Abstract
Coupling Fourier transform infrared spectroscopy with focal plane array detectors at synchrotron radiation sources (SR-FTIR-FPA) has provided a rapid method to simultaneously image numerous biochemical markers in situ at diffraction limited resolution. Since cells and nuclei are well resolved at this spatial resolution, a direct comparison can be made between FTIR functional group images and the histology of the same section. To allow histological analysis of the same section analyzed with infrared imaging, unfixed air-dried tissue sections are typically fixed (after infrared spectroscopic analysis is completed) via immersion fixation. This post fixation process is essential to allow histological staining of the tissue section. Although immersion fixation is a common practice in this filed, the initial rehydration of the dehydrated unfixed tissue can result in distortion of subcellular morphology and confound correlation between infrared images and histology. In this study, vapor fixation, a common choice in other research fields where postfixation of unfixed tissue sections is required, was employed in place of immersion fixation post spectroscopic analysis. This method provided more accurate histology with reduced distortions as the dehydrated tissue section is fixed in vapor rather than during rehydration in an aqueous fixation medium. With this approach, accurate correlation between infrared images and histology of the same section revealed that Purkinje neurons in the cerebellum are rich in cytosolic proteins and not depleted as once thought. In addition, we provide the first direct evidence of intracellular lactate within Purkinje neurons. This highlights the significant potential for future applications of SR-FTIR-FPA imaging to investigate cellular lactate under conditions of altered metabolic demand such as increased brain activity and hypoxia or ischemia.
Keywords: Multi-beam, SR-FTIR-FPA, imaging, Purkinje neuron, nucleus, lactate, protein
Knowledge of neuronal biochemistry at the cellular and subcellular level is critical to elucidate mechanistic pathways involved in both normal brain function and neurodegeneration. However, few methods are available to study the complex biochemical composition of individual neurons in situ. Analysis of neurons at the subcellular level with visible, fluorescence, or electron microscopy is limited to morphology and the distribution of molecules that can be identified with antibodies or tagged with a suitable radioisotope, fluoro- or chromophore. Although such methods have provided many insightful contributions, they cannot simultaneously image an appreciable fraction of the neuronal metabolome, including important metabolites such as lactate. Many analytical methods provide information at the tissue level but usually not at the cellular or subcellular level, and as a direct consequence of this much remains unknown about global neuronal biochemistry.
Illumination of a focal plane array detector with infrared light generated by a synchrotron source provides a rapid method to image across the entire mid-infrared range at diffraction limited spatial resolution.1−6 In the first study to demonstrate subcellular resolution of neurons with SR-FTIR-FPA imaging, FTIR images were coupled with histology of the same tissue section to resolve individual pyramidal neurons within the hippocampus.1
In this study, SR-FTIR-FPA imaging was used to investigate the cellular and subcellular biochemical composition of Purkinje neurons, in situ. Purkinje neurons form the only output pathways in the cerebellar cortex, and are crucial for balance and muscle movement, which is controlled by the cerebellum.7−9 Purkinje neurons are vulnerable to damage (particularly ischemic damage), and a large amount of research has been conducted on the role, content, and distribution of specific proteins and enzymes that could account for this vulnerability.8,10,11 The relative protein content within the cytosol of the Purkinje neuron is somewhat controversial,12,13 due to the difficulties associated with performing semiquantitative analyses from routine histology, such as differential penetration of protein binding dyes through sections, variable dye binding to different protein structures, and nonspecific binding of dyes to other macromolecules (e.g., carbohydrate residues, nucleic acids).14,15 Moreover, an accurate understanding of the total protein content within a cell is important for normalization of the measured expression of individual proteins by immunocytochemistry and autoradiography.12 Our understanding of Purkinje neuron metabolism is far from complete; the large number of connections between Pukinje cells and glial cell processes (particularly Bergmann glia) suggest transport of metabolites into or out of the Purkinje neuron.7,16 High localized levels of lactate dehydrogenase in Purkinje neurons suggests that lactate is produced or consumed in large quantities within these cells.17,18 Unfortunately, the methods used to date have lacked the capability of studying the cellular or subcellular metabolite distribution, and in particular lactate distribution. Nuclear magnetic resonance spectroscopy can be used to study the in vivo alterations in cerebral lactate concentrations, but it lacks the spatial resolution needed to resolve individual neurons.19−21 Likewise, biochemical assays of microdissected tissue for which Purkinje neurons were the major cell type have revealed elevated lactate levels, but the Purkinje cell layer is too small to dissect only the Purkinje cells.22 In the study reported herein, we aim to resolve some long-standing questions regarding intracellular protein content, and to visualize intracellular lactate within Purkinje neurons. Crucial to this work was a detailed examination of the protocols required for subsequent (after spectroscopic analysis) fixation and histological staining of the same tissue section analyzed with FTIR imaging. To the best of our knowledge, this is both the first study to report the true relative protein content of Purkinje neurons, and the first attempt to image the relative lactate content at the cellular level.
Results and Discussion
This study was driven by four specific aims: (1) replication of published work to validate the FTIR spectroscopic markers used to resolve individual neuron soma and subcellular features for this study (nucleus and cytoplasm); (2) comparison and optimization of postfixation (chemical fixation performed after SR-FTIR-FPA imaging) for accurate histology of the same tissue section analyzed with SR-FTIR-FPA imaging; (3) SR-FTIR-FPA imaging investigation of the relative protein content within Purkinje neurons (cytosol and nucleus) and surrounding tissue (cerebellum molecular and granular layer); (4) SR-FTIR-FPA investigation of the relative lactate distribution in Purkinje neurons.
1. Validation of FTIR Spectroscopic Markers to Resolve Neuron Soma and Nucleus
Recently, SR-FTIR-FPA imaging of murine hippocampal tissue was combined with histology of the same tissue section to resolve the soma of individual neurons, and to demonstrate the subcellular spatial resolution capabilities of the technique.1 Specifically, absorbance of the ester carbonyl band was used as a spectroscopic marker to generate semiquantitative images of lipid distribution. Due to the lack of lipid within the neuron soma relative to the surrounding neuropil, resolution of individual pyramidal neurons was achieved. As can be seen in Figure 1, false-color functional group images generated from the area underneath the lipid carbonyl band provide discrimination of the soma of individual rat hippocampal pyramidal neurons from the surrounding neuropil. The same marker band was applied to rat cerebellum tissue (Figure 2), which also allowed identification of the soma of individual Purkinje neurons. A line map of the area under the lipid carbonyl band also clearly indicates the location of the neuronal soma (Figure 3C, D). However, it should be noted that the resolving power of this approach is highly dependent on the lipid content of the neuron and surrounding tissue. Due to the very low lipid content of the granular layer, and the higher relative lipid content of the molecular layer, both of which surround the Purkinje neuron soma in the cerebellum, visual discrimination of the Purkinje neuron soma based on this univariate analytical approach was not as obvious compared to the hippocampal example.
Figure 1.
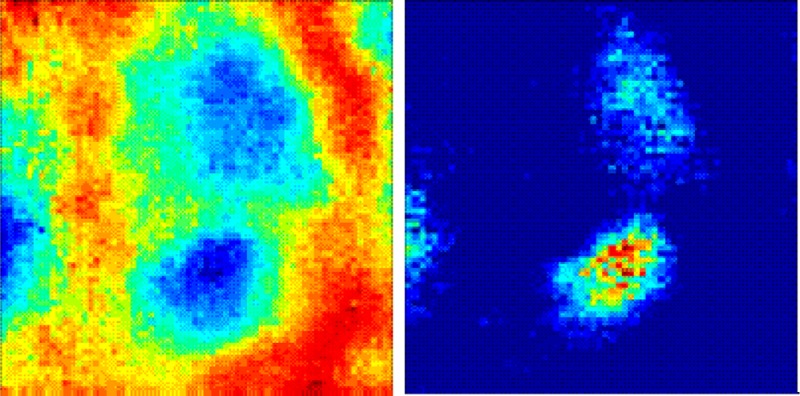
SR-FTIR-FPA imaging of pyramidal neurons within the hippocampus. (A) Optical image of the unstained tissue section with locations of neuron soma and nuclei drawn free hand in black and white, respectively. White square indicates region analyzed with SR-FTIR-FPA imaging. (B) Optical image of (A) without annotations. (C) Optical image of H&E stained section analyzed with SR-FTIR-FPA imaging (black box defines region analyzed). H&E stain performed after spectroscopic analysis. (D) False-color functional group image generated from area underneath the lipid ν(C=O) band (1750–1710 cm–1), to reveal the presence of neuron soma. (E) False-color functional group image generated from the ratio of the area underneath the νs(CH3) band (2880–2865 cm–1) and νs(CH2) band (2860–2845 cm–1), to reveal the presence of neuron nuclei. Scale bars = 10 μm.
