Proteins were identified by immunoaffinity purification of MSI1–GFP and mass spectrometry. The experiment was performed with two biological replicates (IP5c-6c) using inflorescences. Shown are all identified plant PcG proteins.
Unexpectedly, the well-characterized Arabidopsis PcG protein CLF (Goodrich et al, 1997) was not found among the MSI1-binding partners. CLF plays a major role during sporophytic plant development (Goodrich et al, 1997; Chanvivattana et al, 2004; Katz et al, 2004; Wood et al, 2006; Jiang et al, 2008; Doyle and Amasino, 2009) and interacts with EMF2 in vitro and in yeast two-hybrid assays (Chanvivattana et al, 2004), suggesting that CLF is part of the EMF complex. Identification of proteins by mass spectrometry is affected by many protein-specific factors including protein abundance (Lubec and Afjehi-Sadat, 2007), and it is possible that CLF interacts with MSI1 but failed to be detected under our experimental conditions. This notion was supported by the considerably weaker expression of CLF compared with SWN at both transcript and protein levels (Zimmermann et al, 2004; Baerenfaller et al, 2011). Therefore, we tested whether MSI1 interacts with CLF in vivo using an alternative approach. AcV5-tagged CLF (AcV5–CLF) and HA–MSI1 were transiently co-expressed in tobacco leaves, AcV5–CLF was immunoaffinity-purified, and the presence of the co-precipitating proteins was analysed on protein immunoblots. HA–MSI1 was co-precipitated with AcV5–CLF (Figure 1B). This result demonstrates that MSI1 and CLF can associate into a common complex in vivo.
Together, these experiments establish that MSI1, EMF2 and FIE, together with SWN or CLF, constitute the EMF complex. In contrast, there is no strong evidence for functions of MSI2–5 in the EMF complex.
MSI1 is essential for the function of the EMF complex
To establish whether MSI1 is required for the function of the EMF complex, we determined the expression levels of EMF target genes in an MSI1 co-suppression line (msi1–cs) in which the MSI1 protein level is reduced to less than 10% (Hennig et al, 2003). We compared the transcriptional profiles of msi1–cs (Alexandre et al, 2009) and emf2 plants (Liu et al, 2012) and found that transcriptional changes were strongly and significantly correlated between plants of the two genotypes (Pearson correlation=0.44, P<2.2e−16) (Figure 1C). Note that this strong correlation was observed despite considerable differences in experimental conditions (rosette leaves of 23-day-old msi1–cs plants that retain ∼5% MSI1 protein and 7-day-old emf2-null mutant seedlings). The global similarity of transcriptional changes caused by reduced MSI1 or EMF2 loss of function strongly suggests that the biochemical interaction of MSI1 and EMF2 is of functional relevance. The data also confirm that redundancy among MSI1 homologues is limited and that MSI2–5 can only partially, if at all, substitute MSI1 in the EMF complex.
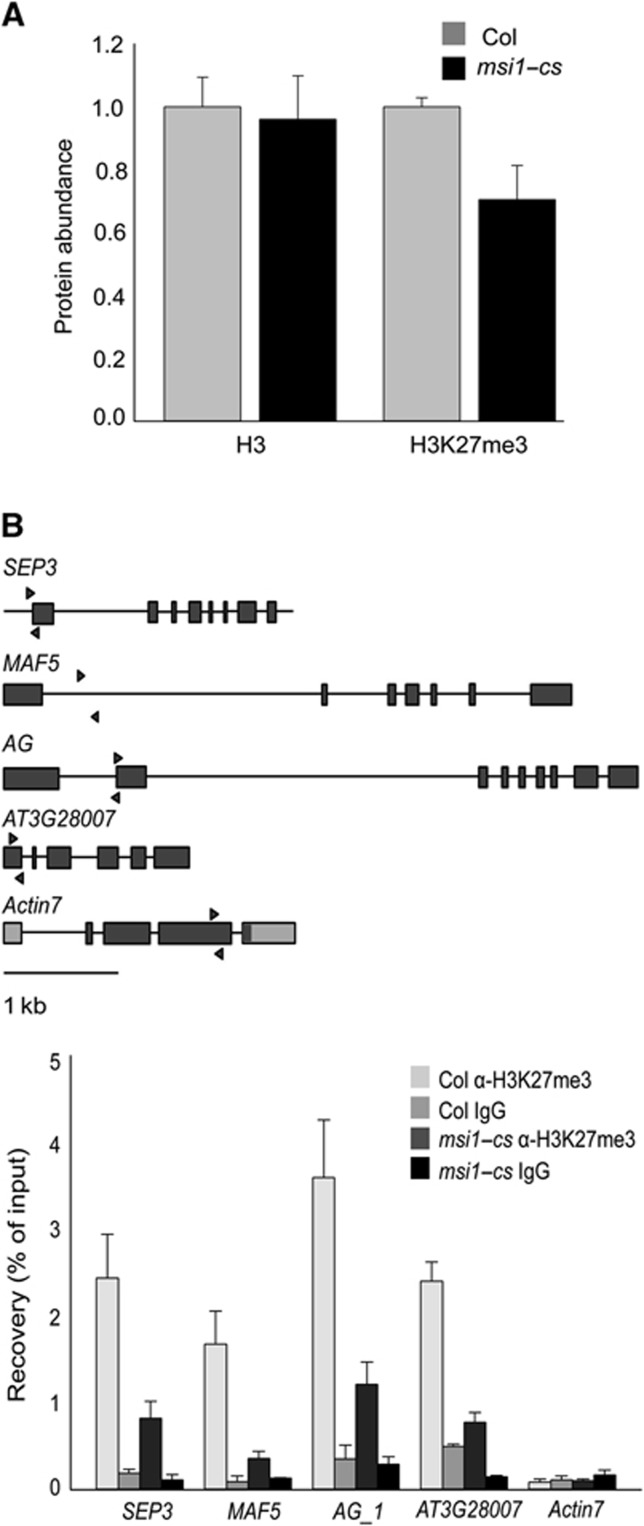
To confirm the microarray data on deregulation of EMF target genes in msi1–cs plants, we tested the expression of some known PcG target genes in leaves (Lafos et al, 2011) by RT–qPCR using independent samples (Figure 1D). Ten of 11 tested PcG target genes were upregulated in msi1–cs plants, demonstrating that the presence of MSI1 in the EMF complex is necessary for the repression of many EMF target genes. Next, we used ChIP to test whether MSI1 binds to EMF target genes. The results show an enrichment of MSI1 at the previously described EMF target genes PISTILLATA (PI), AG and MADS AFFECTING FLOWERING 5(MAF5) (Figure 1E), demonstrating that MSI1 is recruited to at least some EMF target genes. Because PRC2 complexes trimethylate H3K27 in target chromatin, we tested whether MSI1 is needed for this PRC2 function. We found that global H3K27me3 levels were reduced to 70% in msi1–cs plants (Figure 2A, Supplementary Figure S1). Similarly, ChIP results also showed that H3K27me3 is highly reduced in EMF target genes in msi1–cs plants (Figure 2B). Notably, At3g28007 has no increase in expression in msi1–cs but has reduced H3K27me3 demonstrating that loss of H3K27me3 is not a consequence of increased transcription. Together, these results demonstrate that MSI1 is required for full PRC2 function and normal H3K27me3 levels in vivo. Because the MSI1-like subunit was found to be dispensable for PRC2 catalytic activity in vitro (Schmitges et al, 2011), our findings suggest that MSI1 functions in PRC2 regulation or targeting in vivo.
Figure 2.
MSI1 is needed for trimethylation of H3K27. (A) Global H3K27me3 levels are reduced in msi1–cs plants. Total protein levels were analysed by quantitative immunoblotting using anti-H3K27me3 and anti-H3 antibodies in Col and msi1–cs plants. Shown are mean ±s.d. (n=3). (B) H3K27me3 is reduced at the chromatin of EMF target genes in msi1–cs plants. Top: genomic structure of SEP3, MAF5, AG, AT3G28007 and ACTIN7. Black lines, introns; wide bars, exons. Arrows represent the position of primers used for qPCR. Values are recovery as percent of input; shown are mean ±s.d. (n=3).
