Abstract
We report the first case of pulmonary disease caused by a strain of Mycobacterium avium complex of presumed veterinary origin in an elderly patient. All serial isolates were identified by multilocus sequence analysis based on rpoB, hsp65, and 16S rRNA fragments. Disease persisted despite macrolide-based combination antibiotic therapy.
CASE REPORT
A 77-year-old man was referred to our hospital due to progression of lung disease caused by nontuberculous mycobacteria (NTM). Fourteen months prior, he was diagnosed with Mycobacterium avium lung disease and treated with clarithromycin, ethambutol, and rifampin. Drug susceptibility testing (DST) was not performed. Despite >12 months of antibiotic therapy, the patient failed to show any clinical improvement.
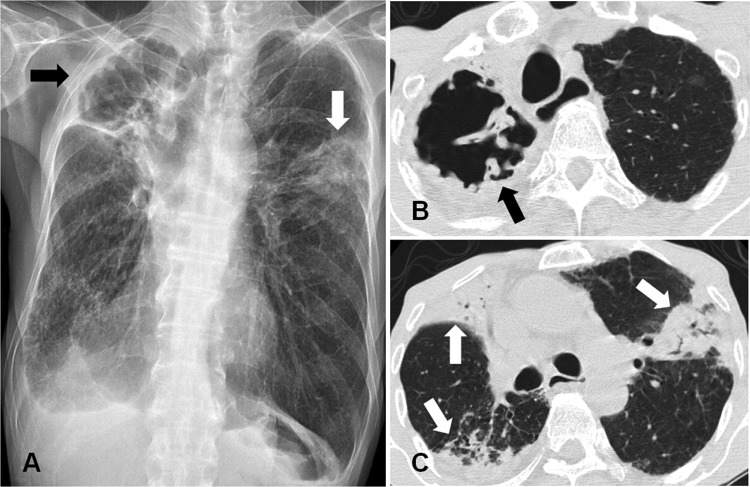
He was a nonsmoker and a retired police officer. He had no history of plenty of time around farm animals. His leukocyte count was 10,160/μl, with a differential of 71% neutrophils. The erythrocyte sedimentation rate was 114 mm/h, and his C-reactive protein (CRP) level was increased to 10.54 mg/dl. A human immunodeficiency virus antibody was negative. A chest radiograph and computed tomography (CT) scan revealed a large cavitary lesion in the right upper lobe and multiple consolidations in both lungs (Fig. 1).
Fig 1.
A 77-year-old man with Mycobacteriumavium avium complex strain infection of presumed veterinary origin. (A) Chest radiograph shows a large cavity in the right upper lobe (black arrow) and a segmented consolidation in the left upper lobe (white arrow). (B) Chest computed tomography (CT) scan shows a large cavity in the right upper lobe (black arrow). (C) Chest CT scan shows multiple segmented consolidations in both lungs (white arrows).
Numerous (4+) acid-fast bacilli (AFB) were detected with both auramine-rhodamine fluorescent staining and the Ziehl-Neelsen staining method according to standard guidelines (1). A 4+ AFB smear was defined as >9 AFB per field (1). NTM were isolated more than five times from sputum specimens in a liquid culture system (Bactec MGIT 960 system; BD Diagnostics, Sparks, MD). To identify the etiological agent, bacteria grown in the MGIT 960 culture system were initially propagated in 7H9 broth (Difco Laboratories, Detroit, MI) supplemented with 10% (vol/vol) oleic acid-albumin-dextrose-catalase (OADC; BD Diagnostics) for 14 days at 37°C and subcultured in egg-based 3% Ogawa solid medium (Shinyang, Seoul, South Korea), and genomic DNA was extracted from the cultured bacteria. Isolate SMC1 was initially identified as M. avium using a reverse line blot hybridization assay (REBA Myco-ID; M&D, Inc., Wonju, South Korea) based on the rpoB gene (2).
To confirm the accuracy of this identification, sequencing analyses of rpoB, hsp65, and 16S rRNA were performed (3–5). The hsp65 and 16S rRNA sequences were 100% identical to those of the M. avium strain (GenBank accession no. CP000479 and GQ153272, respectively). The rpoB sequences showed 99.7% similarity, with only a 2-base mismatch, to that of the M. avium subsp. avium strain (GenBank accession no. GQ153306). Additionally, the rpoB sequences showed 99.6% similarity, with only a 3-base mismatch, to those of the M. avium subsp. hominissuis strain (GenBank accession no. EF521911), M. avium subsp. paratuberculosis strain (GenBank accession no. EF521906), and M. avium subsp. silvaticum strain (GenBank accession no. JN935808). Interestingly, the rpoB sequences were 100% similar to those of the Mycobacterium spp. belonging to the M. avium complex (MAC) of veterinary origin (GenBank accession no. JF327744 and JF327745).