Figure 2.
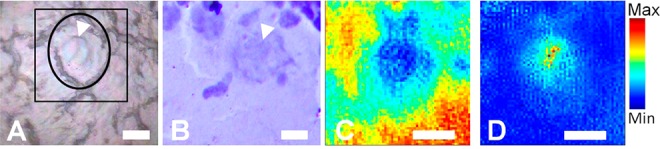
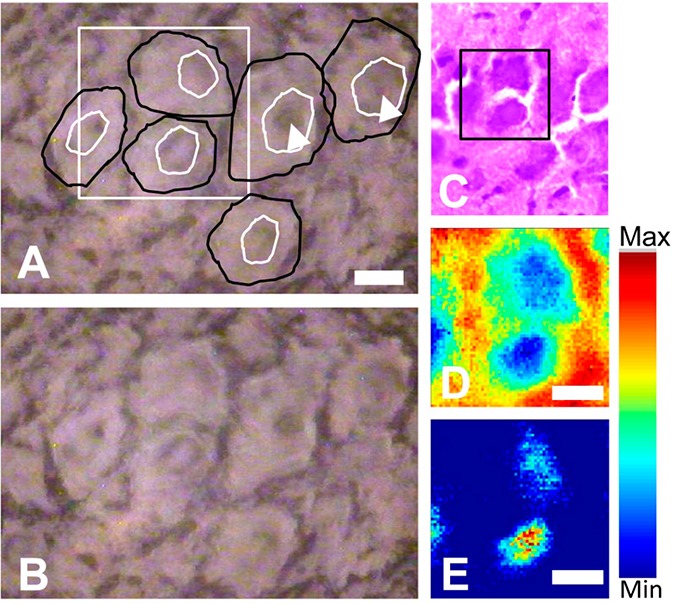
SR-FTIR-FPA imaging of a Purkinje neuron within the cerebellum. (A) Optical image of the unstained tissue section with location of neuron soma drawn free hand in black, the nucleus shown with the white arrowhead. Black square indicates region analyzed with SR-FTIR-FPA imaging. (B) Optical image of cresyl biolet stained section analyzed with SR-FTIR-FPA imaging. Cresyl violet stain performed after spectroscopic analysis. (C) False-color functional group image generated from area underneath the lipid ν(C=O) band (1750–1710 cm–1), to reveal the presence of neuron soma. (D) False-color functional group image generated from the ratio of the area underneath the νs(CH3) band (2880–2865 cm–1) and νs(CH2) band (2860–2845 cm–1), to reveal the presence of neuron nuclei. Scale bars = 10 μm.
Figure 3.
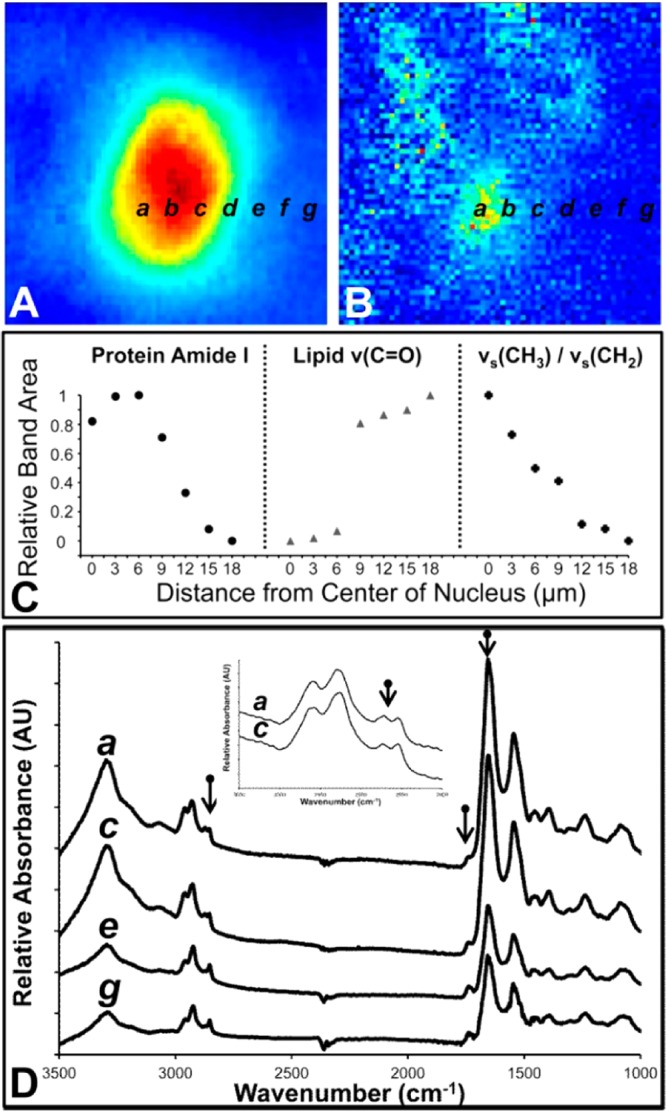
Representative line map of a cross section through a Purkinje neuron and surrounding molecular layer tissue. (A) Relative protein distribution from integrated area under the amide I band. (B) Ratio of methyl to methylene groups ν(CH3):ν(CH2), which highlights the location of the Purkinje neuron nucleus. (C) Line maps generated from integrated areas underneath the amide I band (protein), carbonyl band (lipids), and the methyl to methylene ratio from spectra extracted at 3 μm intervals from the center of the nucleus (a–g). (D) Representative spectra from locations a, c, e, and g from parts (A) and (B). The inset highlights the increased methyl to methylene ratio observed in the nucleus relative to the cytoplasm within the Purkinje neuron.
In addition to resolution of neuron soma, a characteristic marker for the nucleus (the ratio of the area underneath the methyl and the methylene bands) was identified for both rat hippocampal pyramidal neurons, and cerebellar Purkinje neurons. Identification of the nucleus could be achieved from the optical images of the unstained tissue section, and good correlation was observed with the false color functional group maps generated from the νs(CH3):νs(CH2) band ratio (Figure 1E, Figure 2D). The ability to resolve the soma of individual neurons and their nuclei with SR-FTIR-FPA imaging allows the biochemistry of the soma as a whole, the nucleus, or cytoplasm to be studied. Representative examples of spectra collected from the nucleus and cytoplasm, which highlights the increased methyl to methylene ratio within the nucleus, are presented in Figure 3. As the nucleus is a major site of methylation within the cell, this finding is not unexpected, and is likely to exist across all cells and not only neurons.23−25 We have also observed the same increase in relative concentration of methyl groups in the nuclei of neurons of the dentate gyrus (hippocampus), deep cerebellar nuclei (cerebellum) and brain stem (data not shown). The ability to resolve the nucleus based on relative methyl content, may find significant application in future studies regarding brain epigenetics, where neuronal DNA methylation is thought to play critical roles.26 Independent from revealing any chemical information, the choice of the νs(CH3) to νs(CH2) ratio to visualize the cell nucleus is useful due to the relatively short wavelengths of these bands (approximately 3.5 μm), allowing images with diffraction limited resolution for this wavelength to be generated (conservative approximation of 2.7 μm).2,27
2. Post Fixation: Immersion versus Vapor Fixation
The ability of SR-FTIR-FPA array imaging to generate biochemical maps at cellular or subcellular resolution demands concomitant high quality histological images of the same tissue section for accurate correlation between biochemical distribution and morphology of a specific cell or subcellular feature (i.e., nucleus). It is well established that chemical fixation drastically alters the elemental and biochemical composition of brain tissue.28−31 Hence, unfixed flash frozen samples are required for biospectroscopic investigations of this nature, with post fixation performed after spectroscopic analysis to allow the tissue section to be then stained. The literature on FTIR spectroscopic studies of brain tissue reports almost exclusively immersion-formalin-fixation of tissue sections which is performed after spectroscopic analysis and prior to histological staining (typically hematoxylin and eosin or cresyl violet).1,30−35 Although immersion fixation will preserve morphology at the tissue level, it is known that immersion fixation of unfixed tissue sections results in significant alterations to subcellular morphology (dilation of organelles) and loss of proteins and nucleic acids upon immersion in the aqueous fixation medium.28,36−39 As such, when analysis of flash frozen unfixed tissue is required (as is the case for biospectroscopic analysis of diffusible ions and molecules), vapor fixation is recommended post spectroscopic analysis, for histological analysis of the same, or serial sections.36−39 The process of vapor fixation stabilizes unfixed thin and semithin sections prior to immersion in aqueous fixative or staining reagents, which would otherwise instantaneously degrade tissue morphology.36−39 However, this approach has yet to be adopted following FTIR spectroscopic analyses of unfixed tissue sections.