MSI1 regulates FLC expression and the vernalization response
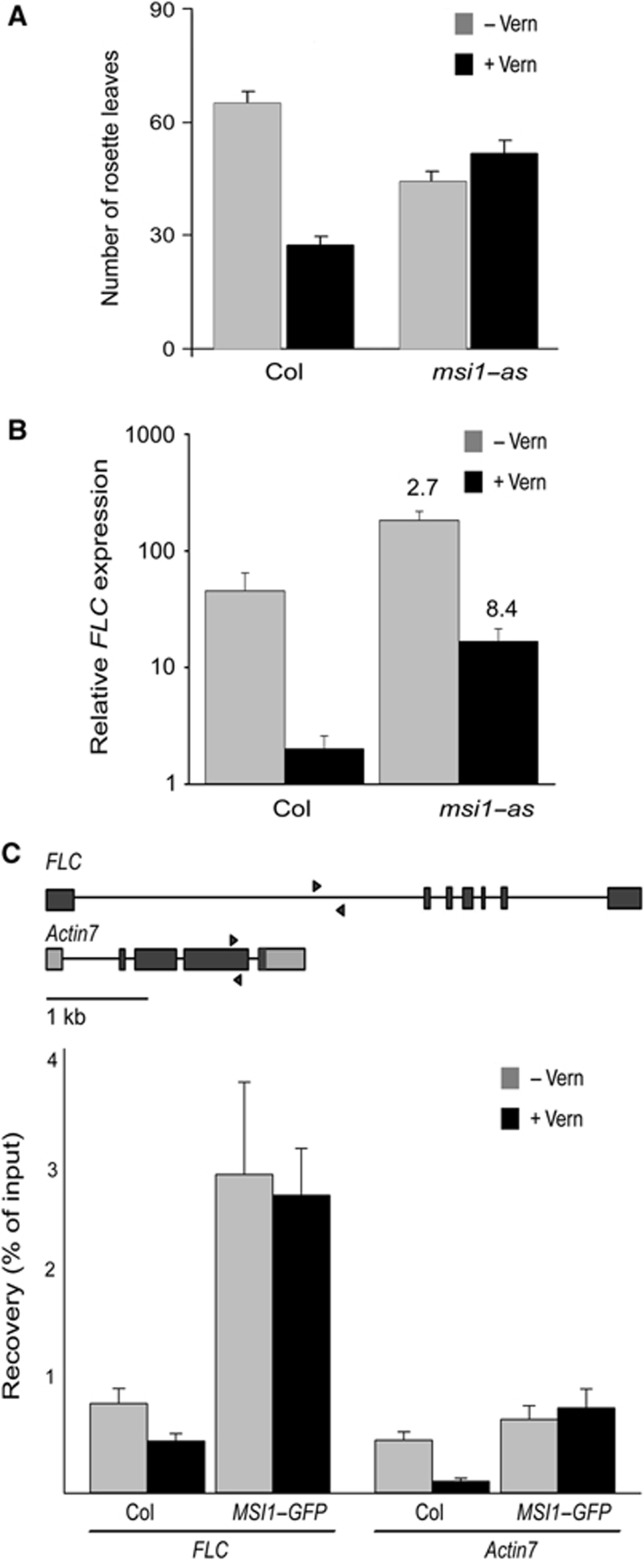
MSI1 is a subunit of the VRN–PHD complex (Table II and De Lucia et al, 2008), which represses FLC after vernalization, but the function of MSI1 in this complex has not been addressed so far. The msi1–cs line showed MSI1 protein reduction and developmental alterations only at the rosette stage (Hennig et al, 2003) and thus did not appear suitable for testing MSI1 function in seedling vernalization. In contrast, MSI1 anti-sense (msi1–as) lines contain about 30–50% of wild-type MSI1 levels in seedlings and exhibit developmental alterations at seedling and rosette stages (Exner et al, 2006). To test whether MSI1 also functions in the vernalization response, we analysed flowering time and FLC expression with and without vernalization in msi1–as and wild-type plants. Vernalized wild-type plants flowered earlier than non-vernalized plants, forming only about half the number of rosette leaves (Figure 3A). Consistent with the phenotype, FLC transcript levels were strongly reduced in vernalized wild-type plants compared with non-vernalized controls (Figure 3B). In contrast, vernalized msi1–as plants flowered similarly to non-vernalized msi1–as plants (Figure 3A), revealing that a normal vernalization response requires MSI1. Non-vernalized msi1–as plants flowered earlier than non-vernalized wild type, possibly because of a partial loss of repression of floral activators that are under PcG protein control. Without vernalization, FLC levels were increased in msi1–as (Figure 3B). Under such conditions, FLC is controlled by the EMF complex (Jiang et al, 2008), and the increased FLC expression in msi1–as is consistent with the requirement for MSI1 in EMF complex function. More importantly, vernalization was less effective in reducing FLC transcript levels in msi1–as than in wild-type plants (11-fold versus 22-fold reduction) (Figure 3B). The reduced efficiency of vernalization treatments to repress FLC and accelerate flowering in msi1–as demonstrates that MSI1 is required for a normal vernalization response.
Figure 3.
MSI1 functions in the vernalization response via regulation of FLC expression. (A) The vernalization response is strongly impaired in MSI1 anti-sense plants (msi1–as). Plants were vernalized for 6 weeks followed by cultivation in SD. Flowering time was measured as the number of rosette leaves produced before bolting. Shown are means±SE (n≤14). (B) FLC is only partially repressed by vernalization in msi1–as plants. Quantitative RT–PCR was performed on cDNA from vernalized (6 weeks at 4°C and 10 days at 23°C) and non-vernalized (10 days at 23°C) plants grown in SD. Relative expression values are shown as mean ±SE (n=3). Values were normalized to a PP2A gene. Values shown above bars represent fold change relative to the wild-type control. (C) MSI1 is recruited to the FLC locus. Top: Genomic structure of FLC and ACTIN7. Black lines, introns; wide bars, exons. Arrows represent the position of primers used for qPCR. Values are recovery as percent of input; shown are mean ±s.d. (n=3).
Next, we tested whether regulation of FLC by MSI1 is direct. In ChIP experiments, MSI1 was enriched at FLC both without and after vernalization (Figure 3C), demonstrating that MSI1 is indeed recruited to FLC. The core VRN complex is present at the FLC locus already without vernalization (De Lucia et al, 2008) and EMF2 also regulates FLC (Jiang et al, 2008), suggesting that MSI1 can bind to FLC as part of the EMF complex and as part of the VRN complex. Together, these results demonstrate that MSI1 is needed for VRN complex function and a normal vernalization response.
MSI1 connects LHP1 to PRC2
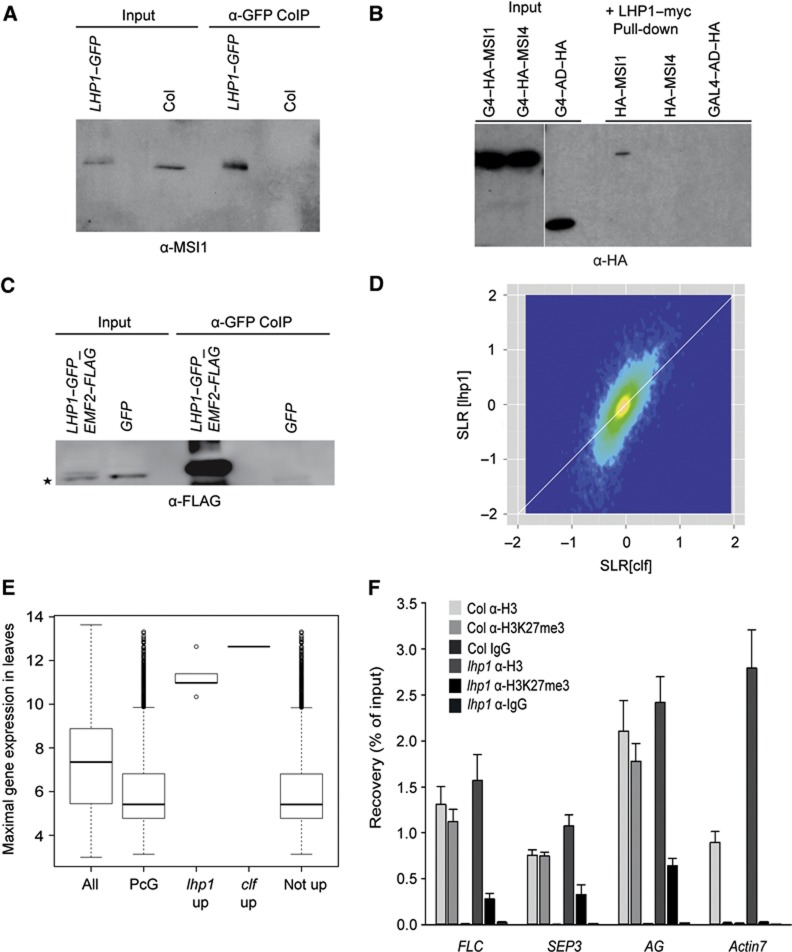
The cross-linked immunoaffinity purification of MSI1–GFP identified LHP1 among the interacting proteins (Table III). To confirm the interaction between MSI1 and LHP1 in vivo, we performed a co-immunoprecipitation (CoIP) assay. Immunoblot analyses revealed co-immunoprecipitation of MSI1 with LHP1, demonstrating that LHP1 and MSI1 indeed coexist in shared complex(es) in vivo (Figure 4A).
Figure 4.
