Abstract
To understand how mitotic kinesins contribute to the assembly and function of the mitotic spindle, we need to purify these motors and analyze their biochemical and ultrastructural properties. Here we briefly review our use of microtubule (MT) affinity and biochemical fractionation to obtain information about the oligomeric state of native mitotic kinesin holoenzymes from eggs and early embryos. We then detail the methods we use to purify full length recombinant Drosophila embryo mitotic kinesins, using the baculovirus expression system, in sufficient yields for detailed in vitro assays. These two approaches provide complementary biochemical information on the basic properties of these key mitotic proteins, and permit assays of critical motor activities, such as MT-MT crosslinking and sliding, that are not revealed by assaying motor domain subfragments.
1. Introduction
Chromosome segregation during mitosis depends upon the action of the mitotic spindle, a subcellular machine whose assembly and function depends upon forces generated by microtubule (MT) polymer ratchets, together with kinesin and dynein motors [1-5]. Tubulin, the major subunit of MTs, is a relatively abundant protein, and consequently it is feasible to purify tubulin from its native host cell in yields suitable for assaying its dynamics and force-generating properties [6, 7]. In contrast, mitotic kinesins are less abundant, and consequently there are relatively few reports describing the purification and assay of these motors from their native host cells [8, 9]. As an alternative approach, many investigators have used bacterial or eukaryotic expression systems to purify sufficient amounts of mitotic kinesins or their subfragments for detailed analysis (e.g. [10-16] ). In this paper, we first present a brief historical perspective outlining the advantages and limitations that we have encountered in purifying mitotic kinesins as native complexes from egg/embryo extracts. We then focus on recent work done in our laboratory on the purification and assay of full length, recombinant Drosophila embryo mitotic kinesin complexes from the bacculovirus system, commenting on the complementary information that can be obtained by comparing information from native versus recombinant proteins.
2. Identification and purification of native embryonic mitotic kinesins
2.1. Development of General Strategy for Identification and Purification of Mitotic Kinesins in Embryonic Systems Using Echinoderms
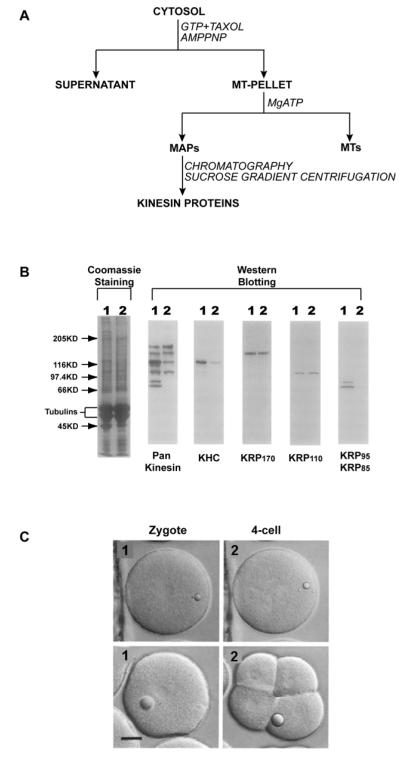
To identify motor proteins that might participate in mitotic movements we, like several other groups, analyzed MTs purified from dividing cell extracts for co-purifying ATPase activities (Fig. 1A) (see [17] for detailed methods). We used sea urchin eggs/embryos which are a model system for studying mitosis and cell division (e.g. [18]), because they stockpile the MT proteins required to support multiple mitoses in the early embryo [19]. We found that taxol-stabilized MTs purified from MgAMPPNP-supplemented sea urchin egg/embryo extracts contained both dynein and kinesin motors [19-21]. Using “pan-kinesin” peptide antibodies that were designed to recognize a broad range of kinesins on immunoblots, we found that these MT preparations in fact contain multiple distinct kinesins [22], albeit only a subset of the 35 kinesins encoded by the sea urchin genome [23] (Fig 1B). Two kinesin holoenzymes, namely kinesin-1 and heterotrimeric kinesin-2 were purified to homogeneity and characterized, but they were found to play roles in the delivery of exocytic vesicles to the cell surface and in ciliogenesis, respectively, rather than in mitosis per se (Fig. 1C)[24-27].
Figure. 1. Pan-kinesin antibody/MT affinity screen for native mitotic kinesins.
(A) General strategy for purification of kinesin holoenzymes from native tissues. Kinesins are co-precipitated with MTs, and subsequently eluted from the MT-pellet by ATP. The resulting MAPs are further separated by conventional fractionation methods. (B) Detection of kinesin polypeptides that copurify with sea urchin egg microtubule precipitates by immunoblotting. Microtubules were prepared from AMPPNP (lane 1) or ATP (lane 2) treated cytosol. The microtubule proteins were analyzed on SDS-PAGE (left-hand panel) or on immunoblots probed with pan-kinesin antibodies (panel 2) that react with multiple kinesins. Using kinesin family-specific antibodies these were identified as kinesin-1 (KHC; panel 3), kinesin-5 (KRP170; panel 4), kinesin-6 (KRPl10; panel 5), and heterotrimeric kinesin-2 (KRP85 and KRP95; panel 6). (C) Effects of antibody microinjection on mitosis in fertilized sea urchin eggs. Embryos were shown 2hr (pane 1) and 4hr (pane 2) after fertilization. In these cells, injection of antibodies to some kinesins (e.g. kinesin-5, -6 and -12) altered spindle morphogenesis and inhibited normal cell division (upper panels), whereas antibodies to others (e.g. kinesin-1 and -2) had no effect on cell division (lower panels). Bar, 25μm.
This biochemical approach was less productive when applied to sea urchin egg/embryo mitotic kinesins due to the relatively low abundance of these motors. Starting from AMPPNP-MTs, we were able to partially purify low yields of three kinesins that may participate in mitosis, based on observations that the inhibition of their function by the microinjection of kinesin isotype-specific antibodies led to defects in mitosis and cell division (Fig. 1B,C) [28, 29]. However, using hydrodynamic analysis [30], we were able to make estimates of the oligomeric states of several motors, specifically; kinesin-5 (aka KRP170; Stokes radius (Rs) = 16.5nm, S-value = 9.0S, M.W. = 610 kDa; 4 × 170 kDa subunits); kinesin-6 (aka KRP110; Rs = 11.5 nm; S-value = 9.8S; M.W. = 464 kDa; 4 × 110kDa subunits); and kinesin-12 (aka KRP180; Rs = 10.1nm; S-value = 8.3S; M.W. = 331Kda; 2 × 167kDa subunits). Based on this work, we conclude that the use of MT affinity combined with pankinesin antibody immunoblotting and hydrodynamic analysis of crude subfractions of AMPPNP-MT eluates can yield useful information about the oligomeric state of mitotic kinesins that complements work with recombinant proteins (Fig. 1 and [17]), but typically it does not yield enough pure protein for detailed biochemical analysis.