In this investigation, two sets of tissue sections (10 μm thick) from unfixed flash frozen cerebellum tissue were either immersion fixed in 4% buffered formalin or were vapor fixed in the gas generated from dry formaldehyde heated to 80 °C, prior to H&E staining (Figure 4). Both sets (immersion and vapor fixed) of H&E stained tissue sections allowed identification of individual Purkinje neurons in addition to the molecular, granular, and inner white matter tissue layers of the cerebellum (Figure 4A,B,D,E). Due to the large size of the Purkinje neuron soma (25 – 40 μm), with a large nucleus of approximately 15 μm,7 a nucleus was not and would not be expected to be visible in each neuron, for both types of fixation protocol. However, visible identification of Purkinje nuclei was enhanced in vapor-fixed tissue relative to immersion-fixed tissue (Figure 4B,E). In smaller cells, for example granule neurons, the nuclei were clearly visible for both fixation protocol, however, the nuclei were slightly larger in immersion-fixed tissue relative to vapor-fixed tissue (Figure 4A,D). Immersion fixation will result in a chemically and morphologically stable sample after fixation is complete. However, these results indicate that the initial immersion of an unfixed tissue section in the aqueous formalin solution results in increased organelle size (most likely due to the dehydration–rehydration process on unfixed tissue) and leaching of cellular proteins and nucleic acids, contributing to degradation of histology quality.
Figure 4.
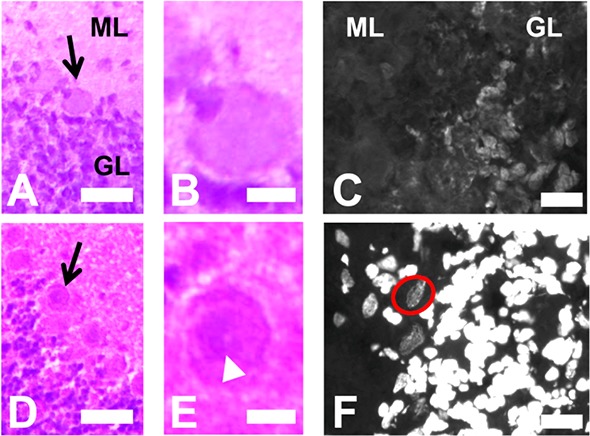
Immersion-formalin-fixation versus vapor fixation. (A,B) Optical images of an immersion-formalin-fixed H&E stained cerebellum section, showing a representative example of a Purkinje neuron (black arrow) with no identifiable nucleus. (C) Fluorescence image of a DAPI stained immersion-formalin-fixed cerebellum section showing poor labeling of nuclear DNA, and no visible Purkinje neurons. (D,E) Optical images of a formaldehyde-vapor-fixed H&E stained cerebellum section, showing a representative example of a Purkinje neuron (black arrow) with an identifiable nucleus (white arrowhead). (F) Fluorescence image of a DAPI stained formaldehyde-vapor-fixed cerebellum section showing strong labeling of nuclear DNA, and visible Purkinje neuron nuclei (highlighted by red circle). ML = molecular layer, GL = granular layer. Scale bars: (A, D) = 50 μm; (B,C,E,F) = 10 μm.
A similar effect was observed in a subsequent fluorescence microscopy investigation using 4′,6-diamidino-2-phenylindole (DAPI) which binds specifically to DNA. Two sets of tissue sections (10 μm thick) from unfixed flash frozen cerebellum tissue were either vapor fixed or immersion-fixed, and DNA stained with DAPI. As shown in Figure 4E, the immersion fixed sample displayed poor morphology from DAPI fluorescence, with no identification of the Purkinje cell nucleus. However, the vapor-fixed tissue displayed greater overall fluorescence intensity with clear identification of the nucleus for both granular and Purkinje neurons (Figure 4F). Therefore, these results highlight that although sample morphology at the cellular level can be achieved in immersion fixed samples, for correlative comparison between subcellular features (i.e., nucleus) and biochemical images generated by SR-FTIR-FPA imaging, vapor fixation of the tissue sections will provide more accurate correlation. It must be noted that the dispersion of nuclear material observed in this study is likely to be most pronounced in large cells (i.e., the Purkinje neurons which are 25–40 μm in size are completely sliced open in a 10 μm thick section),7,9 and will be less pronounced in small cells, which may retain a larger proportion (or all) of their cell membrane, minimizing distortions. In addition, the vapor-fixation method used in this study allowed the application of standard immunohistochemical methodology, and successful identification of astrocytes through antigenicity to glial fibrillary acidic protein (GFAP) was achieved (Supporting Information Figure 1). This highlights the potential for future studies to correlate subcellular SR-FTIR-FPA images with immunocytochemistry of the same tissue section, particularly for studies relating to free cytosolic proteins which cannot be otherwise detected in immersion-fixed tissue.36−39
3. Protein Content of Purkinje Neurons
It is well established that variation in the content and distribution of specific proteins and enzymes within the Purkinje neuron correlates with diminished cell function or cell death in numerous cerebellar ataxia’s and disorders.8 However, it has also been reported that great variance in total protein, enzyme, and RNA content exists between individual Purkinje neurons,9,13 and that normalization of the content of a specific protein or enzyme within a cell against the total protein content of that cell may be required.12 To address this issue, Coomassie blue (a protein-specific stain) is often used to quantify the total protein in tissue sections at the cellular level prior to autoradiography, so that the antigen binding observed for each cell can be normalized against total cellular protein content in the tissue section.12 It has been reported that Coomassie blue stained cerebellum tissue indicates relatively low protein within the cytosol of Purkinje neurons compared to the surrounding molecular and granular layer tissue.12 The Coomassie blue stain offers advantages relative to previous studies which normalized against the average protein content of the whole tissue (calculated from tissue homogenization and biochemical assay). Although other staining methods for total protein do exist, the stains are often nonspecific (stain nucleic acids or carbohydrates in addition to protein) or require defatting of tissue sections, which is not compatible with autoradiography.
However, the Coomassie blue/autoradiography method reported in the literature was performed on unfixed tissue sections, and the potential for loss of proteins upon immersion of the tissue section in the aqueous staining media was not taken into account.12 Therefore, it seems likely that the low protein content in the cytoplasm reported for Purkinje neurons, relative to the surrounding molecular and granular tissue, may in fact be an artifact of the sample preparation method used.28,36−39
In this study SR-FTIR-FPA imaging was employed to reveal the true protein distribution within the Purkinje neuron relative to the surrounding molecular and granular layer tissue (Figures 5 and 6). As this method is a direct, reagent free method, artifacts from differential penetration or binding of staining reagents does not occur. Further, as the spectroscopic analysis was performed on flash frozen unfixed tissue sections differential leaching of protein is not possible. As discussed in subsection 1, the location of neuronal soma can be visualized by the relatively low lipid content within the soma, or through multivariate clustering algorithms (Figure 5B). The nucleus could be identified within the Purkinje neuron from the νs(CH3) to νs(CH2) ratio, as discussed in subsection 1 (Figure 5D). Using this approach, the location of the soma of the Purkinje neuron, the nucleus, and therefore the cytoplasm (i.e., soma where nucleus is not present) could be identified, and the average protein content determined for the Purkinje neuron cytosol and nucleus. As can be seen from the images presented in Figures 5C and 6C, the line map and spectra presented in Figure 3, and the calculated area under the amide I band presented in Table 1, the area underneath the amide I band (a spectroscopic marker for total protein) was found to be significantly greater within the cytosol of the Purkinje neuron relative to the surrounding molecular and granular layer tissue.
Figure 5.
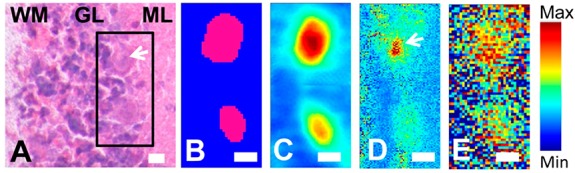
SR-FTIR-FPA imaging investigation of the protein and lactate content of Purkinje neurons (Synchrotron Radiation Center, Wisconsin). (A) Visible light image of the H&E stained tissue section (stained after spectroscopic analysis). Black rectangle indicates region of spectroscopic analysis, white arrow indicates Purkinje neuron with visible nucleus. (B) K-means cluster analysis (2 group) of FTIR spectra revealed two clusters assigned to Purkinje neuron soma (pink) or surrounding tissue (blue). (C) False-color functional group image generated from area underneath the protein amide I band (1690–1610 cm–1). (D) False-color functional group image generated from the ratio of the area underneath the νs(CH3) band (2880–2865 cm–1) and νs(CH2) band (2860–2845 cm–1), to reveal the presence of neuron nuclei (white arrow). (E) False-color functional group image generated from second-derivative intensity at the lactate sensitive region 1127 cm–1 (lactate ν(C–O)). WM = white matter, GL = granular layer, ML = molecular layer. Scale bars: (A–E) = 10 μm.