MSI1 connects LHP1 to PRC2. (A) MSI1 co-purifies with LHP1. LHP1–GFP was immunoprecipitated from inflorescences of 35 S::LHP1–GFP plants, and precipitates were analysed by immunoblotting using anti-MSI1 antibodies. Col wild-type plants served as control. (B) MSI1 interacts directly with LHP1. LHP1–myc was immunoprecipitated from extracts of yeast expressing either HA–GAL4–AD–MSI1 and LHP1–myc or HA–GAL4–AD-–MSI4 and LHP1–myc or HA–GAL4–AD and LHP1–myc. Precipitates were analysed by immunoblotting using anti-HA antibodies. (C) LHP1 co-purifies with EMF2. GFP was immunoprecipitated from inflorescences of plants expressing LHP1–GFP and EMF2–FLAG or GFP, respectively, under the control of the 35S promoter and analysed by immunoblotting using anti-FLAG antibodies. The asterisk marks an unspecific, cross-reacting band. (D) LHP1 function in gene silencing is restricted to the PcG system. Lack of LHP1 and lack of CLF cause similar changes in the transcriptome. Signal log ratios (SLR) for a clf and an lhp1 mutant were plotted. The colour gradient (dark blue to yellow) represents local data point density. The white diagonal line represents identical changes in clf and lhp1. (E) Only PcG target genes with the potential to be expressed in leaves are upregulated in lhp1 and clf mutants. A gene’s potential to be expressed in wild-type leaves was estimated as its maximal expression in wild-type leaves according to the developmental series of AtGenExpress transcriptome data (Schmid et al, 2003). (All) all genes; (PcG) all PcG target genes in leaves (Lafos et al, 2011); (lhp1 up) PcG target genes from leaves that are upregulated in lhp1; (clf up) PcG target genes from leaves that are upregulated in clf; (not up) PcG target genes from leaves that are not upregulated in lhp1 or clf. While most PcG genes have very low leaf expression potentials and are thus inactive throughout wild-type leaf development, PcG target genes that were upregulated in lhp1 or clf had very high expression potentials and thus are active at certain stages of wild-type leaf development. (F) H3K27me3 at PcG target genes is reduced in lhp1 mutants. ChIP was done using roots enriched for dividing cells by 2,4-D treatment. Values are recovery as percent of input; shown are mean ±s.d. (n=3).
To determine whether MSI1 interacts directly with LHP1, we carried out an in vitro pull-down assay. HA–GAL4–AD-tagged MSI1, HA–GAL4–AD-tagged MSI4 and myc-tagged LHP1 were expressed in yeast, and extracts were used for immunoprecipitation with anti-myc antibodies. Immunoblot analyses revealed the presence of MSI1 but not of MSI4 or the negative control in the bound fraction (Figure 4B). The binding of MSI1 and not of MSI4 to LHP1 in the absence of any other plant proteins strongly suggests that the MSI1–LHP1 interaction is specific and direct.
LHP1 fulfils PRC1-like functions in plants (Turck et al, 2007; Zhang et al, 2007; Exner et al, 2009), and we found that LHP1 interacts with MSI1. Thus, it appeared possible that LHP1 interacts with plant PRC2 complexes via MSI1. To test whether LHP1 also interacts with EMF2 in vivo, we performed a CoIP assay using EMF2–FLAG LHP1–GFP double-transgenic plants. Immunoblot analyses clearly showed that EMF2 co-precipitated with LHP1, demonstrating that both proteins coexist in shared complex(es) in vivo (Figure 4C). Because LHP1 interacted with both MSI1 and EMF2, which are present together in the EMF complex, we conclude that LHP1 interacts with the EMF complex in vivo. This suggests that in plants PRC1- and PRC2-like functions are closely integrated.
LHP1 function in gene silencing is restricted to the PcG system
Our finding that MSI1 connects LHP1 to plant PRC2 complexes extends previous findings of LHP1 functions in the plant PcG system (Kotake et al, 2003; Libault et al, 2005; Mylne et al, 2006; Sung et al, 2006; Turck et al, 2007; Zhang et al, 2007; Xu and Shen, 2008; Exner et al, 2009; Bratzel et al, 2010; Latrasse et al, 2011). At the same time, our results raise the question regarding the extent to which LHP1 may function independently of the PcG system. LHP1 is a homologue of HP1 and SWI6, which in metazoa and fission yeast, respectively, function in heterochromatic gene silencing (Zeng et al, 2010) and can bind to heterochromatic H3K9me2 in vitro. To search for potential PcG-unrelated functions of LHP1, we profiled transcriptional changes in lhp1 and clf mutants. One concern with transcript profiling experiments in lhp1, clf and other mutants with pleiotropic phenotypes is the confounding effect of secondary transcriptional changes. Because the pleiotropic phenotype of clf is mostly suppressed under short-day photoperiods (SD) (Schatlowski et al, 2010), the experiment was carried out in SD. The pleiotropic phenotype of lhp1 is less repressed by SD but greatly depends on FT (Kotake et al, 2003). Therefore, we used a lhp1 ft double mutant. Together, we expect that these conditions will considerably reduce secondary transcriptional changes. The transcriptional changes between lhp1 and clf were strongly and significantly correlated (Pearson correlation=0.725, P=2.2e−16) (Figure 4D). There was no considerable subpopulation of genes that was miss-expressed in lhp1 and not changed in clf. The amplitude of changes, however, was frequently higher in lhp1 than in clf (cf. the deviation from the diagonal in Figure 4D). Linear regression suggested that fold changes were on average two-fold larger in lhp1 than in clf, which was probably caused by partial redundancy between CLF and SWN. A Venn diagram representation of the most strongly upregulated genes in emf2, clf, msi1–cs and lhp1 plants shows considerable overlap (Supplementary Figure S2). Differences between gene sets are probably caused by false negatives, differences in plant material and assay conditions and by partial redundancy of some of the genes. It is interesting to note that only a subset of PcG target genes lost repression in the mutants. It was possible that only genes that have the potential to be expressed in leaves were detected as upregulated in lhp1 or clf rosette leaves. We tested this hypothesis using gene-specific leaf expression potentials that were based on all wild-type leaf samples in the developmental AtGenExpress data resource, including cotyledons, rosette and cauline leaves of diverse age or harvesting time (Schmid et al, 2003). The leaf expression potential for a gene is the maximal expression of this gene observed in any of the wild-type leaf samples. Genes with low leaf expression potentials are inactive throughout wild-type leaf development, while genes with high leaf expression potentials are active at certain stages of leaf development. Leaf expression potentials of PcG target genes were considerably smaller than the genome average, demonstrating that many PcG target genes were not expressed in the leaf samples (Figure 4E). Similarly, the PcG target genes that were not upregulated in lhp1 or clf had generally low expression potentials, demonstrating that most of them were not expressed in any leaf sample. In contrast, the PcG target genes that were strongly upregulated in lhp1 or clf had a very high expression potential, demonstrating that they were highly expressed in some leaf samples (Figure 4E). This result is in agreement with the proposal that PcG targets become upregulated in PcG mutants only in tissues in which they have a potential to be expressed (Farrona et al, 2011). Together, our data establish that the main function of LHP1 in gene regulation is related to the PcG system and that there is no evidence of a PcG-independent function of LHP1.
LHP1 is needed to establish full H3K27me3 levels
During the S-phase, new histones are incorporated into replicating chromatin, and existing histone modifications are transiently diluted. Cells have various mechanisms for re-establishing local histone modifications during replication. We sought to determine whether LHP1 could recruit plant PRC2 complexes to PcG target genes and contribute to the re-establishment of H3K27me3 in dividing cells. In order to establish the high number of dividing cells needed to test this hypothesis, lateral root outgrowth was induced by the synthetic auxin 2,4-Dichlorophenoxyacetic acid in wild-type and lhp1 seedlings (Supplementary Figure S3). ChIP with anti-H3 and anti-H3K27me3 antibodies was performed using roots. In wild type, H3K27me3 signals were detected at known PcG protein target genes including SEP3 and FLC but not at the negative control gene ACT7. Consistent with our hypothesis of a contribution of LHP1 to the establishment of H3K27me3 in dividing cells, H3K27me3 of the PcG target genes was significantly lower in lhp1 than in wild type (Figure 4F). Thus, the discovered physical link between LHP1 and PRC2 is highly relevant for the function of the plant PcG protein system.
Discussion
MSI1-like proteins form an evolutionarily conserved family of proteins that is present in all organisms except prokaryotes. Some organisms such as Drosophila have only one MSI1 homologue, while others such as Arabidopsis have several (for review, see Hennig et al, 2005). It has been suggested that MSI1-like proteins function via their histone H3 and H4 binding pockets as it has been demonstrated in detail for the Drosophila MSI1 homologue p55 (Song et al, 2008; Nowak et al, 2011; Schmitges et al, 2011). Drosophila p55 is involved in many chromatin-remodelling complexes: CHROMATIN ASSEMBLY FACTOR1 (CAF-1), histone deacetylase complexes, histone acetyl–transferase complex, the nucleosome-remodelling factor NURF complex and PRC2 (Tyler et al, 1996; Martinez-Balbas et al, 1998; Tie et al, 2001; Czermin et al, 2002; Müller et al, 2002; Nekrasov et al, 2005). Notably, conflicting results have been published about the role of p55 in Drosophila PRC2: although p55 is not required for PRC2 enzymatic activity in vitro (Cao and Zhang, 2004; Ketel et al, 2005) and complete loss of the p55 gene did not affect global H3K27me3 levels in fly larvae (Wen et al, 2012), sectorial loss of p55 caused reduced H3K27me3 in eye discs (Anderson et al, 2011). In organisms that have multiple MSI1-like genes, it is not clear how much functional redundancy exists between them. In Arabidopsis, there are three main clades of MSI1-like genes, MSI1, MSI2/MSI3 and MSI4/MSI5, which evolved before the divergence of monocots and dicots (Hennig et al, 2005). The existence of five MSI1-like proteins in Arabidopsis raises the question of whether only one or different MSI1-like proteins are subunits of the various PRC2 complexes and whether they are functionally redundant. Earlier work had revealed that MSI1 has an essential function in the FIS–PRC2 complex during seed development that is not redundant with MSI2–5 (Köhler et al, 2003; Guitton et al, 2004; Guitton and Berger, 2005; Leroy et al, 2007). However, knowledge about the predicted MSI1-like subunit in the sporophytic PRC2 complexes in Arabidopsis has remained fragmented. Because MSI1-like proteins are not needed for in vitro PRC2 enzymatic activity in animals, it was even possible that trimeric PRC2 complexes lacking an MSI1-like subunit exist.