2.2. Identification and Purification of Mitotic Kinesins from Drosophila melanogaster embryos
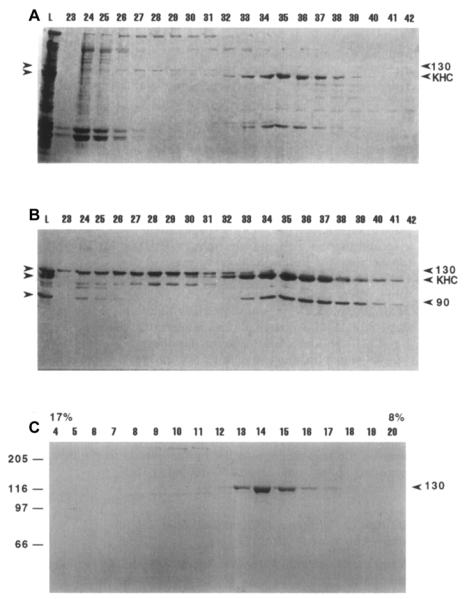
The genome of Drosophila melanogaster encodes 24 kinesins, of which at least 10 are involved in mitosis, and the syncytial embryo is a useful system for analyzing native mitotic kinesins [1, 31]. We applied the MT affinity, peptide antibody screen to Drosophila embryo extracts (see next section), allowing us to partially purify and estimate the oligomeric state of a handful of mitotic kinesins, and to purify and characterize one of them [8, 16, 17, 32, 33]. These are kinesin-5 (aka KLP61F; Rs = 16.2 nm; S-value = 7.6s; M.W. = 490 kDa; 4 × 130kDa subunits); kinesin-14 (aka Ncd; Rs = 7.6 nm; S-value = 4.8S; M.W. = 147 kDa; 2 × 90kDa subunits); and kinesin-6 (aka PavKLP; Rs = 7.6 nm; S-value = 5.8S; M.W. = 186kDa; 2 × 93 kDa subunits). It is worth noting that kinesin-6s from fly and sea urchin behaved as dimers and tetramers, respectively. Again, the low abundance of kinesin-6 and kinesin-14 prohibited extensive purification, but we were able to purify the kinesin-5, KLP61F to homogeneity in sufficient yield for limited biochemical and structural study (Fig. 2).
Figure 2. Purification of the kinesin-5, KLP61F from Drosophila embryos.
(A) SDS PAGE of fractions from gel filtration of Drosophila MAPs (L = loaded MAPs). Numbers above indicate gel filtration fractions. 130 on the right indicates KLP61F, KHC stands for kinesin heavy chain. (B) Corresponding immunoblot probed with pan-kinesin antibody. 90 on the right indicates the 90 kDa subunit of Ncd. (C) Coomassie-stained gel of sucrose density gradient fractions. Arrowhead indicates the peak fraction of KLP61F, 17% and 8% indicate percentage of sucrose at top and bottom of gradient, and molecular weight markers are indicated on the left.
2.3. Purification of the Kinesin-5, KLP61F from Drosophila Melanogaster Embryos
Details of the purification of homotetrameric kinesin-5 from ATP eluates of Drosophila embryo AMPPNP MTs are provided in [17] and are outlined here.
Drosophila Embryo Collection and Cytosol Production
Drosophila embryos were collected cleaned, and homogenized in an equal volume of cold buffer A (100mM PIPES, PH6.9, 0.5mM EDTA, 5.0mM EGTA, 2.5 mM MgSO4, 150mM KCl, 1.0 mM NaN3, 1.0 mM DTT, 0.1mM ATP, protease inhibitors). The homogenate was centrifuged at 15,000g for 40min at 4°C. The clear intermediate supernatant was collected and centrifuged again at 150,000g for 30 min at 4°C, yielding the high speed supernatant (HSS) which was used for protein purification.
Purification of Polymerized MTs and MT-Associated Proteins
Drosophila cytosol was incubated with 1mM GTP and 20 μM taxol at RT for 20min to form MTs. Binding of motors to MTs was accomplished by adding 1mM AMPPNP and incubating the suspension for an additional 20min at RT. The MT-motor complexes were pelleted through a 15% sucrose cushion (35,000g × 1hr, 10°C).
Elution of KLP61F and Other MAPs
The pellet was gently resuspended in 5 ml of ice-cold bufferA containing 250mM KCl, 10mM ATP, 1mM GTP, 10mM MgSO4, and 20 μM taxol. The resuspended pellet was incubated for at least 4hr on ice. Then the solution was rewarmed to RT and incubated for an additional 20min. Finally the solution was centrifuged (200,000g × 20-30 min, 10°C) and the supernatant (ATP eluate, Fig. 2 A, B lane L) containing kinesin motor proteins was collected and concentrated by Centriprep-30.
Gel-Filtration Chromatography of KLP61F
Concentrated samples containing KLP61F were loaded onto a buffer A-pre-equilibrated Bio-gel A 1.5m column (Fig. 2 A, B) or a Superose 6 FPLC gel filtration column. The KLP61F fraction eluted immediately after the void volume. KLP61F fractions were pooled and concentrated again using a Centriprep-30 centrifugal concentrator.
Rebinding of KLP61F and MTs
To further concentrate KLP61F, MTs polymerized from bovine brain tubulin were mixed with pooled KLP61F in the presence of 2 mM AMPPNP. The MT-KLP61F pellet was spun down and KLP61F was eluted using 10mM ATP as described above.
5-20% Sucrose Density Gradient Centrifugation of KLP61F
A 5-20% sucrose density gradient in buffer A containing 0.1 mM ATP was made using a gradient mixer in a 5ml centrifuge tube. 200 μl of the KLP61F eluate was loaded onto the gradient. The gradient was centrifuged at 300,000g for 9 hr at 4°C (Beckman SW55 Ti). Finally the gradient was fractionated with 200 μl per fraction. The presence of KLP61F was monitored by SDS-PAGE (Fig. 2C).
Typically, about 150μg of pure KLP61F was obtained from 200 ml cytosol. Although such yields are not suitable for detailed biochemistry, they were sufficient to allow us to determine that the protein we had purified is the product of the KLP61F gene, rather than being encoded by another kinesin gene using mass spectrsoscopy and protein sequencing [33-35], as well as to examine its homotetrameric, bipolar ultrastructure, and to assay its motility [8, 32]. This provided useful information for subsequent complementary, detailed assays of pure protein obtained in high yield from the baculovirus system (section 3). For example, in the absence of this work, we would not have any information with which to evaluate whether the homotetrameric structure of purified recombinant kinesin-5 (below) is relevant to the native holoenzyme present in embryos.