Figure 6.
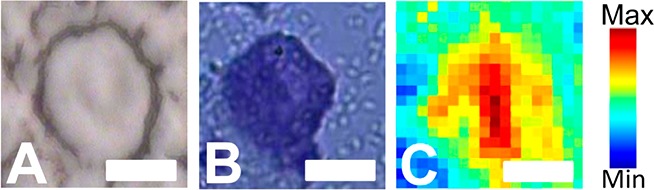
SR-FTIR-FPA imaging investigation of the protein content of Purkinje neurons (Canadian Light Source). (A) Visible light image of the unstained tissue section. (B) Visible light image of the cresyl violet stained tissue section. (stained after spectroscopic analysis). (C) False-color functional group image generated from area underneath the protein amide I band (1690–1610 cm–1). Scale bar = 10 μm.
Table 1. Relative Protein and Lactate Content in Purkinje Neuron Cytoplasm and Surrounding Granular and Molecular Layera.
| Purkinje neuron cytoplasm | .granular layer | molecular layer | |
|---|---|---|---|
| amide I band area (relative protein content) | 9.3 ± 1.9bc | 5.0 ± 1.0 | 5.6 ± 0.77 |
| amide I band area normalized to tissue thickness (relative protein content) | 0.53 ± 0.086bc | 0.25 ± 0.051 | 0.30 ± 0.049 |
| second-derivative intensity at 1127 cm–1 (relative lactate content) | –2.5 × 10–5 ± 1.3 × 10–5bc | –3.6 x10–6 ± 4.0 × 10–6 | –6.2 × 10–6 ± 9.5 × 10–7 |
| second-derivative intensity at 1127 cm–1 normalized to tissue thickness (relative lactate content) | –1.38 × 10–6 ± 5.7 × 10–7bc | 1.82 × 10 –7 ± 2.0 × 10–7 | –3.3 × 10–7 ± 3.3 × 10–8 |
| area underneath lactate band (curve fitting) | 0.14 ± 0.070bc | 0.044 ± 0.0033 | 0.032 ± 0.018 |
| area underneath lactate band (curve fitting) normalized to tissue thickness | 0.0077 ± 0.0030bc | 0.0022 ± 0.00017 | 0.0017 ± 0.0010 |
Values given are the average integrated area under the curve or second-derivative intensity ± standard deviation from n = 4 animals (the units are arbitrary).
Significant increase in Purkinje cell soma relative to molecular layer (p < 0.05).
Significant increase in Purkinje cell soma relative to granular layer (p < 0.05).
To account for local variations in tissue thickness which will affect the apparent protein concentration if constant thickness is assumed, the amide I band area was normalized to tissue section thickness determined from confocal fluorescence microscopy. The detection method employed to measure section thickness (eosin fluorescence) is less sensitive to nucleus thickness (which is not eosinophillic), and it is acknowledged that local variations in the thickness of the nuclei will affect protein content determined for the nucleus. However, significant elevation in the amide I band area for the Purkinje neuron cytosol in both normalized and un-normalized data demonstrates conclusively that the cytosol of the Purkinje neuron has elevated protein relative to the surrounding molecular and granular layer tissue. Therefore, it seems likely that the previous report of low protein content in the cytosol of the Purkinje neuron was due to leaching of protein from the neuron during staining of unfixed tissue sections. Replication of the Coomassie blue staining protocol used in the previous study12 on vapor-fixed tissue sections revealed increased Coomassie blue staining within the cytosol of Purkinje neurons in vapor-fixed tissue (Supporting Information Figure 1). A previous FTIR imaging investigation determined that an elevated protein to lipid ratio exists at the interface of the molecular and granular layers, but did not have the spatial resolution to identify increased protein within individual Purkinje neurons.40 Therefore, to the best of our knowledge this is the first study to reveal the true protein content of Purkinje neurons relative to surrounding tissue and is an important result for all studies which seek to normalize antigen binding or the localized concentration of a specific protein against the localized total protein concentration.
4. Lactate Content of Purkinje Neurons
Mounting evidence suggests lactate is a vital energy substrate for neurons under both anaerobic and aerobic conditions, and astrocytes are the most likely location of lactate production which is then shuttled to neurons (i.e., astrocyte neuron lactate shuttle hypothesis, ANLSH).41−50 However, due to the lack of a suitable technique to image the cellular location of lactate, the ANLSH hypothesis is still much debated.42−44,46−48 It has long been known that the fine structure of the Purkinje neuron has subcellular features consistent with those required to shuttle of metabolites into or out of the cell, however, there is little direct evidence for the identity of these metabolites.7 It has been shown that Purkinje neurons are rich in lactate dehydrogenase, and that Bergmann glia have numerous processes which contact the soma of Purkinje neurons.16−18,22 These findings are consistent with the supply of lactate derived from glia metabolism to Purkinje neurons and suggest a role of lactate in the function of Purkinje neurons.16−18,22 Previous studies with microdissection and biochemical assay have revealed increased lactate in microdissected tissue within which Purkinje neurons were the dominant cell type, however, the Purkinje cell layer is too small to isolate without contamination from surrounding tissue (i.e., molecular and granular layer tissue).22 Therefore, a technique capable of directly imaging the relative lactate concentration at the cellular level (<10 μm spatial resolution) would be of great benefit to this field.
Fortuitously, the ν(C–O) of lactate hydroxyl group (1127 cm–1) in infrared spectra displays a blue shift of approximately 50–100 cm–1 from the abundant carbohydrate hydroxyl groups, allowing calculation of relative lactate concentration with reduced error due to overlapping bands.51−58 However, even with this shift, the lactate band is not fully separated from other bands in mid-IR spectra of brain tissue, and a band narrowing/deconvolution or curve fitting approach is required to determine the relative lactate concentration.51−58 Curve fitting has previously been employed to quantify lactate concentration in biological fluids, or to profile (but not image) the relative lactate distribution in tissue.51−58 However, this procedure has not previously been applied to generate infrared images from several thousand of spectra. Further, due to the possibility of propagated fitting errors, especially under the invalid assumption that the uncountable number of overlying bands which contribute to FTIR spectra can be modeled by a small number of curves,59 and the impossibility of manually inspecting several thousand fitted spectra required in a single FTIR image, curve fitting was not employed to generate images of the relative lactate content in this investigation. Instead, the relative lactate content and distribution was determined from the second-derivative intensity at 1127 cm–1. Second-derivative spectra have previously been employed to study leaching of lactate from tissues during fixation,28 and to image the relative change in lactate concentration between the hypoxic and necrotic core of tumor spheroids.60 Further, variation in second-derivative intensity at the position of absorbance of characteristic protein secondary-structures has been used extensively to determine relative alterations in the content of various protein secondary structures.35,61−69 However, second-derivative intensity is only proportional to band area on the assumption of constant bandwidth at half height. Under conditions of varying bandwidth, the area underneath a band is not conserved in second-derivative spectra, and therefore, second-derivative intensity is no longer proportional to concentration. Therefore, increased or decreased second-derivative intensity is either due to an increase or decrease in analyte concentration, or an increase or decrease in the bandwidth of the absorbing molecule. Unless the latter can be ruled out analysis of second-derivative intensity may result in serious experimental errors, and is a major limitation of this method of analysis However, previous studies which have performed curve fitting analysis to determine the relative lactate concentration demonstrated that constant width at half height of the lactate band is likely to exist in a variety of biological samples (as a constant lactate bandwidth was required for accurate curve fitting results).52−58 Further, in this investigation we have used curve fitting of the average spectra from the regions under analysis (molecular layer, granular layer, and Purkinje neuron) to validate the results from analysis of second-derivative intensities.