MSI1 functions in the EMF and VRN complexes
On the basis of genetic and in vitro protein–protein interaction data, the EMF complex is considered to be the major sporophytic plant PRC2 complex (Yoshida et al, 2001; Chanvivattana et al, 2004; Katz et al, 2004; Schönrock et al, 2006; Jiang et al, 2008). Here, we provide biochemical in vivo evidence for the presence and composition of the EMF complex. We found that this complex comprises MSI1, EMF2, FIE and SWN. MSI1 and FIE are also known to be subunits of the FIS (Köhler et al, 2003) and VRN complexes (De Lucia et al, 2008) and are thus conserved among all known Arabidopsis PRC2 complexes. Interestingly, the histone methyltransferase SWN was well represented in the EMF and VRN complexes, while its homologue CLF was not or was only weakly represented (this work and De Lucia et al, 2008). The role of CLF as a key PcG protein in sporophyte development has been well established (Goodrich et al, 1997; Chanvivattana et al, 2004; Katz et al, 2004; Wood et al, 2006; Jiang et al, 2008; Doyle and Amasino, 2009), and we confirmed that CLF can form a complex with MSI1 in vivo. Several explanations exist for the low representation of CLF in PRC2 complexes analysed by MS/MS: first, identification of CLF and SWN in MS/MS assays could differ, as it has been observed for other proteins (for review, see Lubec and Afjehi-Sadat, 2007). Second, SWN could associate with other PRC2 subunits stronger than CLF, resulting in a preferential loss of CLF during purification. Third, stronger expression of SWN as evident from transcript and protein abundance compendia (Zimmermann et al, 2004; Baerenfaller et al, 2011) could lead to higher levels of SWN- than CLF-containing PRC2 complexes. Genetic analysis showed that CLF is partially redundant with SWN (Chanvivattana et al, 2004) but that only clf and not swn mutants have obvious developmental defects. The strong developmental alterations upon loss of the less abundant CLF may be caused by a subset of CLF-specific PcG protein target genes not shared with SWN-containing PRC2 complexes. Notably, most of the strongest developmental alterations in clf depend on misexpression of a few genes, including AG and FT (Goodrich et al, 1997; Lopez-Vernaza et al, 2012), and it is possible that these genes specifically depend on a CLF complex. Future experiments will have to establish the differential roles of CLF and SWN in plant PRC2 complexes; here, we conclude from our own and other data (De Lucia et al, 2008) that SWN is a major histone methyltransferase subunit in the EMF and VRN complexes in vivo.
Unlike the p55 function in Drosophila PRC2 that remains controversial (Anderson et al, 2011; Schmitges et al, 2011; Wen et al, 2012), we found that Arabidopsis MSI1 is required for EMF complex function in vivo and that it is recruited to the chromatin of at least some EMF target genes. MSI1 has four homologues in Arabidopsis, of which MSI4 and MSI5 are known to act redundantly in the repression of FLC and its homologues MAF4 and MAF5, promoting floral transition (Ausin et al, 2004; Kim et al, 2004; Gu et al, 2011). Both proteins associate with HISTONE DEACETYLASE 6 (HDA6) and are recruited to the FLC locus, leading to de-acetylation of histones and silencing of FLC (Gu et al, 2011). MSI4 was also implicated in the silencing of FLC and FT through its association with the CLF–PRC2 complex and the cullin-RING ubiquitin ligase (CUL4 DDB) (Pazhouhandeh et al, 2011). Here, we show that MSI1 is recruited to the FLC locus and that it is required for FLC repression, demonstrating that MSI4 and MSI5 cannot substitute for MSI1 function in FLC silencing. MSI4 and MSI5 were also not found in the EMF or VRN complexes in vivo (this work; De Lucia et al, 2008) and MSI4 failed to interact with EMF2 in vitro. Together, we suggest that MSI1 functions as the subunit of the EMF and VRN complexes that is homologous to p55 in Drosophila PRC2, while MSI4 interacts with PRC2 as part of histone deacetylase and/or CUL4 DDB complexes.
MSI1 functions in the VRN complex and is required for the vernalization response
A vernalization response is the increase in capacity to flower after long exposure to cold. It ensures that plants start flowering only in the spring when conditions are optimal. The main effect of vernalization at the molecular level is a cold-induced epigenetic silencing of FLC by the VRN PRC2 complex, which is maintained through further plant development (Michaels and Amasino, 1999; Sheldon et al, 1999; Gendall et al, 2001; Bastow et al, 2004). The core VRN complex is already associated with the FLC locus before cold treatment. After exposure to cold, the VRN–PHD complex, which contains the additional three PHD-finger proteins VRN5, VIL2 and VIN3, promotes spreading of H3K27me3 over FLC (De Lucia et al, 2008). MSI1 co-purified with the VRN–PHD complex in vivo (this work and De Lucia et al, 2008), but it was not known whether this reflects a function of MSI1 in the vernalization response. Here, we report that MSI1 is present at the FLC locus and is required for normal repression of FLC and for accelerated flowering after vernalization. Thus, MSI1 is essential for the function of the core VRN complex and of the VRN–PHD complex in vivo and is needed for vernalization response in plants.
MSI1 bridges LHP1 to PRC2
Parts of the PcG system are conserved between animals and plants, but it has remained unclear how far the functional similarities extend (Hennig and Derkacheva, 2009). In particular, repression by PcG proteins in animals requires the coordinated function of PRC2 and PRC1, but the identity of PRC1 complexes in plants has not yet been fully established. In Arabidopsis, LHP1 is considered to be one of the main proteins with a PRC1-like function (Turck et al, 2007; Zhang et al, 2007; Exner et al, 2009). We found that LHP1 can directly bind MSI1 in vitro and can be co-immunoprecipated with MSI1 and EMF2 in vivo, establishing that LHP1 and the EMF–PRC2 complex interact. Although the function of LHP1 in the plant PcG system has been established before, it remained possible that LHP1 had not only PcG-related but also PcG-independent functions. This idea was supported by the homology of LHP1 to animal HP1 and fission yeast SWI6, by some reports of LHP1 targeted to heterochromatin (Zemach et al, 2006) and by the finding that LHP1 binds methylated H3K9 (Turck et al, 2007; Zhang et al, 2007). Our genome-wide transcript profiling established that LHP1 and CLF have very similar effects in the transcriptome. Thus, we conclude that the main function of LHP1 in gene regulation is restricted to the PcG system.
Possible functions of an LHP1–PRC2 interaction
It is possible that the LHP1–PRC2 interaction facilitates recruitment of PRC2 to target genes. In animals and plants, targeting of PcG proteins is poorly understood. It has been suggested for Drosophila PcG proteins that they are recruited by DNA-binding proteins and also by non-coding RNAs (ncRNAs) (for review, see Sawarkar and Paro, 2010). At least in some cases, ncRNAs contribute to PcG targeting in Arabidopsis as well, such as during FLC repression by the VRN complex (Heo and Sung, 2011). Because LHP1 is recruited by the transcription factors SCARECROW and SHORT VEGETATIVE PHASE to the MAGPIE and SEP3 loci, respectively (Cui and Benfey, 2009; Liu et al, 2009), it is possible that Arabidopsis PRC2 recruitment to some target genes depends on transcription factor—LHP1 interactions.
Histone demethylation, as well as incorporation of newly synthesized, non-methylated histones during DNA replication and histone exchange in interphase, causes a continuous loss of H3K27me3 from target chromatin, and stable PcG silencing requires re-establishment of full H3K27me3 levels at target loci. In mammals, the ESC subunit of PRC2 can bind to H3K27me3, suggesting a self-recruiting mechanism to counteract loss of H3K27me3 during DNA replication, histone exchange or demethylation (Hansen et al, 2007; Margueron et al, 2009), while Drosophila PcG proteins remain associated with DNA during replication independent of H3K27me3 (Petruk et al, 2012). For plants, our findings suggest a model in which LHP1 assists in the recruitment of PRC2 to target sites for re-establishing reduced H3K27me3 levels. This model is consistent with the high LHP1 expression in proliferating cells (Kotake et al, 2003; Baerenfaller et al, 2011) and the interaction of LHP1 with the POL2a subunit of DNA polymerase epsilon (DNA Pol ε) (del Olmo et al, 2010). Because LHP1 interacts with plant POL2a, it is possible that LHP1 functions during S-phase in the re-establishment of full H3K27me3 levels after replication (Figure 5). This model is strongly supported by the H3K27me3 ChIP experiment in lhp1 mutants. The model we propose for inheritance of the repressed state of PcG target genes in plants has striking similarity to a model of inheritance of heterochromatic states in yeast and animals (Bannister et al, 2001). In the latter, the LHP1 homologues SWI6 and HP1 bind to H3K9me2 containing chromatin and recruit the H3K9 methyltransferase CLR4/SUV39H1 to re-establish H3K9me2 after replication. Similar to plant LHP1, which interacts with DNA Pol ε, SWI6 and HP1 are recruited to DNA replication forks (Lewis, 1978; Murzina et al, 1999). Thus, it is possible that a common function of HP1 homologues is based on their association with DNA-replication forks to recruit effector proteins such as histone methyltransferases to target chromatin.