3. Expression and purification of recombinant Drosophila mitotic kinesins using the baculovirus expression system
3.1. General strategy for purifying kinesins using the Baculovirus Expression System
Although we were able to prepare sufficient native KLP61F from embryos for limited analysis, the yield of mitotic kinesins from fly embryos is usually too low to perform detailed functional assays. While we had some success with the purification of recombinant KLP61F using bacterial expression systems [36], in our hands the full length protein purified this way tended to aggregate non-specifically. As an alternative, therefore, we explored the expression and purification of active recombinant proteins from baculovirus-infected cell cultures [16]. The baculovirus expression system is one of the most powerful eukaryotic expression systems available. Baculovirus has a strong polyhedrin promotor that ensures a high level of heterologous gene expression. In addition, this system exploits the ability of cultured insect cell lines (e.g. sf9) to carry out post-translational modification of expressed proteins similar to those that occur in the natural host cell.
Baculoviruses belong to a group of large double-stranded DNA virus that infect different species of insects [37]. Recombinant baculoviruses are used to express heterologous genes in insect cells. Benefits of protein expression with baculovirus include safe use, high expression levels of soluble protein, the possibility of multiple gene expression, easy scale up with suspension cultures, and proper protein folding as well as appropriate eukaryotic post-translational modifications. Autographa californica nuclear polyhedrosis virus (AcNPV) is the most common baculovirus used in laboratories. AcNPV has a strong polyhedrin promoter which is expressed during the late stages of infection. Polyhedrin is the main matrix protein that embeds the occluded virus particles [38]. Deletion of this nonessential gene is not lethal to the virus and it can be replaced with a gene of interest to generate viable recombinant baculoviruses. The recombinant baculoviruses can either produce recombinant protein or generate additional viruses by infecting additional insect cells. As an extra bonus, replacing the polyhedrin gene results in the production of occlusion body-negative viruses, which have an obviously different plaque morphology from the wild-type. This makes the recombinant viruses easy to identify.
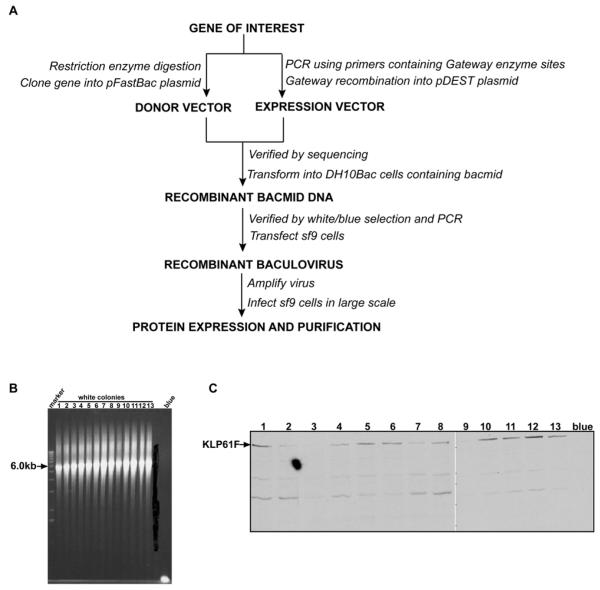
Currently there are two main ways to generate recombinant baculoviruses. One utilizes the “baculovirus expression vector system” [39], where a transfer vector containing the gene of interest is co-transfected with AcNPV DNA into insect cells. By homologous recombination, the foreign gene is inserted into the baculovirus DNA to form recombinant viruses. The positive recombinant baculoviruses can be selected based on plaque phenotype selection, color selection, or positive survival selection. This method, however, is inefficient and time consuming. An improved method employs the “bac-to-bac baculovirus expression system” (Invitrogen). In this system, recombinant baculoviruses are generated by site-specific transposition to insert foreign genes into bacmid DNA propagated in E. coli. Colonies containing recombinant bacmids are identified by antibiotic selection and blue/white screening. The steps used to generate a recombinant baculovirus by the “bac-to-bac baculovirus expression system” are outlined in figure 3A. In the next paragraphs, we will describe our work on the expression of several Drosophila mitotic kinesins as examples to illustrate the detailed methods.
Figure 3. Generation of recombinant baculoviruses and gene expression using the BAC-to-BAC expression system.
(A) General scheme for expression mitotic kinesins by bac-to-bac expression system. (B) PCR analysis to check the insertion of KLP61F into Bacmid DNA. PUC/M13 amplification primers are directed at sequences on either side of the transposition site within the lacZα -complementation region of the bacmid. The expected results from PCR are as follows: Bacmid alone, ~300bp; Bacmid transposed with pFastBacHT, ~2430bp; Bacmid transposed with pFastBacHT containing KLP61F, ~6.0kb (2430bp+3502bp). (C) Western-blot screen of Sf9 cells transfected with baculovirus bearing KLP61F gene. Cell lysates were subjected to 7.5% SDS-PAGE. After protein bands were transferred onto a nitrocellulose membrane, KLP61F was detected by immunoblotting with mouse monoclonal anti-polyhistidine antibody(1:2000), and a secondary rabbit anti-mouse antibody (1:5000). Lane 1-13, cell lysates from 13 white colonies. Blue, cell lysate from blue colony.
3.2. Expression and purification of rKLP61F and rNcd from the Bac-to-Bac baculovirus expression System
Cloning KLP61F/Ncd cDNA into Bacmid
Full length KLP61F and Ncd cDNAs were sequenced, excised with restriction enzymes, then subcloned into a transfer vector pFastHTA (Invitrogen) with 6His tags on their N-termini. The recombinant transfer vector (donor vector) was transformed into DH5α for screening. The verified transfer vector containing the KLP61F/Ncd gene was tansformed into DH10Bac E.coli that contains bacmid DNA, a baculovirus shuttle vector. Since the insertion of the gene of interest will disrupt the expression of the lacZα peptide, colonies containing the recombinant bacmid are white in a background of blue colonies containing unaltered bacmid. We usually obtained 10-20 white colonies in a background of about 100 blue colonies. To further verify the insertion of KLP61F/Ncd, we used PUC/M13 forward / reverse amplification primers to run a PCR in order to estimate the recombinant bacmid’s size (Fig. 3B).
Transfecting Sf9 Cells with Recombinant Bacmid
Sf9 cells in Sf-900/SFM medium(Invitrogen) were seeded into 35mm 6-well plates, 9×105 cells /2ml/well. 10μl bacmid DNA mixed with 6μl CellFECTIN (Invitrogen) was added to the sf9 cells. Cells were assayed for KLP61F protein activity 72h after the start of transfection. The supernatant contains the first generation of recombinant baculovirus (P1 virus).