The relative distribution of lactate was visualized through maps of second-derivative intensity at 1127 cm–1 (Figure 5E) and the average relative lactate content of the Purkinje neuron, molecular layer, and granular layer determined both from curve fitting nonderivative data (Figure 7A, Table 1), and from second-derivative intensity (Figure 7B, Table 1) revealed increased relative lactate concentration within Purkinje neurons relative to surrounding tissues. To the best of our knowledge, this provides the first direct evidence of an intracellular lactate pool within the Purkinje neuron. It should be noted that lactate is not absent from the molecular and granular layer tissue surrounding the Purkinje neuron, and lactate “hot spots” are observed, which is consistent with the generation of lactate in surrounding glial cells. As reported in the literature the use of SR-FTIR-FPA imaging with high numerical aperture Schwarzschild objectives in this study (74× objective, N.A. 0.65), would yield diffraction limited resolution imaging at the wavelength of the lactate sensitive band (6.8 μm at 1127 cm–1).2 However, resolution beyond this simple approximation from the Rayleigh criterion has recently been reported, and hence, the true spatial resolution for the lactate sensitive band may in fact be better than this (approximated as 5 um based on reported calculations).27 Future studies will aim to determine the exact resolution for which lactate can be imaged with this method. However, even with the conservative estimate of 6.8 μm, this is an improvement of 6.7 μm compared to the diffraction limited spatial resolution which could theoretically be achieved with a conventional globar source with lower numerical aperture (15× objective, N.A. 0.4).
Figure 7.
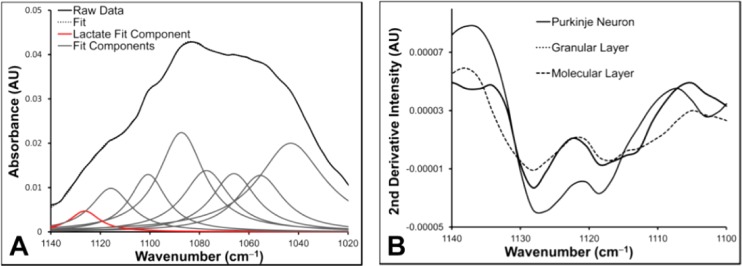
Determination of relative lactate concentration from FTIR spectra. (A) A representative example of the curve fitting approach used to determine the integrated area underneath the band centered at 1127 cm–1. This approach was used to demonstrate the significant increase in integrated band area for the band centered at 1127 cm–1 from the average spectra collected from Purkinje neurons relative to the surrounding molecular and granular layer tissue presented in Table 1. (B) Representative second-derivative spectra from a Purkinje neuron and surrounding molecular and granular layer tissue, highlighting the increase intensity at 1127 cm–1 from spectra collected within the Purkinje neuron.
These results highlight the exciting potential of SR-FTIR-FPA imaging to study in situ lactate distribution at the cellular level, which may be of great benefit in future studies of the production, metabolism and shuttling of lactate between astrocytes and neurons. However, it must be noted that the lactate concentration within the brain is highly dependent on many variables. In particular, animal anesthesia, and the time period following animal death greatly affect localized lactate concentration.21,70 Therefore, the lactate observed within the Purkinje neuron in this investigation may not reflect an in vivo state, but rather one induced during animal anesthesia and death. A thorough investigation of how sample preparation effects lactate concentration, and other biochemical markers effected by hypoxia-ischemia, which can be detected with novel spectroscopic techniques (i.e., FTIR, XAS, XRF), has been undertaken by this research group, and will be reported in a separate manuscript.
Conclusions
This study has demonstrated that vapor-fixation, a common protocol for post fixation of flash-frozen tissue in the fields of histology and cytochemistry, could offer significant advantages to future studies combining FTIR spectroscopic imaging with histology or immunocytochemistry of the same tissue section. The ability of vapor fixation to transfer structural stability to an unfixed tissue section without prior immersion of the section in aqueous media was shown to largely prevent dispersion or dilation of nuclear material in this study. Further, correlation between subcellular SR-FTIR-FPA images, which reveal the nucleus, with accurate histology provided the first reported true relative distribution of total protein within the Purkinje neuron. This result revealed that these cells are in fact rich in protein, not depleted as previously reported. It is important to note that the main aim of this study was not to discredit the use of immersion-fixation in future studies. Immersion fixation of tissue sections post FTIR spectroscopic analysis does yield accurate morphology at the tissue and cellular level. Further, an exhaustive study of different fixatives, and fixative concentrations was not performed in this study. It is possible that higher fixative concentration, employed over a shorted immersion time period, with reduced volume of fixation medium (i.e., drop fixation), may minimize protein loss from the nucleus, allowing successful preservation of subcellular features, in which case histology from immersion-fixed tissue would be comparable with vapor-fixed tissue. As such, this study serves to highlight artifacts that can result through the use of immersion fixation, and to encourage investigators to carefully consider the choice of postfixation protocol following FTIR spectroscopic analysis, such that the best possible histology, and correlation between FTIR images and histological images, can be achieved.
Finally, the ability to colocalize biochemical distribution revealed by SR-FTIR-FPA imaging with accurate histological images of the same tissue section successfully demonstrated subcellular resolution of Purkinje neuron cytoplasm and nucleus, revealing the first direct in situ evidence of intracellular lactate within Purkinje neurons. Due to the inability of neurons to store glycogen, but the known high content of lactate dehydrogenase within Purkinje neurons, this suggests the strong likelihood of a lactate shuttle between Purkinje neurons and glia. This novel ability to study cellular lactate distribution with SR-FTIR-FPA, which has never previously been achieved by any technique is expected to find widespread use in future metabolic studies of the role of lactate as a neuronal energy substrate during normal and hypoxic or ischemic conditions.
Methods
Animal Models
Cerebellum was obtained from four (n = 4) control (i.e., healthy) 6 week old male Sprague–Dawley rats. Rats were housed with a 12 h light/12 h dark cycle with ad libitum access to chow and water. This work was approved by the University of Saskatchewan’s Animal Research Ethics Board, and adhered to the Canadian Council on Animal Care guidelines for humane animal use.
Sample Preparation
Animals were anesthetized with isoflurane, and humanely sacrificed through decapitation, and the head immediately flash frozen in liquid nitrogen. Frozen brains were chiseled out from the frozen head on dry ice. Ten micrometer thick sections of the cerebellum were cut on a cryomicrotome and melted onto CaF2. Triplicate sections were cut from each animal (i.e., 12 sections were analyzed in total). Due to the known effect of storage conditions on infrared spectra collected from biological samples,71 sections were stored in a desiccator, in the dark, and at room temperature prior to analyses. All FTIR spectroscopic analysis was performed within 72 h of tissue sectioning.
SR-FTIR-FPA Spectroscopic Imaging at the Canadian Light Source (CLS)
SR-FTIR-FPA spectroscopic images were collected at the CLS with a Hyperion 3000 microscope fitted with an upper objective of 52× magnification and a numerical aperture of 0.6, and a lower condenser of 15× magnification and 0.4 numerical aperture. This arrangement yielded a pixel size of 0.77 μm. The incident infrared beam was focused and aligned to the center of the array, then defocused such that an array area of approximately 28 μm × 28 μm (36 × 36 pixels) was illuminated by light. During data processing individual images were cropped to a region consisting of 36 × 36 pixels, and 2 × 2 pixel binning performed to yield a final image of 18 × 18 pixels (1.54 μm effective pixel size), with adequate spectral signal-to-noise within each pixel.
SR-FTIR spectroscopic images were collected with a spectral resolution of 4 cm–1 and the coaddition of 1024 scans, and a background image was collected from blank substrate using 1024 coadded scans. The background was collected immediately prior to each sample. Normalization to beam current was not performed.
SR-FTIR-FPA Spectroscopic Imaging at the Synchrotron Radiation Center (SRC)
SR-FTIR-FPA spectroscopic images were collected at the SRC as previously described.2 In general, images were collected from a Hyperion 3000 microscope fitted with an upper objective of 74× magnification and a numerical aperture of 0.65, and a lower condenser of 15× magnification and 0.6 numerical aperture. This arrangement yielded an effective pixel size of 0.54 μm × 0.54 μm. The incident infrared beam was focused and aligned to the center of the array, then defocused to cover the complete array (34.6 μm × 34.6 μm).
SR-FTIR spectroscopic images were collected with a spectral resolution of 4 cm–1 and the coaddition of 512 scans. A background image was collected from blank substrate using 1064 coadded scans. The background was collected immediately prior to each sample. Normalization to beam current was not performed.
The spatial resolution for the bands analyzed were calculated as previously reported for this method, using a Rayleigh criterion factor of 0.51 (for Schwarzschild objectives) and a numerical aperture of 0.65.2
Data Processing and Data Analysis
All data processing and image generation was performed using Cytospec software (Cytospec, version 1.2.04) and Opus software (Version 6.5, Bruker, Ettlingen, Germany). False color functional group images were generated from band areas in un-normalized spectra. Areas were integrated from 2880–2865 cm–1 for νs(CH3), 2860–2845 cm–1 for νs(CH2), and 1690–1610 cm–1 for the amide I band. A linear baseline was subtracted across the same range as integration.