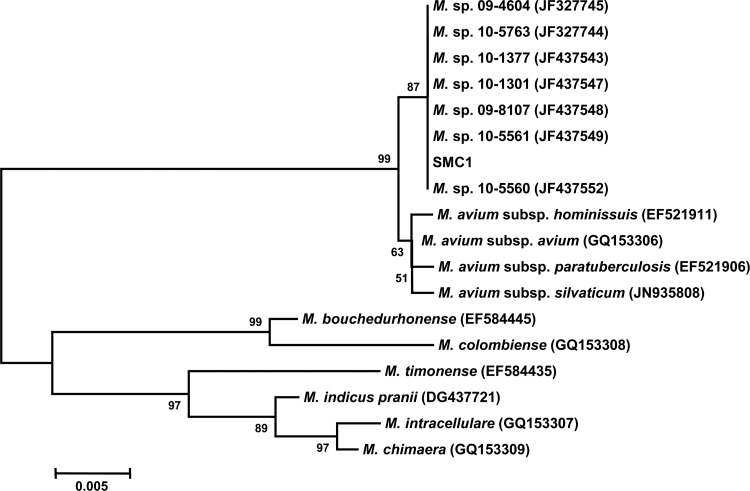
A phylogenetic tree, based on rpoB gene sequences from the isolate SMC1 and from those of closely related species within the MAC, is shown in Fig. 2. The sequences were compared with those obtained from the GenBank sequence database. The rpoB gene sequences of the 18 strains were aligned to those of the type strains of closely related mycobacteria by using CLUSTAL_X software (6). The topological results and tree were inferred via the neighbor-joining method, visualized by using the MEGA 5.0 software package (7), and evaluated by bootstrap analyses based on 1,000 resamplings. Phylogenetic analysis based on partial rpoB sequences showed that this strain was a novel MAC isolate of veterinary origin (Fig. 2).
Fig 2.
The phylogenetic position of isolate SMC1 and other species belonging to the Mycobacterium avium complex based on partial rpoB gene sequences. This tree was constructed using the neighbor-joining method. The percentages indicated at nodes represent bootstrap levels supported by 1,000 resampled data sets. Scale bars indicate evolutionary distance in base substitutions per site.
Drug susceptibility testing was performed by using a broth microdilution method according to the guidelines (8), which revealed that SMC1 was resistant to clarithromycin (MIC, >64 μg/ml) and linezolid (MIC, 64 μg/ml) and was susceptible to moxifloxacin (MIC, 1 μg/ml). SMC1 had an expected point mutation of A→C at position 2059 in the 23S rRNA gene, which is known as the major mechanism of acquired macrolide resistance (9).
The patient was treated with antibiotics, including isoniazid, rifampin, ethambutol, and streptomycin, in our hospital. Despite this combination antibiotic therapy for more than 12 months, the patient's symptoms and radiographic findings worsened. Sputum AFB staining and culture examinations were persistently positive.
Among NTM, MAC is a common cause of disease (10, 11). M. avium is ubiquitous in nature and an opportunistic pathogen for both animals and humans (12). M. avium is subdivided into four subspecies: avium, hominissuis, paratuberculosis, silvaticum (13). M. avium subsp. avium is known to be a bird-type strain that causes a contagious disease among birds, while M. avium subsp. hominissuis can infect humans, pigs, cattle, and other animals (13). M. avium subsp. avium has been detected in human patients, pigs, and other animals; however, these appear to be rare cases (14).
The rpoB gene was sequenced using Myco-F/Myco-R primers, which amplify a 711-bp fragment, the hypervariable region of the rpoB gene, and permits one-step identification of MAC isolates at the species or subspecies level, as well as the detection of potentially novel MAC species otherwise unable to be determined by 16S rRNA gene sequence analysis (15). The isolate SMC1 was clustered among novel MAC isolates from veterinary hosts, including elk, cats, cattle, and pigs, although it did not demonstrate 100% identity with the rpoB sequence for any of the clades containing M. avium subsp. avium, M. avium subsp. hominissuis, M. avium subsp. paratuberculosis, or M. avium subsp. silvaticum. The insertion element profiles of these Mycobacterium spp. of veterinary origin were negative for IS900, IS901, and DT1 and positive for IS1245, which is similar to the results for M. avium subsp. hominissuis (16). The taxonomic position of these isolates within the MAC was not defined. SMC1 is an undefined Mycobacterium spp. of veterinary origin but is genetically close to MAC. We report the first case of MAC strain infection, of presumed veterinary origin, in an elderly adult patient.
The high genetic relatedness among M. avium subsp. hominissuis isolated from pigs and humans was reported in the Netherlands, Sweden, Germany, and Finland (17–20). Like these strains, isolate SMC1 may infect humans infrequently. Direct transmission of NTM between animals and humans has not yet been proven. It remains to be determined whether humans are infected directly from animals or if they are infected from common environmental sources.
ACKNOWLEDGMENTS
This work was supported by the Midcareer Researcher Program through a National Research Foundation grant funded by the Ministry of Education, Science, and Technology (2011-0015546).