Testing the Expression of Protein by Western-Blotting
Cell lysates or purified protein was analyzed by 7.5% SDS-PAGE. The protein bands were transferred onto a nitrocellulose membrane, then KLP61F/Ncd was detected by immunoblotting with mouse monoclonal anti-polyhistidine antibody (Sigma, 1:2000 dilution), and a secondary rabbit anti-mouse antibody (Sigma, 1:5000 dilution). The KLP61F/Ncd protein was detected at a molecular weight of 138KD/80 KD but was not detected in cells infected with virus derived from a blue colony (Fig. 3C).
Purification of KLP61F/Ncd with Ni-NTA resin
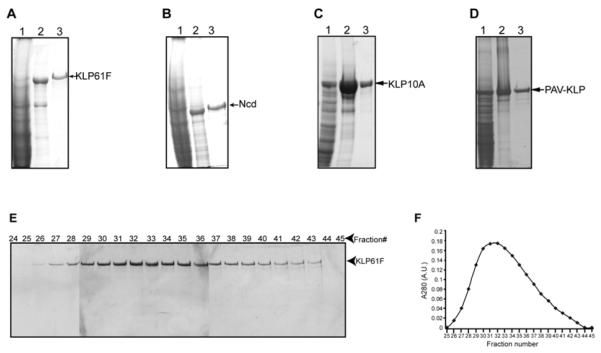
The P1 viruses were amplified and then used to infect sf9 cells in large scale cultures. Infected sf9 cells were harvested by centrifugation at 1000g×15mins and resuspend in lysis buffer (50mM Tris base, pH8.0, 300mM NaCl, 2mM MgCl2, 20mM Imidazole, 10mM 2-marcaptoethanol, 0.1mM ATP, Protease inhibitors). After 2 rounds of French pressing or sonication, the lysate was centrifuged at 35k rpm × 37mins (Beckman Ti 50.2 rotor), and the supernatant was collected for purification on a Ni-NTA column since there is a 6xhis tag fusion on the N-terminus of the protein. The supernatant was passed through Ni-NTA resins that were equilibrated with balance buffer (50mM Tris base, pH8.0, 300mM NaCl, 2mM MgCl2, 20mM Imidazole), then washed with washing buffer (20mM Tris, pH8.0, 1M NaCl, 2mM MgCl2, 20mM Imidazole, Protease inhibitors). Finally, KLP61F/Ncd was eluted with elution buffer (20mM Tris, pH8.0, 100mM NaCl, 2mM MgCl2, 300mM Imidazole, 10mM 2-marcaptoethanol, 0.1mM ATP, Protease inhibitors) (Fig. 4 A, B ).
Figure 4. Purification of Recombinant KLP61F and Ncd from the Baculovirus Expression System.
(A-D) Commassie-blue-stained SDS-polyacrylamide gels show typical recombinant-motor-protein fractions obtained from the infected Sf9 cells. (A) Purification of rKLP61F. The following are shown: lane 1, Sf9 cell high-speed supernatant; lane 2, Ni-NTA affinity-column eluate; and lane 3, gel-filtration (Biogel A-15M, Biorad) fractionated and concentrated rKLP61F. (B) Purification of rNcd. The following are shown: lane 1, Sf9 cell high-speed supernatant; lane 2, Ni-NTA affinity-column eluate; and lane 3, gel-filtration fractionated and concentrated Ncd. (C) Purification of rKLP10A ; 1, Sf9 cell high-speed supernatant; lane 2, Ni-NTA affinity-column eluate; and lane3, gel-filtration (Superose 6 FPLC, GE Pharmacia) column eluate. (D) Purification of rPAV-KLP. The following are shown: lane 1, Sf9 cell high-speed supernatant; lane 2, Ni-NTA affinity-column eluate; and lane3, gel-filtration (Superose 6 FPLC, GE Pharmacia) column eluate. (E,F) Purification of KLP61F from Ni-NTA eluate by Bio-gel A15m Gel filtration. (E) Commassie blue-stained SDS-polyacrylamide gel shows the composition of protein fractions obtained during the purification of KLP61F from the gel-filtration chormotography. Void volume occurs at fractions 22-25 and included volume occurs at fractions 62-66 (F) Corresponding plot of A280 versus fraction number shows one peak of monodisperse KLP61F.
Gel filtration Chromatography of Baculovirus expressed KLP61F/Ncd
A bio-gel A15m column (Biorad, 200ml) was equilibrated with buffer L (20 mM Tris, PH8.0, 75 mM KCl, 2mM MgCl2, supplemented right before use with 2mM DTT, protease inhibitors, and 0.1mM ATP). Then, 5ml Ni-NTA column-purified KLP61F protein was loaded onto the column. The column was run at a flow rate of 0.3ml/min and eluate was collected in 3ml /fraction. The presence of rKLP61F was monitored by SDS-PAGE and Commassie staining (Fig. 4E). If necessary, the gel filtration fractions can be pooled and concentrated by Centriprep 30. 2-4 mg protein was purified in a monodisperse, active state from 5g cell pellet and as far as could be discerned based on hydrodynamic analysis, the oligomeric state of the recombinant proteins was identical to the native embryonic proteins [16].
Purification of KLP61F subfragments
In order to study the domains required for homotetramerization as well as MT-MT crosslinking and sliding by full length KLP61F, we prepared several KLP61F subfragments using the same approach as described above for the intact motor [16, 40]. It was of particular interest to compare the full length tetrameric rKLP61F (Rs = 16.7 nm; S-value = 7.4S; native M.W. = 520kDa) with a motorless KLP61F homotetramer (K354-1066; Rs = 16.4 nm; S-value = 5.5S; M.W. = 378kDa) and a homotetrameric stalk subfragment lacking the motor and tail domains (K354-923; Rs = 13.3 nm; S-value = 4.9S; M.W. = 272 kDa) as discussed in section 4.