The curve fitting procedure used to estimate lactate concentration used the same approach as previously reported.52−58 All second-derivative spectra were derived with a 13 point Savitsky-Golay smoothing function.
To account for variations in tissue thickness band area and second-derivative intensities were normalized to local tissue thickness. Tissue thickness was determined via confocal microscopic analysis of eosin fluorescence (514 nm excitation) of the H&E stained sections with 1 μm “z” steps. A significant difference between the integrated area underneath bands, or second-derivative intensity was calculated with a Student’s t test, with a 95% confidence limit.
Data was collected in transmission mode to minimize electric field standing wave interference.72,73 To rule out resonant mie scattering as a possible cause of the biochemical conclusions drawn in this study, the RMieS-EMSC algorithm was applied to images as previously described.74,75 Representative examples of functional group images generated from the integrated area underneath the amide I band from uncorrected raw data and corrected data are shown in Supporting Information Figure 2. Although the images are not identical, the overall relative protein distribution did not change as a consequence of mie scattering correction.
Post Fixation of Flash-Frozen Tissue Sections and Histology and Fluorescence Microscopic Analyses
Following FTIR spectroscopic analyses, tissue sections were vapor fixed through incubation with formaldehyde gas release from heating dry paraformaldehyde powder at 80 °C for 2 h. The tissue sections were allowed to equilibrate back to room temperature and remained sealed in the presence of formaldehyde vapor for a further 2 h. Examples of immersion fixed tissue were prepared from serial sections to those analyzed with FTIR spectroscopic analysis. The sections were immersed in 4% phosphate buffered formalin solution for 4 h. The tissue sections were then subjected to staining with Mayers hematoxylin and counter stained with eosin for routine histology. Fluorescence microscopy, used to image the location of DNA, was performed on serial sections of both vapor-fixed and immersion-fixed tissue coverslipped with Prolong Gold Antifade Reagent with DAPI (P36931, Molecular Probes, Invitrogen).
Coomassie Blue Staining
Ten micrometer thick sections were cut from the same cerebellum blocks used for FTIR spectroscopic analyses, air-dried, and then vapor-fixed as described above. Tissue sections were briefly immersed in Coomassie blue (2 min), then rinsed in PBS, and optical images immediately taken from the wet section (coomaassie blue is removed via washing in ethanol solution, hence tissue dehydration and coverslipping was not performed).
Glial Fibrillary Acidic Protein (GFAP) Immunohistochemistry
Ten micrometer thick sections were cut from the same cerebellum blocks used for FTIR spectroscopic analyses, air-dried, and then vapor-fixed as described above. Sections were then washed in phosphate buffered saline (PBS) and exposed to 1.0% H2O2. Sections were blocked in normal goat serum (5%; Invitrogen, Carlsbad, CA), followed by overnight incubation at 4 °C with polyclonal rabbit anti-GFAP (1:1000, Z0344; DaloCytomation, ON, Canada). Slides were washed in PBS and incubated in goat antirabbit biotinylated secondary antibodies (1:500; Vector Laboratories, Burlingame, CA). Sections were treated with diaminobenzidine.
Acknowledgments
Research described in this paper was performed at the Canadian Light Source, which is supported by the Natural Sciences and Engineering Research Council of Canada, the National Research Council Canada, the Canadian Institutes of Health Research, the Province of Saskatchewan, Western Economic Diversification Canada, and the University of Saskatchewan.
Supporting Information Available
Additonal figures as described in the text. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
M.J.H., F.B., C.H., P.G.P., H.N., I.J.P., and G.N.G. designed the experiments and wrote the paper. M.J.H. performed the experiments with assistance from F.B., D.B., C.H., and S.E.S. M.J.H. performed data analysis with assistance from F.B. and C.H.
This work is based in part upon research conducted at the Synchrotron Radiation Center, which is primarily funded by the University of Wisconsin—Madison with supplemental support from facility Users and the University of Wisconsin—Milwaukee. This work was supported by a joint Canadian Institutes of Health Research (CIHR)/Heart and Stroke Foundation Synchrotron of Canada grant to the Team in Synchrotron Medical Imaging, grant number CIF99472, awarded to H.N., P.G.P., I.J.P., G.N.G., and others. M.J.H. is a Saskatchewan Health Research Foundation postdoctoral fellow, an SMI postdoctoral fellow and a fellow in the CIHR-Training grant in Health Research Using Synchrotron Techniques (CIHR-THRUST) Fellow (I.J.P. and others). G.N.G. and I.J.P. are Canada Research Chairs.
The authors declare no competing financial interest.
Supplementary Material
References
- Kastyak-Ibrahim M. Z.; Nasse M. J.; Rak M.; Hirschmugl C.; Del Bigio M. R.; Albensi B. C.; Gough K. M. (2012) Biochemical label-free tissue imaging with subcellular-resolution synchrotron FTIR with focal plane array detector. NeuroImage 60, 376–383. [DOI] [PubMed] [Google Scholar]
- Nasse M. J.; Walsh M. J.; Mattson E. C.; Reininger R.; Kajdacsy-Balla A.; Macias V.; Bhargava R.; Hirschmugl C. J. (2011) High-resolution Fourier-transform infrared chemical imaging with multiple synchrotron beams. Nat. Methods 8, 413–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petibois C.; Cestelli-Guidi M.; Piccinini M.; Moenner M.; Marcelli A. (2010) Synchrotron radiation FTIR imaging in minutes: a first step towards real-time cell imaging. Anal. Bioanal. Chem. 397, 2123–2129. [DOI] [PubMed] [Google Scholar]
- Miller L. M.; Dumas P. (2006) Chemical imaging of biological tissue with sycnhrotron infrared light. Biochem. Biophys. Acta 1758, 846–857. [DOI] [PubMed] [Google Scholar]
- Carr G. L., Chubar O., and Dumas P. (2005) Multichannel detection with a synchrotron light source: design and potential. In Spectrochemical Analysis Using Infrared Detectors (Bhargava R. and Levin I., Ed.), pp 56–84, Blackwell Publishing, Oxford. [Google Scholar]
- Moss D.; Gasharova B.; Mathis Y.-L. (2006) Practical tests of a focal plane array detector microscope at the ANKA-IR beamline. Infrared Phys. Technol. 49, 53–56. [Google Scholar]
- Herndon R. M. (1963) The fine structure of the Purkinje cell. J. Cell Biol. 18, 167–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J.; Hao T.; Shaw C.; Patel A. J.; Szabó G.; Rual J.-F.; Fisk C. J.; Li N.; Smolyar A.; Hill D. E.; Barabási A.-L.; Vidal M.; Zoghbi H. Y. (2006) A Protein Protein Interaction Network for Human Inherited Ataxias and Disorders of Purkinje Cell Degeneration. Cell 125, 801–814. [DOI] [PubMed] [Google Scholar]
- McKay B. E.; Turner R. W. (2005) Physiological and morphological development of the rat cerebellar Purkinje cell. J. Physiol. 567, 829–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber F. E.; Palmon S. C.; Traystman R. J.; Martin L. J. (1995) Global Incomplete Cerebral Ischemia Produces Predominantly Cortical Neuronal Injury. Stroke 26, 2091–2096. [DOI] [PubMed] [Google Scholar]
- Diemer N. H.; Siemkowicz E. (1981) Regional neurone damage after cerebral ischemia in the normo- and hypoglycaemic rat. Neuropathol. Appl. Neurobiol. 7, 217–227. [DOI] [PubMed] [Google Scholar]