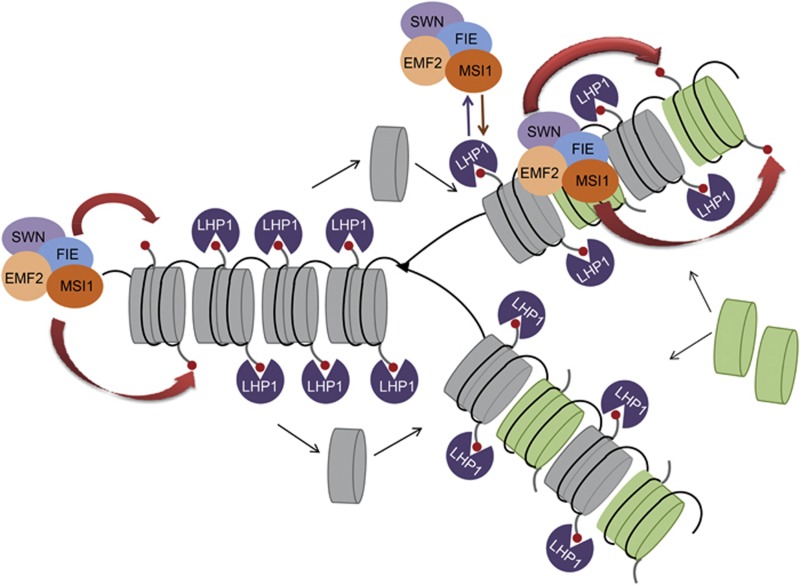
Figure 5.
Model of LHP1 function in semi-conservative inheritance of H3K27me3. During DNA replication, new histones are incorporated into chromatin diluting epigenetic marks. We propose that LHP1 binds to nucleosomes with old histones that carry H3K27me3 and via binding to MSI1 recruits the EMF complex, which trimethylates H3K27 of newly incorporated histones. H3K27me3 is symbolized by red circles, old and new nucleosomes are grey and green, respectively.
Together, we propose that MSI1 functions in Arabidopsis PRC2 complexes to link PRC2 to LHP1, which then serves to tether PRC2 to target chromatin and maintain full H3K27me3 levels after DNA replication, histone exchange or demethylation.
Materials and methods
Plant material and growth conditions
Wild-type plants were Arabidopsis thaliana accession Columbia (Col). Transgenic plants were generated by floral dip with Agrobacterium tumefaciens (strain GV 3101) (Logemann et al, 2006). To generate constructs for tagged EMF2, CLF, MSI1 and MSI4 proteins, cDNAs were cloned into vectors pEarleyGate 201, 202 and 204 (Earley et al, 2006), which were transformed into Col plants or infiltrated into leaves of N. benthamiana as previously described (Goodin et al, 2002). The MSI1–GFP, LHP1–GFP, MSI1 co-suppression (msi1–cs) and MSI1 anti-sense (msi1–as) plant lines have been described earlier (Hennig et al, 2003; Exner et al, 2006; Alexandre et al, 2009; Exner et al, 2009). The lhp1-6, clf1-29 and ft-10 alleles were used (Yoo et al, 2005; Schönrock et al, 2006; Exner et al, 2009). EMF2–FLAG, LHP1–GFP and lhp1-6 ft-10 plants were obtained by crossing. Plant growth conditions were as described previously (Exner et al, 2009). Vernalization treatments and measuring of flowering time were carried out as described earlier (Bouveret et al, 2006; Möller-Steinbach et al, 2010). For induction of root cell division, seedlings were grown vertically on ½ MS, 1% sucrose and 0.8% agar plates under constant light conditions. After 5 days, seedlings were transferred to plates supplemented with 750 nM 2,4-dichlorophenoxyacetic acid (2,4-D) and grown for an additional 3 days. Roots were separated from the shoots and used for ChIP and gene expression analyses.
Immunoprecipitation and protein immunoblot analyses
For immunoprecipitation (IP) followed by mass spectrometry 10 g of plant material was ground in a mortar with liquid nitrogen; for co-immunoprecipitation (CoIP) 2–4 g of plant material was used. Soluble proteins were extracted in 2 volumes of extraction buffer (10 mM Tris–HCl (pH 7.5), 150 mM NaCl, 0.5% Igepal, 1% Triton -00x and protease inhibitors (Roche, Basel, Switzerland)) at 4°C for 30 min with gentle rocking. For protein cross-linking 2 mM DTSSP was added to the extraction buffer, in which Tris was replaced with 20 mM HEPES, and the extracts were incubated at 4°C for 2 h. To stop cross-linking, 50 mM Tris–HCl (pH 7.5) was added, followed by incubation at 4°C for 20 min. To extract non-soluble proteins, NaCl was added to the final concentration of 2.5 M, followed by incubation at 4°C for 1 h. The centrifuged supernatant (4500 g) was filtered through four layers of Miracloth (Calbiochem, San Diego, CA, USA) and desalted using PD-10 desalting columns (GE Healthcare, Little Chalfont, UK). The centrifuged (4500 g) supernatant was pre-cleared with 30 μl of pre-washed protein A sepharose beads (GE Healthcare, Little Chalfont, UK) at 4°C for 20 min with gentle rocking. An input aliquot was taken from the pre-cleared centrifuged (2000 × g) supernatant before the rest of the supernatant was subjected to IP with 50 μl of bead-coupled antibodies at 4°C for 2 h with gentle rocking. The precipitate was washed six times in extraction buffer and eluted in 2 × Laemmli buffer. The following antibodies were used for IP: anti-FLAG magnetic beads (Sigma, #M8823), anti-GFP Trap_A (Chromotek, Planegg-Martinsried, Germany), anti-HA antibodies (Sigma, #H3663) and anti-AcV5 antibodies (Sigma, #A2980) coupled to protein A agarose beads and anti-myc beads (Sigma, #A7470). For protein immunoblots, proteins were separated by 12% SDS–PAGE and transferred to a PVDF membrane (Roth, Karlsruhe, Germany) by semi-dry blotting in 25 mM Tris–HCl (pH 8.3), 150 mM Glycin and 10% methanol for 1 h at 15 V. Enhanced chemiluminescence detection was performed as recommended by the manufacturer (GE Healthcare). The following antibodies and dilutions were used for immunoblotting: anti-HA (Sigma, #H3663), 1:1000; anti-V5 (Sigma, #V8012), 1:1000; anti-MSI1 (Hennig et al, 2003), 1:1000; anti-FLAG (Sigma, #A8592), 1:1000; anti-H3K27me3 (Millipore, Billerica, Massachusetts, USA, #07-449), 1:1000; and anti-H3 (Abcam, Cambridge, USA, # ab24834), 1:1000.
Histone extraction and quantitative immunoblotting
Approximately 2 g of frozen rosette leaves were ground to a fine powder and homogenized for 15 min in histone extraction buffer (0.25 M sucrose, 1 mM CaCl2, 15 mM NaCl, 60 mM KCl, 5 mM MgCl2, 15 mM PIPES, pH 7, 0.5% Triton X-100 including protease inhibitors (Roche) and 10 mM sodium butyrate). Extracts were cleared by centrifugation and pellets were dissolved in 0.2 N H2SO4. Total histones were precipitated with 33% Trichloroacetic acid, washed twice with acetone containing 0.1% HCl and once with acetone, briefly air-dried and dissolved in 1 x Laemmli buffer. The histone extract was run on 15% SDS–PAGE gels and transferred onto PVDF membranes (Roth). Proteins were probed with rabbit anti-H3K27me3 mixed with mouse anti-H3 (Abcam) antibodies. Goat anti-mouse IgG-IRDye 800CW (LI-COR, #926-32210) and goat anti-rabbit IgG-IRDye 680LT (LI-COR, #926-68021) were used as secondary antibodies. Membranes were scanned using an Odyssey Fc Imager (LI-COR Biosciences, Bad Homburg, Germany), and band intensities were quantified using Odyssey quantification software.