3.3 Expression and purification of other mitotic kinesins (the kinesin-13 rKLP10A, and the kinesin-6 rPAV-KLP)
We adopted the same strategy as in section 3.2 to express and purify the kinesin-13 KLP 10A and the kinesin-6 PAV-KLP using the Baculovirus system with a few modifications. Instead of restriction enzyme digestion, the cDNA of KLP10A and PAV-KLP were specifically amplified by PCR using primers containing gateway attB enzyme sites and 6×His tags, which were engineered onto the protein’s C-terminus. By gateway BP and LR reactions (Invitrogen), the genes were cloned into pDEST 8 vector. The recombinant pDEST 8 vector (expression vector) was verified by sequencing, then transformed into DH10Bac E.coli. The recombinant baculovirus was generated and protein expression was checked as described in 3.2. Both proteins were purified from Sf9 cell high-speed supernatants with Ni-NTA affinity chromatography, followed by superpose 6 gel-filtration FPLC chromatography (GE Pharmacia), and pure proteins in milligram yields were obtained (3-5 mg protein from 5g cell pellet) (Fig. 4C,D). The same Tris buffer system as in section 3.2 was used for affinity and gel-filtration columns, except that the KCl concentration in buffer L was increased from 75mM to 300 mM for maximal yield of PAV-KLP, as our results showed that PAV-KLP was not soluble when the salt concentration was lower than 200mM.
4. In vitro assays for mitotic motor proteins
4.1. General comments
Microtubule-based motor proteins are fascinating as they use the chemical energy from ATP to generate force to translocate MT vectorially. In vitro biochemistry, often done using motor domain subfragments, has addressed very important questions about motor activity, such as how the motor uses ATP hydrolysis to perform work, and the force-velocity relationship of single motors. Our focus on purifying the full length proteins has allowed us to assay motor ultrastructure and activities such as MT-MT crosslinking and sliding which cannot be studied using protein subfragments. An obvious advantage of in vitro systems for all this work is that investigators can reconstitute mitotic movements and can also directly observe the interactions between different motors.
MT gliding assay is a basic motility assay that can be applied to both full length motors and subfragments thereof. Motors are adsorbed onto a coverslip and are immobilized. When MTs plus ATP in buffer are introduced to the motors, MTs bind to the adsorbed motors and are transported by them along the coverslip surface (Fig. 5A). Since the diameter of the MT (25nm) is below the limit of resolution of the light microscope, video-enhanced differential interference contrast (VE-DIC) microscopy or fluoresecent microscopy of fluorescent dye-labeled MTs are commonly used (Fig. 5B). Both DIC and flurescence techniques provide the same information on motor’s directionality, gliding velocities, and the kinetic parameters. If more than one motor is introduced to the system, the gliding assay can be used to test functional interactions between these motors [16]. For example, we have analyzed antagonistic motility between the “plus end-directed” kinesin-5, KLP61F and the “minus end-directed” kinesin-14, Ncd, by plotting MT gliding rate versus mole fraction of Ncd (where mole fraction is [Ncd]/([Ncd]+[KLP61F]), ignoring solvent and buffer concentrations). The results revealed the range of mole fractions within which the two motors could potentially balance one another in vivo to maintain the prometaphase spindle at a steady state length.
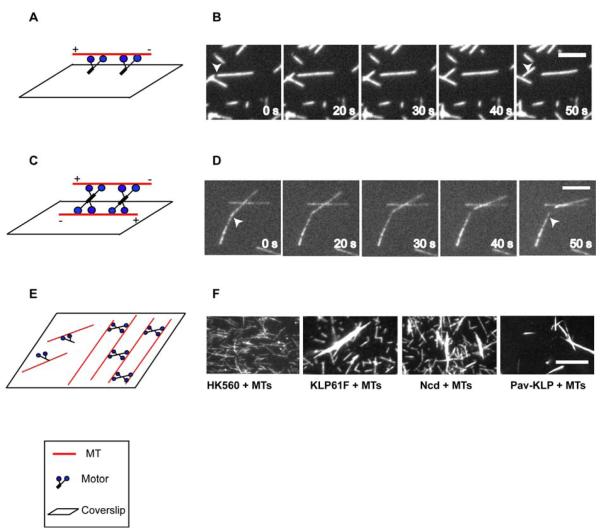
Figure 5. Microtuble gliding assay (A, B) sliding assay (C, D) and bundling assay (E, F).
(A) Diagram of MT gliding assay. MTs are moved by motor proteins absorbed to the coverslip. (B) Fluorescence images of MTs moved by KLP61F immobilized on a coverslip. The scale bar represents 5μm. (C) Diagram of MT sliding assay. MTs attach via motor proteins to surface-immobilized MTs, and motors drive the MTs in solution to slide over the fixed MTs in an anti-parallel orientation. (D) Fluorescence images of one MT being slid by KLP61F over another fixed MT. The scale bar represents 5μm. (E) Diagram of Bundling Assay. Mtitotic motors crosslink adjacent MTs into bundles. (F) Purified rKLP61F, rNcd, and rPav-KLP can bundle MTs in 1mMATP. Fluorescence microscopy shows that purified rKLP61F, rNcd and rPav-KLP have obvious bundling activity. In the presence of 1 mM ATP, free MTs cannot be bundled by HK560 but can form robust MT bundles in the presence of rKLP61F, rNcd, and rPav-KLP. The salt condition for HK560, KLP61F and Ncd is 75 mM KCl, while for Pav-KLP is 200 mM KCl. The scale bar represents 10 μm.
MT-MT sliding assay is a modified gliding assay, pioneered by the elegant studies of Peterman, Schmidt and colleagues [12, 41], which is designed for specifically assaying the ability of full-length motors to slide adjacent MTs in relation to one another. For example, to test the idea that bipolar Kinesin-5 can crosslink anti-parallel MTs and slide them apart, both ends of the motor have to be attached to MTs. Instead of sticking the motors onto the coverslip, a set of MTs are pre-fixed onto the coverslip, and the motors and fluorescent MTs from solution bind to the fixed MTs (Fig. 5 C). To prevent the motors from binding to the coveslip, it is blocked using a polymer brush [12]. In this way, motor domains on both ends of the protein can freely move on MTs. Using these assays, rKLP61F has been shown to move on both MTs that it crosslinks [40].
MT bundling assay
Some kinesins have MT-binding sites at both ends of the intact holoenzyme, so they can crosslink MTs into bundles under physiological MgATP concentrations. To test this idea, pure full-length kinesin proteins are mixed with fluorescent MTs in the presence of ATP and are examined by fluorescence light microscopy. MTs will form bundles if the proteins can crosslink MTs [16]. In addition, when the concentrations of motor and MTs are decreased to generate bundles only consisting of two MTs, the parallel and anti-parallel orientations of MTs within bundles can be determined by using polarity marked-MTs. By this way we can test if the kinesin proteins display any polarity preference when crosslinking MTs [40].
4.2. MT gliding assay using purified rKLP61F
MTs polymerization
To make regular fluorescent MTs, unlabeled bovine tubulin and rhodamine-labeled tubulin (Cytoskeleton, ratio = 15:1) were mixed in BRB80 (80 mM PIPES, 1 mM MgCl2, 1 mM EGTA, pH 6.8). Use centrifugation to get rid of any aggregation (Beckman TLA 100, 80K rpm, 15 mins, 4°C). The supernatant was diluted to 2 mg/ml with BRB80, and then GTP was added to 1mM. Incubate the mixture at 37 °C for 20 min. Taxol was added stepwise to 10μM and NaN3 to 2μM.