- Miller J. A.; Curella P.; Zahniser N. R. (1988) A new densitometric procedure to measure protein levels in tissue slices used in quantitative autoradiography. Brain Res. 447, 60–66. [DOI] [PubMed] [Google Scholar]
- Tandler C. J.; RÃ-os H.; Pellegrino de Iraldi A. (1997) Differential staining of two subpopulations of Purkinje neurons in rat cerebellum with acid dyes. Biotech. Histochem. 72, 231–239. [DOI] [PubMed] [Google Scholar]
- Horobin R. W. (2002) Biological staining: mechanisms and theory. Biotech. Histochem. 77, 3–13. [PubMed] [Google Scholar]
- PrentÃ̧ P. (2001) A contribution to the theory of biological staining based on the principles for structural organization of biological macromolecules. Biotech. Histochem. 76, 137–161. [PubMed] [Google Scholar]
- Kasischke K. A. (2008) A new pathway for lactate production in the CNS. J. Physiol. 586, 1207–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughton J. D.; Charnay Y.; Belloir B.; Pellerin L.; Magistretti P. J.; Bouras C. (2000) Differential messenger RNA distribution of lactate dehydrogenase LDH-1 and LDH-5 isoforms in the rat brain. Neuroscience 96, 619–625. [DOI] [PubMed] [Google Scholar]
- Manocha S. L.; Bourne G. H. (1968) Histochemical mapping of lactate dehydrogenase and monoamine oxidase in the medulla oblongata and cerebellum of squirrel monkey (saimiri sciureus). J. Neurochem. 15, 1033–1040. [DOI] [PubMed] [Google Scholar]
- Bouzier-Sore A. K.; Serres S.; Canioni P.; Merle M. (2003) Lactate involvement in neuron-glia metabolic interaction: 13C-NMR spectroscopy contribution. Biochimie 85, 841–848. [DOI] [PubMed] [Google Scholar]
- Behar K. L.; den Hollander J. A.; Stromski M. E.; Ogino T.; Shulman R. G.; Petroff O. A. C.; Prichard J. W. (1983) High Resolution H nuclear magnetic resonance study of cerebral hypoxia in vivo. Proc. Natl. Acad. Sci. U.S.A. 80, 4945–4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funfschilling U.; Supplie L. M.; Mahad D.; Boretius S.; Saab A. S.; Edgar J.; Brinkmann B. G.; Kassmann C. M.; Tzvetanova I. D.; Mobius W.; Diaz F.; Meijer D.; Suter U.; Hamprecht B.; Sereda M. W.; Moraes C. T.; Frahm J.; Goebbels S.; Nave K.-A. (2012) Glycolytic oligodendrocytes maintain myelin and long-term axonal integrity. Nature 485, 517–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCandless D. W.; Feussner G. K.; Lust W. D.; Passonneau J. V. (1979) Metabolite levels in brain following experimental seizures: The effects of maximal electroshock and phenytoin in cerebellar layers. J. Neurochem. 32, 743–753. [DOI] [PubMed] [Google Scholar]
- Allfrey V. G.; Faulkner R.; Mirsky A. E. (1964) Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 51, 786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedar H.; Bergman Y. (2009) Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet. 10, 295–304. [DOI] [PubMed] [Google Scholar]
- Klose R. J.; Zhang Y. (2007) Regulation of histone methylation by demethylimination and demethylation. Nat. Rev. Mol. Cell. Biol. 8, 307–318. [DOI] [PubMed] [Google Scholar]
- Urdinguio R. G.; Sanchez-Mut J. V.; Esteller M. (2009) Epigenetic mechanisms in neurological diseases: genes, syndromes, and therapies. Lancet Neurol. 8, 1056–1072. [DOI] [PubMed] [Google Scholar]
- Mattson E. C.; Unger M.; Manandhar B.; Alavi Z.; Hirschmugl C. J. (2013) Multi-beam Synchrotron FPA Chemical Imaging: Impact of Schwarzchild Objective on Spatial Resolution. J. Phys. Conf. Ser. 425, 142001–6. [Google Scholar]
- Hackett M. J.; McQuillan J. A.; El-Assaad F.; Aitken J. B.; Levina A.; Cohen D. D.; Siegele R.; Carter E. A.; Grau G. E.; Hunt N. H.; Lay P. A. (2011) Chemical alterations to murine brain tissue induced by formalin fixation: implications for biospectroscopic imaging and mapping studies of disease pathogenesis. Analyst 136, 2941–2952. [DOI] [PubMed] [Google Scholar]
- Hackett M. J.; Smith S. E.; Paterson P. G.; Nichol H.; Pickering I. J.; George G. N. (2012) X-ray Absorption Spectroscopy at the Sulfur K-Edge: A New Tool to Investigate the Biochemical Mechanisms of Neurodegeneration. ACS Chem. Neurosci. 3, 178–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett M. J.; Lee J.; El-Assaad F.; McQuillan J. A.; Carter E. A.; Grau G. E.; Hunt N. H.; Lay P. A. (2012) FTIR Imaging of Brain Tissue Reveals Crystalline Creatine Deposits Are an ex Vivo Marker of Localized Ischemia during Murine Cerebral Malaria: General Implications for Disease Neurochemistry. ACS Chem. Neurosci. 3, 1017–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallant M.; Rak M.; Szeghalmi A.; Del Bigio M. R.; Westaway D.; Yang J.; Julian R.; Gough K. M. (2006) Focally elevated creatine detected in amyloid precursor protein (APP) transgenic mice and alzheimer disease brain tissue. J. Biol. Chem. 281, 5–8. [DOI] [PubMed] [Google Scholar]
- Chwiej J.; Dulinska J.; Janeczko K.; Dumas P.; Eichert D.; Dudala J.; Setkowicz Z. (2010) Synchrotron FTIR micro-spectroscopy study of the rat hippocampal formation after pilocarpine-evoked seizures. J. Chem. Neuroanat. 40, 140–147. [DOI] [PubMed] [Google Scholar]
- Dulinska J.; Setkowicz Z.; Janeczko K.; Sandt C.; Dumas P.; Uram L.; Gzielo-Jurek K.; Chwiej J. (2012) Synchrotron radiation Fourier-transform infrared and Raman microspectroscopy study showing an increased frequency of creatine inclusions in the rat hippocampal formation following pilocarpine-induced seizures. Anal. Bioanal. Chem. 402, 2267–2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leskovjan A. C.; Kretlow A.; Miller L. M. (2010) Fourier Transform Infrared Imaging Showing Reduced Unsaturated Lipid Content in the Hippocampus of a Mouse Model of Alzheimer’s Disease. Anal. Chem. 82, 2711–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneipp J.; Miller L. M.; Joncic M.; Kittel M.; Lasch P.; Beekes M.; Naumann D. (2003) In situ identification of protein structural changes in prion-infected tissue. Biochim. Biophys. Acta, Mol. Basis Dis. 1639, 152–158. [DOI] [PubMed] [Google Scholar]
- Frederik P. M.; Bomans P. H.; Busing W. M.; Odselius R.; Hax W. M. (1984) Vapor fixation for immunocytochemistry and X-ray microanalysis on cryoultramicrotome sections. J. Histochem. Cytochem. 32, 636–642. [DOI] [PubMed] [Google Scholar]
- Pekki A.; Tuohimaa P. (1989) New freeze-dry and vapor fixation method for immunohistochemistry of soluble proteins: subcellular location of the progesterone receptor. J. Histochem. Cytochem. 37, 1207–1213. [DOI] [PubMed] [Google Scholar]
- Jockusch H.; Voigt S.; Eberhard D. (2003) Localization of GFP in Frozen Sections from Unfixed Mouse Tissues: Immobilization of a Highly Soluble Marker Protein by Formaldehyde Vapor. J. Histochem. Cytochem. 51, 401–404. [DOI] [PubMed] [Google Scholar]
- Fauchtbauer E. M.; Rowlerson A. M.; Gatz K.; Friedrich G.; Mabuchi K.; Gergely J.; Jockusch H. (1991) Direct correlation of parvalbumin levels with myosin isoforms and succinate dehydrogenase activity on frozen sections of rodent muscle. J. Histochem. Cytochem. 39, 355–361. [DOI] [PubMed] [Google Scholar]
- Lewis E. N.; Kidder L. H.; Levin I. W.; Kalasinsky V. F.; Hanig J. P.; Lester D. S. (1997) Applications of Fourier Transform Infrared Imaging Microscopy in Neurotoxicity. Ann. N.Y. Acad. Sci. 820, 234–247. [DOI] [PubMed] [Google Scholar]
- Barros L. F.; Deitmer J. M. (2010) Glucose and lactate supply to the synapse. Brain Res. Rev. 63, 149–159. [DOI] [PubMed] [Google Scholar]
- Bonvento G.; Herard A.-S.; Voutsinos-Porche B. (2005) The astrocyte-neuron lactate shuttle: a debated but still valuable hypothesis for brain imaging. J. Cereb. Blood Flow Metab. 25, 1394–1399. [DOI] [PubMed] [Google Scholar]