Tandem mass spectrometry analyses
After IP, the proteins were separated by 12% SDS–PAGE, and in-gel digestion was performed (Shevchenko et al, 1996). Mass spectrometry measurements were recorded on an LTQ Orbitrap-XL (Thermo Finnigan, Vernon Hills, IL, USA). MS/MS spectra were searched with MASCOT (Matrix Science, London, UK) against the Arabidopsis TAIR9 protein database with a concatenated decoy database (download on 19 June 2009) supplemented with contaminants. The search parameters were as follows: requirement for tryptic ends, one missed cleavage allowed, peptide tolerance±5 p.p.m., MS/MS tolerance±0.6 Da. Carbamidomethylation of cysteine was set as fixed modification, and oxidation of methionine was set as a variable modification. The processed data were imported into Scaffold (Proteome Software). The cutoff for data analyses was set to a minimum confidence of 90% for protein identification and to a minimum confidence of 95% for peptide identification. The spectrum false-discovery rate was calculated by dividing the number of decoy database spectrum assignments by the number of spectrum assignments. The false-positive rate was below 1% in all measured experiments. Proteins identified with at least two unique peptides in at least two replicates but never in control samples were taken into account.
Protein expression in Saccharomyces cerevisiae and immunoprecipitation assays
The MSI1 and MSI4 cDNAs were cloned into vector pGADT7 (Clonetech, Mountain View, CA); the LHP1 cDNA was cloned into vector pFLAG–attR (Stanyon et al, 2003). Proteins were expressed in S. cerevisiae strain BY4741 (Brachmann, 1998 #11297). If a constitutive promoter was used (pGADT7), cells were grown until OD600=1.1; they were then harvested and frozen in liquid nitrogen. If an inducible promoter was used (pFLAG–attR, pYES2), protein expression was induced at OD600=1.1 by 2% galactose; the cells were then harvested after 6 h and frozen in liquid nitrogen. After re-suspension in extraction buffer (20 mM HEPES pH 7.6, 10% glycerol, 200 mM potassium acetate, 1 mM EDTA, 1 mM DTT, protease inhibitors (Roche), 1 mM PMSF), cells were disrupted using a French Press (20 K, 1200, p.s.i., three times). The input sample was taken from the centrifuged supernatant (10 min at 4000 g followed by 10 min at 14.000 g); the rest of the supernatant was used for immunoprecipitation as described above.
Chromatin immunoprecipitation (ChIP)
ChIP assays were performed as described before (Exner et al, 2009) using the LowCell# ChIP kit (Diagenode, Liège, Belgium) according to the manufacturer’s instructions. Antibodies used in ChIP were anti-GFP (Molecular Probes Invitrogen, #A11122), IgG (Sigma-Aldrich, #I5006), anti-histone H3 (Millipore, #07690) and anti-H3K27me3 (Millipore, #07690). qPCR with gene-specific primers (Supplementary Table. S3) was performed using a MyiQ system (BIO-RAD, Hercules, CA, USA) and Sybr Green master mix (Fermentas) according to the manufacturer’s instructions.
RNA isolation and RT–qPCR
RNA extraction and RT–qPCR were performed as described previously (Leroy et al, 2007; Alexandre et al, 2009) with some modifications: qPCR with gene-specific primers (Supplementary Table S4) was performed using a MyiQ system and either the Sybr Green master mix (Fermentas) or the Fast Start Universal Probe Master (Rox) reagent and the Universal Probe Library set (Roche Diagnostics, Rotkreuz, Switzerland) according to the manufacturer’s instructions.
Microarray analysis
Plants were grown for 48 days in short-day photoperiods. Leaf number 6 was harvested at ZT (zeitgeber time)=7 h, and RNA was isolated, labelled using the GeneChip WT Sense Target Labeling Assay and hybridized to Affymetrix AGRONOMICS1 Arabidopsis tiling arrays as described (Rehrauer et al, 2010; Müller et al, 2012). Data were normalized and analysed as described (Rehrauer et al, 2010; Müller et al, 2012), based on TAIR10 annotations (http://www.arabidopsis.org). Leaf-specific expression potentials were estimated as the maximum expression measured in any of the leaf samples from the AtGenExpress reference set for development (Schmid et al, 2005). PcG targets in leaves were taken from Lafos et al (2011). Gene expression data for msi1–cs and emf2 plants were taken from Alexandre et al (2009) and Liu et al (2012).
Data availability
The microarray raw data from this publication were submitted to the ArrayExpress (www.ebi.ac.uk/arrayexpress) database (accession number E-MTAB-1412). The protein interactions from this publication have been submitted to the IMEx (Orchard et al, 2012) consortium through IntAct (Aranda et al, 2010) and assigned the identifier IM-18782. Processed microarray data are enclosed as Supplementary Table S5.
Supplementary Material
Acknowledgments
We thank Drs. Gilles Beck and Nicole Schatlowski for critical reading of the manuscript. We thank Dr. R. L. Finley Jr (Wayne State University) for providing plasmid pFLAG–attR. This work was supported by grants from the Swiss National Research Foundation, ETH Zurich, the Swedish Research Council and the Sixth Framework Program of the European Commission through the AGRON-OMICS Integrated Project (grant no. LSHG–CT–2006–037704).
Author contributions: MD, YMS, TW, IM and WM performed the experiments; MD, TW, PN and SB performed the MS/MS analyses; MD, YMS, TW, WM, WG and LH planned the experiments; and MD, LH and WG wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Alexandre C, Möller-Steinbach Y, Schönrock N, Gruissem W, Hennig L (2009) Arabidopsis MSI1 is required for negative regulation of the response to drought stress. Mol Plant 2: 675–687 [DOI] [PubMed] [Google Scholar]
- Anderson AE, Karandikar UC, Pepple KL, Chen Z, Bergmann A, Mardon G (2011) The enhancer of trithorax and polycomb gene Caf1/p55 is essential for cell survival and patterning in Drosophila development. Development 138: 1957–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda B, Achuthan P, Alam-Faruque Y, Armean I, Bridge A, Derow C, Feuermann M, Ghanbarian AT, Kerrien S, Khadake J, Kerssemakers J, Leroy C, Menden M, Michaut M, Montecchi-Palazzi L, Neuhauser SN, Orchard S, Perreau V, Roechert B, van Eijk K et al. (2010) The IntAct molecular interaction database in 2010. Nucleic Acids Res 38: D525–D531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausin I, Alonso-Blanco C, Jarillo JA, Ruiz-Garcia L, Martinez-Zapater JM (2004) Regulation of flowering time by FVE, a Retinoblastoma-associated protein. Nat Genet 36: 162–166 [DOI] [PubMed] [Google Scholar]
- Baerenfaller K, Hirsch-Hoffmann M, Svozil J, Hull R, Russenberger D, Bischof S, Lu Q, Gruissem W, Baginsky S (2011) pep2pro: a new tool for comprehensive proteome data analysis to reveal information about organ-specific proteomes in Arabidopsis thaliana. Integr Biol 3: 225–237 [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120–124 [DOI] [PubMed] [Google Scholar]
- Bastow R, Mylne JS, Lister C, Lippman Z, Martienssen RA, Dean C (2004) Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427: 164–167 [DOI] [PubMed] [Google Scholar]
- Bouveret R, Schönrock N, Gruissem W, Hennig L (2006) Regulation of flowering time by Arabidopsis MSI1. Development 133: 1693–1702 [DOI] [PubMed] [Google Scholar]
- Bratzel F, Lopez-Torrejon G, Koch M, Del Pozo JC, Calonje M (2010) Keeping cell identity in Arabidopsis requires PRC1 RING-finger homologs that catalyze H2A monoubiquitination. Curr Biol 20: 1853–1859 [DOI] [PubMed] [Google Scholar]
- Butenko Y, Ohad N (2011) Polycomb-group mediated epigenetic mechanisms through plant evolution. Biochim Biophys Acta 1809: 395–406 [DOI] [PubMed] [Google Scholar]
- Cao R, Zhang Y (2004) Suz12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol Cell 15: 57–67 [DOI] [PubMed] [Google Scholar]
- Chanvivattana Y, Bishopp A, Schubert D, Stock C, Moon YH, Sung ZR, Goodrich J (2004) Interaction of polycomb-group proteins controlling flowering in Arabidopsis. Development 131: 5263–5276 [DOI] [PubMed] [Google Scholar]
- Cui H, Benfey PN (2009) Interplay between SCARECROW, GA and LIKE HETEROCHROMATIN PROTEIN 1 in ground tissue patterning in the Arabidopsis root. Plant J 58: 1016–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V (2002) Drosophila Enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal polycomb sites. Cell 111: 185–196 [DOI] [PubMed] [Google Scholar]
- De Lucia F, Crevillen P, Jones AM, Greb T, Dean C (2008) A PHD-polycomb repressive complex 2 triggers the epigenetic silencing of FLC during vernalization. Proc Natl Acad Sci USA 105: 16831–16836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, Koseki H, Brockdorff N (2004) Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell 7: 663–676 [DOI] [PubMed] [Google Scholar]