To make polarity-marked MTs, “bright seed mix” (2mg/ml bright tubulin (rhodamine-tubulin: unlabeled tubulin = 1:2), 1mM DTT and 1mM GMPCPP in BRB80) was incubated at37°C for 15min, then was mixed with “dim elongation mix” (1.5mg/ml dim tubulin (rhodamine-tubulin: unlabeled tubulin = 1:8), 1.2 mg/ml NEM-tubulin, 1mMDTT and 1mM GTP in BRB80) and incubated at 37°C for an additional 30mins. Finally taxol was added stepwise to 20 μM.
(Optional) To get rid of free tubulin, pellet microtubules over a 40% glycerol cushion (Beckman TLA100, 30K, 15min, RT). Rinse pellet and resuspend it in BRB80 + 10μM taxol + 2μM NaN3. The MTs can be stored at RT for 2-3 weeks.
Making flow chambers
Two strips of double-stick tape are cut, then put onto a glass slide. An acid-cleaned coverslip is then pressed down onto the tape to make a flow chamber.
MT gliding assay
The flow chamber was filled with 1mg/ml casein or BSA and incubated for 5minutes. Unbound casein was washed out with 3 flow cell volumes of wash buffer. It is not necessary to use BRB 80 in the MT motility assay once MTs are polymerized in BRB80. Since KLP61F with high concentration tends to precipitate in BRB80, a Tris-based buffer, buffer L (20 mM Tris PH8.0, 75 mM KCl, 2mM MgCl2, 2mM DTT, 0.1mMATP, Protease inhibitors,10% Glycerol), was used in KLP61F’s gliding assay. Freshly purified KLP61F (10nM-100nM) was flown into the chamber and incubated on ice for 5 minutes. Unbound KLP61F was washed out with 3 flow cell volumes of wash buffer. MTs were diluted 500-1000 times in buffer L supplied with 10μM taxol, 1mM ATP and antifade. Diluted MTs were flown into the chamber and the MT motility was observed under a fluorescence microscope (Fig. 5B).
4.3. MT sliding assay using purified KLP61F
MT preparation
Biotinylated Cy5-labeled MTs were polymerized from a mixture of 0.1 μM Cy5 labeled tubulin, 0.8 μM biotin-labeled tubulin and 10 μM unmodified tubulin in the presence of 1 mM GMPCPP (Jena Bioscience) and 2 mM DTT at 35 °C for 25 minutes. After stabilization with 10 μM taxol, MTs were pelleted through a glycerol cushion to remove free tubulin and were subsequently resuspended.
Making flow chambers
Cover slips were treated with dimethyl-dichlororsilane, and chambers were prepared by attaching the cover slips to microscope slides using double-stick tape.
MT sliding assay
The chamber was incubated for 5 minutes with BSA-biotin (Sigma-Aldrich, 0.1 mg ml−1) in PEM80 (80 mM K2PIPES, 1 mM EGTA, 2 mM MgCl2, pH 6.8, set with HCl), washed with buffer and incubated for 5 minutes with streptavidin (Biochemika, 0.1 mg ml−1). The surface was blocked by incubating with a watery solution of Pluronic F108 (0.2 %(w/v), BASF) for five minutes. Next, the chambers were incubated with biotinylated Cy5-labeled MTs (5min). After rinsing with buffer, the chambers were flushed with 1 nM KLP61F, 2 mM ATP and rhodamine labeled MTs in motility buffer (PEM80 buffer containing 4 mM DTT, and antifade).
4.4. MT bundling assays of kinesin motors
Bundling assays to determine the crosslinking activity of KLP61F, Ncd and Pav-KLP
For purified full-length rKLP61F, motorless rKLP61F, rKLP61F stalk, rNcd, and rPav-KLP, the gel-filtration fractions were pooled and concentrated by Centriprep 30. Fluorescent MTs were polymerized as described in 4.2. MTs were pelleted and resuspended in buffer L + 10μM taxol. Bundling assays were performed in buffer L with 2.5 μM MTs, 0.2 μM motor proteins, 10 μM taxol, and 1 mM ATP (final concentrations). For Pav-KLP, the KCl concentration in buffer L was changed from 75mM to 200mM for the reason mentioned in 3.3. The mixture was rocked at room temperature for 30 min. Mixtures of 25 μl were transferred into a flow chamber with a 0.02 mg/ml DEAE-Dextran-coated coverslip. The unstuck mixture was washed out with 75 μl buffer L supplied with 1 mM ATP, 10 μM taxol and antifade after 3 min. Bundling of fluorescent MTs was observed by fluorescence microscopy (Nikon E600). Human kinesin-1 neck-motor-domain construct, fragment HK560 (1–560 aa with GFP on the C terminus, Vale lab) was used under the same conditions as a control. Each experiment was repeated more than three times to ensure consistent results. The results showed that purified KLP61F, motorless KLP61F, Ncd and Pav-KLP can all bundle MTs under 1mM ATP (Fig. 5F). However, the rKLP61F stalk and the control HK560 do not bundle MTs. The results suggest that MT cross-linking is not due to the action of two heads at only one end of the protein but requires MT binding activities by domains at opposite ends of the full-length oligomerized motor, and in the case of KLP61F, tetrameric fragments lacking either the motor or the tail domains, but not both, can bundle MTs [16, 40].
Bundling assays to determine the orientation of MTs bundled by KLP61F
In order to determine the crosslinking preference of KP61F, we used cover slips that were positively charged by silanization with 0.1 % (V/V) DETA (3-(2-(2-aminoethylamino)ethyl-amino)propyl-trimethoxysilane, Aldrich) in water (incubated for 10 minutes, subsequently washed in water). Sample chambers were incubated with a mixture of polarity marked MTs, 20 nM KLP61F, 2 mM nucleotide (AMPPNP, ATP/AMPPNP, or ADP) in PEM80 buffer containing 4 mM DTT, and antifade. Fluorescent images were taken, and for all observed bundles consisting of two MTs of which the polarity could be unambiguously assigned, the relative orientation was determined. For each experiment, control experiments without KLP61F and with KLP61F stalk subfragements were performed to exclude the possibility that MT bundling occurred non-specifically. The result showed that KLP61F displayed a three-fold higher preference for crosslinking MTs in the antiparallel, relative to the parallel orientation. This preference is preserved when the motor domains are totally absent as was the case for the motorless KLP61F, suggesting that the C-terminal tail domains might specify the orientation preference of kinesin-5 motors [40].