- Chih C.-P.; Lipton P.; Roberts E. L. (2001) Do active cerebral neurons really use lactate rather than glucose?. Trends Neurosci. 24, 573–578. [DOI] [PubMed] [Google Scholar]
- Chih C.-P.; Roberts E. L. Jr. (2003) Energy Substrates for Neurons During Neural Activity[colon] A Critical Review of the Astrocyte-Neuron Lactate Shuttle Hypothesis. J. Cereb. Blood Flow Metab. 23, 1263–1281. [DOI] [PubMed] [Google Scholar]
- Figley C. R.; Stroman P. W. (2011) The role(s) of astrocytes and astrocyte activity in neurometabolism, neurovascular coupling, and the production of functional neuroimaging signals. Eur. J. Neurosci. 33, 577–588. [DOI] [PubMed] [Google Scholar]
- Mangia S.; Simpson I. A.; Vannucci S. J.; Carruthers A. (2009) The in vivo neuron-to-astrocyte lactate shuttle in human brain: evidence from modeling of measured lactate levels during visual stimulation. J. Neurochem. 109, 55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellerin L.; Bouzier-Sore A.-K.; Aubert A.; Serres S.; Merle M.; Costalat R.; Magistretti P. J. (2007) Activity-dependent regulation of energy metabolism by astrocytes: An update. Glia 55, 1251–1262. [DOI] [PubMed] [Google Scholar]
- Schurr A. (2005) Lactate: the ultimate cerebral oxidative energy substrate?. J. Cereb. Blood Flow Metab. 26, 142–152. [DOI] [PubMed] [Google Scholar]
- Magistretti P. J.; Pellerin L.; Rothman D. L.; Shulman R. G. (1999) Energy on Demand. Science 283, 496–497. [DOI] [PubMed] [Google Scholar]
- Pellerin L.; Pellegri G.; Bittar P. G.; Charnay Y.; Bouras C.; Martin J. L.; Stella N.; Magistretti P. J. (1998) Evidence Supporting the Existence of an Activity-Dependent Astrocyte-Neuron Lactate Shuttle. Dev. Neurosci. 20, 291–299. [DOI] [PubMed] [Google Scholar]
- Liu K.; Mantsch H. H. (1999) Simultaneous quantitation from infrared spectra of glucose concentrations, lactate concentrations and lecithin/sphingomyelin ratios in amniotic fluid. Am. J. Obstet. Gynecol. 180, 696–702. [DOI] [PubMed] [Google Scholar]
- Petibois C.; Deleris G. (2006) Chemical mapping of tumor progression by FT-IR imaging: towards molecular histopathology. Trends Biotechnol. 24, 455–462. [DOI] [PubMed] [Google Scholar]
- Petibois C.; Cazorla G.; Gin H.; Deleris G. (2000) Differentiation of populations with different physiological profiles by plasma Fourier-transform infrared classification. J. Lab. Clin. Med. 137, 184–190. [DOI] [PubMed] [Google Scholar]
- Petibois C.; Gionnet K.; Goncalves M.; Perromat A.; Moenner M.; Deleris G. (2006) Analytical Performances of FT-IR spectrometry and imaging for concentration measurements within biological fluids, cells and tissues. Analyst 131, 640–647. [DOI] [PubMed] [Google Scholar]
- Petibois C.; Drogat B.; Bikfalvi A.; Deleris G.; Moenner M. (2007) Histological mapping of biochemical changes in solid tumours by FT-IR spectral imaging. FEBS Lett. 581, 5469–5474. [DOI] [PubMed] [Google Scholar]
- Petibois C.; Melin A. M.; Perromat A.; Cazorla G.; Deleris G. (2000) Glucose and lactate concentration determination on single microsamples by Fourier-transform infrared spectroscopy. J. Lab. Clin. Med. 135, 210–214. [DOI] [PubMed] [Google Scholar]
- Deleris G.; Petibois C. (2003) Applications of FT-IR spectrometry to plasma contents analysis and monitoring. Vib. Spectrosc. 32, 129–136. [Google Scholar]
- Petibois C.; Desbat B. (2010) Clinical application of FTIR imaging: new reasons for hope. Trends Biotechnol. 28, 495–500. [DOI] [PubMed] [Google Scholar]
- Gough K. M.; Tzadu L.; Kastyak M. Z.; Kuzyk A. C.; Julian R. L. (2010) Theoretical and experimental considerations for interpretation of amide I bands in tissue. Vib. Spectrosc. 53, 71–76. [Google Scholar]
- Zhang J. Z.; Bryce N. S.; Siegele R.; Carter E. A.; Paterson D.; de Jonge M. D.; Howard D. L.; Ryan C. G.; Hambley T. W. (2012) The use of spectroscopic imaging and mapping techniques in the characterisation and study of DLD-1 cell spheroid tumour models. Integr. Biol. 4, 1072–1080. [DOI] [PubMed] [Google Scholar]
- Heraud P.; Caine S.; Campanale N.; Karnezis T.; McNaughton D.; Wood B. R.; Tobin M. J.; Bernard C. C. A. (2010) Early Dectection of the Chemical Changes Occurring During the Induction and Prevention of Autoimmune-mediated Demyelination Detected by FT-IR Imaging. NeuroImage 49, 1180–1189. [DOI] [PubMed] [Google Scholar]
- Dong A.; Huang P.; Caughey W. S. (1990) Protein secondary structures in water from second-derivative amide I infrared spectra. Biochemistry 29, 3303–3308. [DOI] [PubMed] [Google Scholar]
- Kong J.; Yu S. (2007) Fourier Transform Infrared Spectroscopic Analysis of Protein Secondary Structures. Acta Biochim. Biophys. Sinica 39, 549–559. [DOI] [PubMed] [Google Scholar]
- Susi H.; Michael Byler D. (1983) Protein structure by Fourier transform infrared spectroscopy: Second derivative spectra. Biochem. Biophys. Res. Commun. 115, 391–397. [DOI] [PubMed] [Google Scholar]
- Arrondo J. L. R.; Goñi F. M. (1999) Structure and dynamics of membrane proteins as studied by infrared spectroscopy. Prog. Biophys. Mol. Biol. 72, 367–405. [DOI] [PubMed] [Google Scholar]
- Jackson M.; Mantsch H. H. (1995) The use and misuse of FTIR spectroscopy in the determination of protein structure. Crit. Rev. Biochem. Mol. Biol. 30, 95–120. [DOI] [PubMed] [Google Scholar]
- Surewicz W. K.; Mantsch H. H.; Chapman D. (1993) Determination of protein secondary structure by Fourier transform infrared spectroscopy: a critical assessment. Biochemistry 32, 389–394. [DOI] [PubMed] [Google Scholar]
- Ami D.; Natalello A.; Gatti-Lafranconi P.; Lotti M.; Doglia S. M. (2005) Kinetics of inclusion body formation studied in intact cells by FT-IR spectroscopy. FEBS Lett. 579, 3433–3436. [DOI] [PubMed] [Google Scholar]
- Kneipp J.; Lasch P.; Baldauf E.; Beekes M.; Naumann D. (2000) Detection of pathological molecular alterations in scrapie-infected hamster brain by Fourier transform infrared (FT-IR) spectroscopy. Biochem. Biophys. Acta 1501, 189–199. [DOI] [PubMed] [Google Scholar]
- Gatfield P. D.; Lowry O. H.; Schulz D. W.; Passonneau J. V. (1966) Regional energy reserves in mouse brain and changes with ischemia and anaesthesia. J. Neurochem. 13, 185–195. [DOI] [PubMed] [Google Scholar]
- Stitt D. M.; Kastyak-Ibrahim M. Z.; Liao C. R.; Morrison J.; Albensi B. C.; Gough K. M. (2012) Tissue acquisition and storage associated oxidation considerations for FTIR microspectroscopic imaging of polyunsaturated fatty acids. Vib. Spectrosc. 60, 16–22. [Google Scholar]
- Bassan P.; Lee J.; Sachdeva A.; Pissardini J.; Dorling K. M.; Fletcher J. S.; Henderson A.; Gardner P. (2012) The inherent problem of transflection-mode infrared spectroscopic microscopy and the ramifications for biomedical single point and imaging applications. Analyst 138, 144–157. [DOI] [PubMed] [Google Scholar]
- Filik J.; Frogley M. D.; Pijanka J. K.; Wehbe K.; Cinque G. (2012) Electric field standing wave artefacts in FTIR micro-spectroscopy of biological materials. Analyst 137. [DOI] [PubMed] [Google Scholar]
- Bassan P.; Kohler A.; Martens H.; Lee J.; Byrne H. J.; Dumas P.; Gazi E.; Brown M.; Clarke N.; Gardner P. (2009) Resonant Mie scattering (RMieS) correction of infrared spectra from highly scattering biological samples. Analyst 135, 268–277. [DOI] [PubMed] [Google Scholar]
- Bassan P.; Sachdeva A.; Kohler A.; Hughes C.; Henderson A.; Boyle J.; Shanks J. H.; Brown M.; Clarke N. W.; Gardner P. (2012) FTIR microscopy of biological cells and tissue: data analysis using resonant Mie scattering (RMieS) EMSC algorithm. Analyst 137, 1370–1377. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