- del Olmo I, Lopez-Gonzalez L, Martin-Trillo MM, Martinez-Zapater JM, Pineiro M, Jarillo JA (2010) EARLY IN SHORT DAYS 7 (ESD7) encodes the catalytic subunit of DNA polymerase epsilon and is required for flowering repression through a mechanism involving epigenetic gene silencing. Plant J 61: 623–636 [DOI] [PubMed] [Google Scholar]
- Doyle MR, Amasino RM (2009) A single amino acid change in the enhancer of zeste ortholog CURLY LEAF results in vernalization-independent, rapid flowering in Arabidopsis. Plant Physiol 151: 1688–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Exner V, Aichinger E, Shu H, Wildhaber T, Alfarano P, Caflisch A, Gruissem W, Köhler C, Hennig L (2009) The chromodomain of LIKE HETEROCHROMATIN PROTEIN 1 is essential for H3K27me3 binding and function during Arabidopsis development. PLoS ONE 4: e5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exner V, Taranto P, Schönrock N, Gruissem W, Hennig L (2006) Chromatin assembly factor CAF-1 is required for cellular differentiation during plant development. Development 133: 4163–4172 [DOI] [PubMed] [Google Scholar]
- Farrona S, Thorpe FL, Engelhorn J, Adrian J, Dong X, Sarid-Krebs L, Goodrich J, Turck F (2011) Tissue-specific expression of FLOWERING LOCUS T in Arabidopsis is maintained independently of polycomb group protein repression. Plant Cell 23: 3204–3214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S (2003) Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by polycomb and HP1 chromodomains. Genes Dev 17: 1870–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis NJ, Saurin AJ, Shao Z, Kingston RE (2001) Reconstitution of a functional core polycomb repressive complex. Mol Cell 8: 545–556 [DOI] [PubMed] [Google Scholar]
- Gendall AR, Levy YY, Wilson A, Dean C (2001) The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell 107: 525–535 [DOI] [PubMed] [Google Scholar]
- Goodin MM, Dietzgen RG, Schichnes D, Ruzin S, Jackson AO (2002) pGD vectors: versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. Plant J 31: 375–383 [DOI] [PubMed] [Google Scholar]
- Goodrich J, Puangsomlee P, Martin M, Long D, Meyerowitz EM, Coupland G (1997) A polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386: 44–51 [DOI] [PubMed] [Google Scholar]
- Gu X, Jiang D, Yang W, Jacob Y, Michaels SD, He Y (2011) Arabidopsis homologs of retinoblastoma-associated protein 46/48 associate with a histone deacetylase to act redundantly in chromatin silencing. PLoS Genet 7: e1002366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guitton AE, Berger F (2005) Loss of function of MULTICOPY SUPPRESSOR OF IRA 1 produces nonviable parthenogenetic embryos in Arabidopsis. Curr Biol 15: 750–754 [DOI] [PubMed] [Google Scholar]
- Guitton AE, Page DR, Chambrier P, Lionnet C, Faure JE, Grossniklaus U, Berger F (2004) Identification of new members of FERTILISATION INDEPENDENT SEED polycomb group pathway involved in the control of seed development in Arabidopsis thaliana. Development 131: 2971–2981 [DOI] [PubMed] [Google Scholar]
- Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, Rappsilber J, Lerdrup M, Helin K (2007) A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol 10: 1291–1300 [DOI] [PubMed] [Google Scholar]
- Hennig L, Bouveret R, Gruissem W (2005) MSI1-like proteins: an escort service for chromatin assembly and remodeling complexes. Trends Cell Biol 15: 295–302 [DOI] [PubMed] [Google Scholar]
- Hennig L, Derkacheva M (2009) Diversity of polycomb group complexes in plants: same rules, different players? Trends Genet 25: 414–423 [DOI] [PubMed] [Google Scholar]
- Hennig L, Taranto P, Walser M, Schönrock N, Gruissem W (2003) Arabidopsis MSI1 is required for epigenetic maintenance of reproductive development. Development 130: 2555–2565 [DOI] [PubMed] [Google Scholar]
- Heo JB, Sung S (2011) Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331: 76–79 [DOI] [PubMed] [Google Scholar]
- Jiang D, Wang Y, Wang Y, He Y (2008) Repression of FLOWERING LOCUS C and FLOWERING LOCUS T by the Arabidopsis polycomb repressive complex 2 components. PLoS ONE 3: e3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz A, Oliva M, Mosquna A, Hakim O, Ohad N (2004) FIE and CURLY LEAF polycomb proteins interact in the regulation of homeobox gene expression during sporophyte development. Plant J 37: 707–719 [DOI] [PubMed] [Google Scholar]
- Ketel CS, Andersen EF, Vargas ML, Suh J, Strome S, Simon JA (2005) Subunit contributions to histone methyltransferase activities of fly and worm polycomb group complexes. Mol Cell Biol 25: 6857–6868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Hyun Y, Park JY, Park MJ, Park MK, Kim MD, Kim HJ, Lee MH, Moon J, Lee I, Kim J (2004) A genetic link between cold responses and flowering time through FVE in Arabidopsis thaliana. Nat Genet 36: 167–171 [DOI] [PubMed] [Google Scholar]
- Köhler C, Hennig L, Bouveret R, Gheyselinck J, Grossniklaus U, Gruissem W (2003) Arabidopsis MSI1 is a component of the MEA/FIE polycomb group complex and required for seed development. EMBO J 22: 4804–4814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotake T, Takada S, Nakahigashi K, Ohto M, Goto K (2003) Arabidopsis TERMINAL FLOWER 2 gene encodes a LIKE HETEROCHROMATIN PROTEIN 1 homolog and represses both FLOWERING LOCUS T to regulate flowering time and several floral homeotic genes. Plant Cell Physiol 44: 555–564 [DOI] [PubMed] [Google Scholar]
- Lafos M, Kroll P, Hohenstatt ML, Thorpe FL, Clarenz O, Schubert D (2011) Dynamic regulation of H3K27 trimethylation during Arabidopsis differentiation. PLoS Genet 7: e1002040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latrasse D, Germann S, Houba-Herin N, Dubois E, Bui-Prodhomme D, Hourcade D, Juul-Jensen T, Le Roux C, Majira A, Simoncello N, Granier F, Taconnat L, Renou JP, Gaudin V (2011) Control of flowering and cell fate by LIF2, an RNA binding partner of the polycomb complex component LHP1. PLoS One 6: e16592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy O, Hennig L, Breuninger H, Laux T, Köhler C (2007) Polycomb group proteins function in the female gametophyte to determine seed development in plants. Development 134: 3639–3648 [DOI] [PubMed] [Google Scholar]
- Lewis EB (1978) A gene complex controlling segmentation in Drosophila. Nature 276: 565–570 [DOI] [PubMed] [Google Scholar]
- Li W, Wang Z, Li J, Yang H, Cui S, Wang X, Ma L (2011) Overexpression of AtBMI1C, a Polycomb group protein gene, accelerates flowering in Arabidopsis. PLoS One 6: e21364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libault M, Tessadori F, Germann S, Snijder B, Fransz P, Gaudin V (2005) The Arabidopsis LHP1 protein is a component of euchromatin. Planta 222: 910–925 [DOI] [PubMed] [Google Scholar]
- Liu C, Xi W, Shen L, Tan C, Yu H (2009) Regulation of floral patterning by flowering time genes. Dev Cell 16: 711–722 [DOI] [PubMed] [Google Scholar]
- Liu MS, Chen LF, Lin CH, Lai YM, Huang JY, Sung ZR (2012) Molecular and functional characterization of broccoli EMBRYONIC FLOWER 2 genes. Plant Cell Physiol 53: 1217–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logemann E, Birkenbihl RP, Ulker B, Somssich IE (2006) An improved method for preparing Agrobacterium cells that simplifies the Arabidopsis transformation protocol. Plant Meth 2: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Vernaza M, Yang S, Muller R, Thorpe F, de Leau E, Goodrich J (2012) Antagonistic roles of SEPALLATA3, FT and FLC genes as targets of the polycomb group gene CURLY LEAF. PLoS One 7: e30715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubec G, Afjehi-Sadat L (2007) Limitations and pitfalls in protein identification by mass spectrometry. Chem Rev 107: 3568–3584 [DOI] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury Iii WJ, Voigt P, Martin SR, Taylor WR, De Marco V, Pirrotta V, Reinberg D, Gamblin SJ (2009) Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461: 762–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D (2011) The polycomb complex PRC2 and its mark in life. Nature 469: 343–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Balbas MA, Tsukiyama T, Gdula D, Wu C (1998) Drosophila NURF-55, a WD repeat protein involved in histone metabolism. Proc Natl Acad Sci U S A 95: 132–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohd-Sarip A, Venturini F, Chalkley GE, Verrijzer CP (2002) Pleiohomeotic can link polycomb to DNA and mediate transcriptional repression. Mol Cell Biol 22: 7473–7483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möller-Steinbach Y, Alexandre C, Hennig L (2010) Flowering time control. Methods Mol Biol 655: 229–237 [DOI] [PubMed] [Google Scholar]
- Müller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O'Connor MB, Kingston RE, Simon JA (2002) Histone methyltransferase activity of a Drosophila polycomb group repressor complex. Cell 111: 197–208 [DOI] [PubMed] [Google Scholar]