Footnotes
Published ahead of print 3 April 2013
REFERENCES
- 1. American Thoracic Society 2000. Diagnostic standards and classification of tuberculosis in adults and children. Am. J. Respir. Crit. Care Med. 161:1376–1395 [DOI] [PubMed] [Google Scholar]
- 2. Lee H, Bang HE, Bai GH, Cho SN. 2003. Novel polymorphic region of the rpoB gene containing Mycobacterium species-specific sequences and its use in identification of mycobacteria. J. Clin. Microbiol. 41:2213–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Adekambi T, Colson P, Drancourt M. 2003. rpoB-based identification of nonpigmented and late-pigmenting rapidly growing mycobacteria. J. Clin. Microbiol. 41:5699–5708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kim H, Kim SH, Shim TS, Kim MN, Bai GH, Park YG, Lee SH, Chae GT, Cha CY, Kook YH, Kim BJ. 2005. Differentiation of Mycobacterium species by analysis of the heat-shock protein 65 gene (hsp65). Int. J. Syst. Evol. Microbiol. 55:1649–1656 [DOI] [PubMed] [Google Scholar]
- 5. Turenne CY, Tschetter L, Wolfe J, Kabani A. 2001. Necessity of quality-controlled 16S rRNA gene sequence databases: identifying nontuberculous Mycobacterium species. J. Clin. Microbiol. 39:3637–3648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Clinical and Laboratory Standards Institute 2011. Susceptibility testing of mycobacteria, nocardiae, and other aerobic actinomycetes; approved standard—second edition, document no. M24-A2. CLSI, Wayne, PA: [PubMed] [Google Scholar]
- 9. Nash KA, Inderlied CB. 1995. Genetic basis of macrolide resistance in Mycobacterium avium isolated from patients with disseminated disease. Antimicrob. Agents Chemother. 39:2625–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Daley CL, Griffith DE. 2010. Pulmonary non-tuberculous mycobacterial infections. Int. J. Tuberc. Lung Dis. 14:665–671 [PubMed] [Google Scholar]
- 11. Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, Holland SM, Horsburgh R, Huitt G, Iademarco MF, Iseman M, Olivier K, Ruoss S, von Reyn CF, Wallace RJ, Jr, Winthrop K. 2007. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am. J. Respir. Crit. Care Med. 175:367–416 [DOI] [PubMed] [Google Scholar]
- 12. Inderlied CB, Kemper CA, Bermudez LE. 1993. The Mycobacterium avium complex. Clin. Microbiol. Rev. 6:266–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mijs W, de Haas P, Rossau R, Van der Laan T, Rigouts L, Portaels F, van Soolingen D. 2002. Molecular evidence to support a proposal to reserve the designation Mycobacterium avium subsp. avium for bird-type isolates and ‘M. avium subsp. hominissuis’ for the human/porcine type of M. avium. Int. J. Syst. Evol. Microbiol. 52:1505–1518 [DOI] [PubMed] [Google Scholar]
- 14. Thegerstrom J, Marklund BI, Hoffner S, Axelsson-Olsson D, Kauppinen J, Olsen B. 2005. Mycobacterium avium with the bird type IS1245 RFLP profile is commonly found in wild and domestic animals, but rarely in humans. Scand. J. Infect. Dis. 37:15–20 [DOI] [PubMed] [Google Scholar]
- 15. Ben Salah I, Adekambi T, Raoult D, Drancourt M. 2008. rpoB sequence-based identification of Mycobacterium avium complex species. Microbiology 154:3715–3723 [DOI] [PubMed] [Google Scholar]
- 16. Higgins J, Camp P, Farrell D, Bravo D, Pate M, Robbe-Austerman S. 2011. Identification of Mycobacterium spp. of veterinary importance using rpoB gene sequencing. BMC Vet. Res. 7:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Komijn RE, de Haas PE, Schneider MM, Eger T, Nieuwenhuijs JH, van den Hoek RJ, Bakker D, van Zijd Erveld FG, van Soolingen D. 1999. Prevalence of Mycobacterium avium in slaughter pigs in The Netherlands and comparison of IS1245 restriction fragment length polymorphism patterns of porcine and human isolates. J. Clin. Microbiol. 37:1254–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mobius P, Lentzsch P, Moser I, Naumann L, Martin G, Kohler H. 2006. Comparative macrorestriction and RFLP analysis of Mycobacterium avium subsp. avium and Mycobacterium avium subsp. hominissuis isolates from man, pig, and cattle. Vet. Microbiol. 117:284–291 [DOI] [PubMed] [Google Scholar]
- 19. Ramasoota P, Chansiripornchai N, Kallenius G, Hoffner SE, Svenson SB. 2001. Comparison of Mycobacterium avium complex (MAC) strains from pigs and humans in Sweden by random amplified polymorphic DNA (RAPD) using standardized reagents. Vet. Microbiol. 78:251–259 [DOI] [PubMed] [Google Scholar]
- 20. Tirkkonen T, Pakarinen J, Moisander AM, Makinen J, Soini H, Ali-Vehmas T. 2007. High genetic relatedness among Mycobacterium avium strains isolated from pigs and humans revealed by comparative IS1245 RFLP analysis. Vet. Microbiol. 125:175–181 [DOI] [PubMed] [Google Scholar]