5. Conclusions
The purification and assay of kinesin motors involved in mitosis is obviously an important yet challenging endeavour. We have directed our efforts to a study of the biochemical properties of mitotic kinesin holoenzymes, which presents a particular challenge, since they are usually not present in sufficiently high abundance to allow their purification in sufficiently high yield for detailed analysis. On the other hand, while it is possible to express and purify reasonable yields of recombinant full length kinesin proteins using the bacculovirus expression system, it is unclear if the resulting preparations are missing factors such as accessory subunits that are normally present in the native complex. Thus it seems that, at present, the best compromise is to partially purify native holoenzymes via MT affinity in order to gain information about the possible oligomeric state of the native motor complex, and to use highly purified bacculovirus-expressed full length protein for detailed biochemical studies.
Studying the biochemical properties of purified full length mitotic kinesins is important in revealing motor activities that could play critical roles in mitosis that are not illuminated by studies of motor domain subfragments, for example MT-MT crosslinking and sliding. In optimal cases, as with the homotetrameric kinesin-5, KLP61F, the oligomeric state of the native holoenzyme and the bacculovirus-expressed protein appear to be the same (although it is admittedly difficult to be absolutely certain that critical low molecular weight subunits are absent from the recombinant preparations). In the case of KLP61F, the combined study of the native and recombinant protein has allowed us to determine that; (i) it is a bipolar homotetramer, capable of crosslinking both parallel and antiparallel MTs into bundles, with a 3-fold preference for the antiparallel orientation; (ii) it moves toward the plus ends of MTs at a rate of about 0.04μm/s; (iii) it crosslinks and slides adjacent MTs in relation to one another; and (iv) it antagonizes Ncd in motility assays. We hope that the strategies outlined here will be of value as a guide to other workers who are interested in studying such biochemical and structural properties of purified mitotic kinesin complexes.
Acknowledgements
Our work on mitotic motors is supported by NIH grant GM 55507. We thank Dr Frank McNally for invaluable advice in initially setting up the baculovirus expression system in our laboratory. We also acknowledge a very productive collaboration with members of the Peterman and Schmidt laboratories, notably Siet Van den Wildenberg and Lukas Kapitein, who initiated the kinesin-5 MT-MT sliding and orientation preference assays; these assays were applied to KLP61F when LT spent an enjoyable visit in the Peterman laboratory in Amsterdam.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Brust-Mascher I, Scholey JM. Mitotic spindle dynamics in Drosophila. Int Rev Cytol. 2007;259:139–72. doi: 10.1016/S0074-7696(06)59004-7. [DOI] [PubMed] [Google Scholar]
- [2].Mitchison TaES. Mitosis: a history of division. Nat Cell Biol. 2001;3:E17–21. doi: 10.1038/35050656. [DOI] [PubMed] [Google Scholar]
- [3].Walczak CE, Heald R. Mechanisms of mitotic spindle assembly and function. Int Rev Cytol. 2008;265:111–58. doi: 10.1016/S0074-7696(07)65003-7. [DOI] [PubMed] [Google Scholar]
- [4].Mogilner A, Oster G. Polymer motors: pushing out the front and pulling up the back. Curr Biol. 2003;13:R721–33. doi: 10.1016/j.cub.2003.08.050. [DOI] [PubMed] [Google Scholar]
- [5].Sharp DJ, Rogers GC, Scholey JM. Microtubule motors in mitosis. Nature. 2000;407:41–7. doi: 10.1038/35024000. [DOI] [PubMed] [Google Scholar]
- [6].Dogterom M, Yurke B. Measurement of the force-velocity relation for growing microtubules. Science. 1997;278:856–60. doi: 10.1126/science.278.5339.856. [DOI] [PubMed] [Google Scholar]
- [7].Kinoshita K, Arnal I, Desai A, Drechsel DN, Hyman AA. Reconstitution of physiological microtubule dynamics using purified components. Science. 2001;294:1340–3. doi: 10.1126/science.1064629. [DOI] [PubMed] [Google Scholar]
- [8].Cole DG, Saxton WM, Sheehan KB, Scholey JM. A “slow” homotetrameric kinesin-related motor protein purified from Drosophila embryos. J Biol Chem. 1994;269:22913–6. [PMC free article] [PubMed] [Google Scholar]
- [9].DeLuca JG, Newton CN, Himes RH, Jordan MA, Wilson L. Purification and characterization of native conventional kinesin, HSET, and CENP-E from mitotic hela cells. J Biol Chem. 2001;276:28014–21. doi: 10.1074/jbc.M102801200. [DOI] [PubMed] [Google Scholar]
- [10].Desai A, Verma S, Mitchison TJ, Walczak CE. Kin I kinesins are microtubule-destabilizing enzymes. Cell. 1999;96:69–78. doi: 10.1016/s0092-8674(00)80960-5. [DOI] [PubMed] [Google Scholar]
- [11].Helenius J, Brouhard G, Kalaidzidis Y, Diez S, Howard J. The depolymerizing kinesin MCAK uses lattice diffusion to rapidly target microtubule ends. Nature. 2006;441:115–9. doi: 10.1038/nature04736. [DOI] [PubMed] [Google Scholar]
- [12].Kapitein LC, Peterman EJ, Kwok BH, Kim JH, Kapoor TM, Schmidt CF. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 2005;435:114–8. doi: 10.1038/nature03503. [DOI] [PubMed] [Google Scholar]
- [13].McDonald HB, Stewart RJ, Goldstein LS. The kinesin-like ncd protein of Drosophila is a minus end-directed microtubule motor. Cell. 1990;63:1159–65. doi: 10.1016/0092-8674(90)90412-8. [DOI] [PubMed] [Google Scholar]
- [14].Sawin KE, LeGuellec K, Philippe M, Mitchison TJ. Mitotic spindle organization by a plus-end-directed microtubule motor. Nature. 1992;359:540–3. doi: 10.1038/359540a0. [DOI] [PubMed] [Google Scholar]
- [15].Song H, Endow SA. Rapid purification of microtubule motor domain proteins expressed in bacteria. Biotechniques. 1997;22:82–5. doi: 10.2144/97221bm18. [DOI] [PubMed] [Google Scholar]