- Müller M, Patrignani A, Rehrauer H, Gruissem W, Hennig L (2012) Evaluation of alternative RNA labeling protocols for transcript profiling with Arabidopsis AGRONOMICS1 tiling arrays. Plant Meth 8: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzina N, Verreault A, Laue E, Stillman B (1999) Heterochromatin dynamics in mouse cells: interaction between chromatin assembly factor 1 and HP1 proteins. Mol Cell 4: 529–540 [DOI] [PubMed] [Google Scholar]
- Mylne JS, Barrett L, Tessadori F, Mesnage S, Johnson L, Bernatavichute YV, Jacobsen SE, Fransz P, Dean C (2006) LHP1, the Arabidopsis homologue of HETEROCHROMATIN PROTEIN1, is required for epigenetic silencing of FLC. Proc Natl Acad Sci USA 103: 5012–5017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov M, Wild B, Müller J (2005) Nucleosome binding and histone methyltransferase activity of Drosophila PRC2. EMBO Rep 6: 348–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak AJ, Alfieri C, Stirnimann CU, Rybin V, Baudin F, Ly-Hartig N, Lindner D, Muller CW (2011) Chromatin-modifying complex component Nurf55/p55 associates with histones H3 and H4 and polycomb repressive complex 2 subunit Su(z)12 through partially overlapping binding sites. J Biol Chem 286: 23388–23396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orchard S, Kerrien S, Abbani S, Aranda B, Bhate J, Bidwell S, Bridge A, Briganti L, Brinkman FS, Cesareni G, Chatr-aryamontri A, Chautard E, Chen C, Dumousseau M, Goll J, Hancock RE, Hannick LI, Jurisica I, Khadake J (2012) Protein interaction data curation: the international molecular exchange (IMEx) consortium. Nat Meth 9: 345–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazhouhandeh M, Molinier J, Berr A, Genschik P (2011) MSI4/FVE interacts with CUL4-DDB1 and a PRC2-like complex to control epigenetic regulation of flowering time in Arabidopsis. Proc Natl Acad Sci USA 108: 3430–3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruk S, Sedkov Y, Johnston DM, Hodgson JW, Black KL, Kovermann SK, Beck S, Canaani E, Brock HW, Mazo A (2012) TrxG and PcG proteins but not methylated histones remain associated with DNA through replication. Cell 150: 922–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehrauer H, Aquino C, Gruissem W, Henz SR, Hilson P, Laubinger S, Naouar N, Patrignani A, Rombauts S, Shu H, Van de PeY, Vuylsteke M, Weigel D, Zeller G, Hennig L (2010) AGRONOMICS1: a new resource for Arabidopsis transcriptome profiling. Plant Physiol 152: 487–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Pulido L, Devos D, Sung ZR, Calonje M (2008) RAWUL: a new Ubiquitin-like domain in PRC1 Ring finger proteins that unveils putative plant and worm PRC1 orthologs. BMC Genomics 9: 308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawarkar R, Paro R (2010) Interpretation of developmental signaling at chromatin: the polycomb perspective. Dev Cell 19: 651–661 [DOI] [PubMed] [Google Scholar]
- Schatlowski N, Stahl Y, Hohenstatt ML, Goodrich J, Schubert D (2010) The CURLY LEAF interacting protein BLISTER controls expression of polycomb-group target genes and cellular differentiation of Arabidopsis thaliana. Plant Cell 22: 2291–2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Schmid M, Uhlenhaut NH, Godard F, Demar M, Bressan R, Weigel D, Lohmann JU (2003) Dissection of floral induction pathways using global expression analysis. Development 130: 6001–6012 [DOI] [PubMed] [Google Scholar]
- Schmitges FW, Prusty AB, Faty M, Stutzer A, Lingaraju GM, Aiwazian J, Sack R, Hess D, Li L, Zhou S, Bunker RD, Wirth U, Bouwmeester T, Bauer A, Ly-Hartig N, Zhao K, Chan H, Gu J, Gut H, Fischle W et al. (2011) Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell 42: 330–341 [DOI] [PubMed] [Google Scholar]
- Schönrock N, Bouveret R, Leroy O, Borghi L, Köhler C, Gruissem W, Hennig L (2006) Polycomb-group proteins repress the floral activator AGL19 in the FLC-independent vernalization pathway. Genes Dev 20: 1667–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon CC, Burn JE, Perez PP, Metzger J, Edwards JA, Peacock WJ, Dennis ES (1999) The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11: 445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem 68: 850–858 [DOI] [PubMed] [Google Scholar]
- Shu H, Wildhaber T, Siretskiy A, Gruissem W, Hennig L (2012) Distinct modes of DNA accessibility in plant chromatin. Nat Commun 3: 1281. [DOI] [PubMed] [Google Scholar]
- Song JJ, Garlick JD, Kingston RE (2008) Structural basis of histone H4 recognition by p55. Genes Dev 22: 1313–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillane C, MacDougall C, Stock C, Köhler C, Vielle-Calzada J, Nunes SM, Grossniklaus U, Goodrich J (2000) Interaction of the Arabidopsis polycomb group proteins FIE and MEA mediates their common phenotypes. Curr Biol 10: 1535–1538 [DOI] [PubMed] [Google Scholar]
- Stanyon CA, Limjindaporn T, Finley RL Jr. (2003) Simultaneous cloning of open reading frames into several different expression vectors. Biotechniques 35: 522–526 [DOI] [PubMed] [Google Scholar]
- Sung S, He Y, Eshoo TW, Tamada Y, Johnson L, Nakahigashi K, Goto K, Jacobsen SE, Amasino RM (2006) Epigenetic maintenance of the vernalized state in Arabidopsis thaliana requires LIKE HETEROCHROMATIN PROTEIN 1. Nat Genet 38: 706–710 [DOI] [PubMed] [Google Scholar]
- Tie F, Furuyama T, Prasad-Sinha J, Jane E, Harte PJ (2001) The Drosophila polycomb group proteins ESC and E(Z) are present in a complex containing the histone-binding protein p55 and the histone deacetylase RPD3. Development 128: 275–286 [DOI] [PubMed] [Google Scholar]
- Turck F, Roudier F, Farrona S, Martin-Magniette ML, Guillaume E, Buisine N, Gagnot S, Martienssen RA, Coupland G, Colot V (2007) Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of Histone H3 Lysine 27. PLoS Genet 3: 0855–0866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler JK, Bulger M, Kamakaka RT, Kobayashi R, Kadonaga JT (1996) The p55 subunit of Drosophila chromatin assembly factor-1 is homologous to a histone deacetylase-associated protein. Mol Cell Biol 16: 6149–6159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y (2004) Role of histone H2A ubiquitination in polycomb silencing. Nature 431: 873–878 [DOI] [PubMed] [Google Scholar]
- Wen P, Quan Z, Xi R (2012) The biological function of the WD40 repeat-containing protein p55/Caf1 in Drosophila. Dev Dyn 241: 455–464 [DOI] [PubMed] [Google Scholar]
- Wood CC, Robertson M, Tanner G, Peacock WJ, Dennis ES, Helliwell CA (2006) The Arabidopsis thaliana vernalization response requires a polycomb-like protein complex that also includes VERNALIZATION INSENSITIVE 3. Proc Natl Acad Sci USA 103: 14631–14636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Shen WH (2008) Polycomb silencing of KNOX genes confines shoot stem cell niches in Arabidopsis. Curr Biol 18: 1966–1971 [DOI] [PubMed] [Google Scholar]
- Yang CH, Chen LJ, Sung ZR (1995) Genetic regulation of shoot development in Arabidopsis: role of the EMF genes. Dev Biol 169: 421–435 [DOI] [PubMed] [Google Scholar]
- Yoo SK, Chung KS, Kim J, Lee JH, Hong SM, Yoo SJ, Yoo SY, Lee JS, Ahn JH (2005) CONSTANS activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 through FLOWERING LOCUS T to promote flowering in Arabidopsis. Plant Physiol 139: 770–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida N, Yanai Y, Chen L, Kato Y, Hiratsuka J, Miwa T, Sung ZR, Takahashi S (2001) EMBRYONIC FLOWER2, a novel polycomb group protein homolog, mediates shoot development and flowering in Arabidopsis. Plant Cell 13: 2471–2481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, Li Y, Ben-Meir H, Oliva M, Mosquna A, Kiss V, Avivi Y, Ohad N, Grafi G (2006) Different domains control the localization and mobility of LIKE HETEROCHROMATIN PROTEIN1 in Arabidopsis nuclei. Plant Cell 18: 133–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, Ball AR Jr., Yokomori K (2010) HP1: Heterochromatin binding proteins working the genome. Epigenetics 5: 287–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Germann S, Blus BJ, Khorasanizadeh S, Gaudin V, Jacobsen SE (2007) The Arabidopsis LHP1 protein colocalizes with histone H3 Lys27 trimethylation. Nat Struct Mol Biol 14: 869–871 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) Genevestigator. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The microarray raw data from this publication were submitted to the ArrayExpress (www.ebi.ac.uk/arrayexpress) database (accession number E-MTAB-1412). The protein interactions from this publication have been submitted to the IMEx (Orchard et al, 2012) consortium through IntAct (Aranda et al, 2010) and assigned the identifier IM-18782. Processed microarray data are enclosed as Supplementary Table S5.