- [16].Tao L, Mogilner A, Civelekoglu-Scholey G, Wollman R, Evans J, Stahlberg H, et al. A homotetrameric kinesin-5, KLP61F, bundles microtubules and antagonizes Ncd in motility assays. Curr Biol. 2006;16:2293–302. doi: 10.1016/j.cub.2006.09.064. [DOI] [PubMed] [Google Scholar]
- [17].Meyer D, Rines DR, Kashina A, Cole DG, Scholey JM. Purification of novel kinesins from embryonic systems. Methods Enzymol. 1998;298:133–54. doi: 10.1016/s0076-6879(98)98015-6. [DOI] [PubMed] [Google Scholar]
- [18].Rappaport R. Cytokinesis in animal cells. Int Rev Cytol. 1971;31:169–213. doi: 10.1016/s0074-7696(08)60059-5. [DOI] [PubMed] [Google Scholar]
- [19].Scholey JM, Neighbors B, McIntosh JR, Salmon ED. Isolation of microtubules and a dynein-like MgATPase from unfertilized sea urchin eggs. J Biol Chem. 1984;259:6516–25. [PubMed] [Google Scholar]
- [20].Porter ME, Grissom PM, Scholey JM, Salmon ED, McIntosh JR. Dynein isoforms in sea urchin eggs. J Biol Chem. 1988;263:6759–71. [PubMed] [Google Scholar]
- [21].Scholey JM, Porter ME, Grissom PM, McIntosh JR. Identification of kinesin in sea urchin eggs, and evidence for its localization in the mitotic spindle. Nature. 1985;318:483–6. doi: 10.1038/318483a0. [DOI] [PubMed] [Google Scholar]
- [22].Cole DG, Cande WZ, Baskin RJ, Skoufias DA, Hogan CJ, Scholey JM. Isolation of a sea urchin egg kinesin-related protein using peptide antibodies. J Cell Sci. 1992;101(Pt 2):291–301. doi: 10.1242/jcs.101.2.291. [DOI] [PubMed] [Google Scholar]
- [23].Morris RL, Hoffman MP, Obar RA, McCafferty SS, Gibbons IR, Leone AD, et al. Analysis of cytoskeletal and motility proteins in the sea urchin genome assembly. Dev Biol. 2006;300:219–37. doi: 10.1016/j.ydbio.2006.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Bi GQ, Morris RL, Liao G, Alderton JM, Scholey JM, Steinhardt RA. Kinesin- and myosin-driven steps of vesicle recruitment for Ca2+-regulated exocytosis. J Cell Biol. 1997;138:999–1008. doi: 10.1083/jcb.138.5.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Cole DG, Chinn SW, Wedaman KP, Hall K, Vuong T, Scholey JM. Novel heterotrimeric kinesin-related protein purified from sea urchin eggs. Nature. 1993;366:268–70. doi: 10.1038/366268a0. [DOI] [PubMed] [Google Scholar]
- [26].Morris RL, Scholey JM. Heterotrimeric kinesin-II is required for the assembly of motile 9+2 ciliary axonemes on sea urchin embryos. J Cell Biol. 1997;138:1009–22. doi: 10.1083/jcb.138.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Skoufias DA, Cole DG, Wedaman KP, Scholey JM. The carboxyl-terminal domain of kinesin heavy chain is important for membrane binding. J Biol Chem. 1994;269:1477–85. [PubMed] [Google Scholar]
- [28].Chui KK, Rogers GC, Kashina AM, Wedaman KP, Sharp DJ, Nguyen DT, et al. Roles of two homotetrameric kinesins in sea urchin embryonic cell division. J Biol Chem. 2000;275:38005–11. doi: 10.1074/jbc.M005948200. [DOI] [PubMed] [Google Scholar]
- [29].Rogers GC, Chui KK, Lee EW, Wedaman KP, Sharp DJ, Holland G, et al. A kinesin-related protein, KRP(180), positions prometaphase spindle poles during early sea urchin embryonic cell division. J Cell Biol. 2000;150:499–512. doi: 10.1083/jcb.150.3.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Siegel LM, Monty KJ. Determination of molecular weights and frictional ratios of proteins in impure systems by use of gel filtration and density gradient centrifugation. Application to crude preparations of sulfite and hydroxylamine reductases. Biochim Biophys Acta. 1966;112:346–62. doi: 10.1016/0926-6585(66)90333-5. [DOI] [PubMed] [Google Scholar]
- [31].Goshima G, Vale RD. The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J Cell Biol. 2003;162:1003–16. doi: 10.1083/jcb.200303022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kashina AS, Baskin RJ, Cole DG, Wedaman KP, Saxton WM, Scholey JM. A bipolar kinesin. Nature. 1996;379:270–2. doi: 10.1038/379270a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kashina AS, Scholey JM, Leszyk JD, Saxton WM. An essential bipolar mitotic motor. Nature. 1996;384:225. doi: 10.1038/384225a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Barton NR, Pereira AJ, Goldstein LS. Motor activity and mitotic spindle localization of the Drosophila kinesin-like protein KLP61F. Mol Biol Cell. 1995;6:1563–74. doi: 10.1091/mbc.6.11.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Hoyt MA, He L, Loo KK, Saunders WS. Two Saccharomyces cerevisiae kinesin-related gene products required for mitotic spindle assembly. J Cell Biol. 1992;118:109–20. doi: 10.1083/jcb.118.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Sharp DJ, McDonald KL, Brown HM, Matthies HJ, Walczak C, Vale RD, et al. The bipolar kinesin, KLP61F, cross-links microtubules within interpolar microtubule bundles of Drosophila embryonic mitotic spindles. J Cell Biol. 1999;144:125–38. doi: 10.1083/jcb.144.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Classification and nomenclature of viruses. Fourth report of the International Committee on Taxonomy of Viruses. Intervirology. 1982;17:1–199. doi: 10.1159/000149278. [DOI] [PubMed] [Google Scholar]
- [38].Rohrmann GF. Polyhedrin structure. J Gen Virol. 1986;67(Pt 8):1499–513. doi: 10.1099/0022-1317-67-8-1499. [DOI] [PubMed] [Google Scholar]
- [39].Smith GE, Summers MD, Fraser MJ. Production of human beta interferon in insect cells infected with a baculovirus expression vector. Mol Cell Biol. 1983;3:2156–65. doi: 10.1128/mcb.3.12.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Van den Wildenberg S, Tao L, Kapitein LC, Schmidt CF, Scholey JM, Peterman EJ. The Homotetrameric kinesin-5, KLP61F, preferentially crosslinks microtubules into antiparallel orientations. Curr. Biol. 2008;18:1860–64. doi: 10.1016/j.cub.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kapitein LC, Kwok BH, Weinger JS, Schmidt CF, Kapoor TM, Peterman EJ. Microtubule cross-linking triggers the directional motility of kinesin-5. J Cell Biol. 2008;182:421–8. doi: 10.1083/jcb.200801145. [DOI] [PMC free article] [PubMed] [Google Scholar]