Abstract
The early bactericidal activity of antituberculosis agents is usually determined by measuring the reduction of the sputum mycobacterial load over time on solid agar medium or in liquid culture. This study investigated the value of a quantitative PCR assay for early bactericidal activity determination. Groups of 15 patients were treated with 6 different antituberculosis agents or regimens. Patients collected sputum for 16 h overnight at baseline and at days 7 and 14 after treatment initiation. We determined the sputum bacterial load by CFU counting (log CFU/ml sputum, reported as mean ± standard deviation [SD]), time to culture positivity (TTP, in hours [mean ± SD]) in liquid culture, and Xpert MTB/RIF cycle thresholds (CT, n [mean ± SD]). The ability to discriminate treatment effects between groups was analyzed with one-way analysis of variance (ANOVA). All measurements showed a decrease in bacterial load from mean baseline (log CFU, 5.72 ± 1.00; TTP, 116.0 ± 47.6; CT, 19.3 ± 3.88) to day 7 (log CFU, −0.26 ± 1.23, P = 0.2112; TTP, 35.5 ± 59.3, P = 0.0002; CT, 0.55 ± 3.07, P = 0.6030) and day 14 (log CFU, −0.55 ± 1.24, P = 0.0006; TTP, 54.8 ± 86.8, P < 0.0001; CT, 2.06 ± 4.37, P = 0.0020). The best discrimination between group effects was found with TTP at day 7 and day 14 (F = 9.012, P < 0.0001, and F = 11.580, P < 0.0001), followed by log CFU (F = 4.135, P = 0.0024, and F = 7.277, P < 0.0001). CT was not significantly discriminative (F = 1.995, P = 0.091, and F = 1.203, P = 0.316, respectively). Culture-based methods are superior to PCR for the quantification of early antituberculosis treatment effects in sputum.
INTRODUCTION
Several novel antituberculosis drugs and regimens are currently under clinical investigation. The first step in their evaluation is the measurement of early bactericidal activity (EBA) in sputum over up to 2 weeks of treatment in smear-positive, treatment-naive pulmonary tuberculosis patients. The EBA is commonly defined as the mean daily decrease in CFU counted on agar plates (log CFU) per ml of expectorated sputum (1, 2). Alternatively, the change in time to culture positivity (TTP) in broth culture can be measured in the mycobacterial growth indicator tube (MGIT) system (Becton, Dickinson, Sparks, MD) (3). This is based on the inverse relationship of the time a culture requires to develop a critical measure of metabolic activity to the number of viable bacteria initially inoculated into the system. TTP in semiautomated liquid culture has been shown to correlate well with CFU counting in the first 2 weeks of treatment (3–5).
The Xpert MTB/RIF assay (Xpert; Cepheid, Sunnyvale, CA) is a new nucleic acid amplification test (NAAT) detecting M. tuberculosis complex on sputum specimens with real-time PCR. Xpert is rapid (less than 1 h of hands-on time), standardized, and easy to use (6, 7). Its sensitivity to detect M. tuberculosis in sputum of untreated patients suspected of having tuberculosis in high-burden countries is 98% for AFB-positive samples and 72% for AFB-negative specimens with an overall specificity of >99% (8). Xpert is able to detect as few as 100 organisms per ml in vitro suspension (9). It is less well known that Xpert also provides a semiquantitative measurement based on the number of PCR cycles required for detection of a critical amount of DNA (cycle threshold [CT]). It was repeatedly demonstrated on spiked samples and sputum samples collected from untreated tuberculosis patients that the quantitative CT readouts from Xpert correlate with (semi)quantitative results of conventional microbiological tests such as smear microscopy grade, liquid culture TTP, and solid-culture CFU counts (9–13).
As a platform for quantifying M. tuberculosis in sputum, Xpert could make EBA studies less costly, more reproducible, and technically more accessible if the change in CT is a reliable measure of treatment effects. However, it is unclear how DNA load correlates with culture-based quantification of viable bacteria in patients being treated for tuberculosis. To investigate this question, we performed a prospective study to compare the ability of log CFU, TTP, and Xpert CT to monitor and distinguish between treatment effects in a 14-day EBA study with 6 parallel groups receiving different antituberculosis treatments.
MATERIALS AND METHODS
Sample collection and ethics.
Sputum samples were collected during a 14-day early bactericidal activity study from untreated adult pulmonary tuberculosis patients. Participants were required to be smear positive (≥1+; WHO/IUATLD scale [14]) and free of severe comorbidities. HIV-positive patients with a CD4 count of <250/μl and those on antiretroviral therapy were excluded. From December 2010 until August 2011, 90 patients (59% male; 10% HIV positive; median age, 30.4 years) were hospitalized at one of two centers in Cape Town, South Africa (University of Cape Town Lung Institute, Mowbray; Task Applied Science, Intercare, Durbanville) and randomized to one of six equally sized groups that were treated with different dosages and/or combinations of two antituberculosis agents. All patients' isolates were susceptible to the treatment given. Spontaneously expectorated sputum was collected for 16 h overnight, refrigerated after collection, and transported to the central study laboratory (Centre of Clinical Tuberculosis Research, Department of Biomedical Sciences, Stellenbosch University). Specimens were allowed to warm up to room temperature before homogenization for 30 min with a magnetic stirrer and processing. The institutional ethical committees and the Medicines Control Council (MCC) granted approval for the main study and this substudy.
Determination of log CFU.
A maximum of 10 ml of homogenized sputum was digested by addition of an equal volume of 0.1% dithiothreitol (Sputasol; Oxoid, Cambridge, United Kingdom) for 20 min. Sterile saline (0.85%) containing 0.01% Tween was used to generate 10-fold serial dilutions. A volume of 100 μl of every dilution was incubated on each half of two selective agar plates containing Middlebrook 7H11 agar enriched with Middlebrook OADC (oleic acid-albumin-dextrose-catalase; Becton, Dickinson) and Selectatab (Mast; Bootle, Merseyside, United Kingdom) containing polymyxin B sulfate (200,000 units/liter), amphotericin B (10 mg/liter), ticarcillin (100 mg/liter), and trimethoprim (10 mg/liter). The plates were incubated at 37°C for 3 to 4 weeks before colonies were counted on the dilution that yielded between 20 and 200 colonies. These data were log transformed to provide log CFU.
Determination of TTP.
Five ml of digested sputum was mixed with an equal volume of 2% NaOH (BBL Mycoprep; Becton, Dickinson) and decontaminated for 20 min at room temperature. This mixture was neutralized by addition of sterile phosphate-buffered saline (PBS, pH 6.8; Becton, Dickinson) to a final volume of 45 ml and centrifuged for 15 min at 3,000 × g and 4°C. The supernatant was discarded and the pellet resuspended with PBS to a final volume of 2 ml. Two MGITs for each specimen were prepared by addition of 0.8 ml PANTA (polymyxin B, amphotericin B, nalidixic acid, trimethoprim, and azlocillin) mixed with OADC (Becton, Dickinson). Each MGIT was inoculated with 0.5 ml of resuspended pellet and incubated at 37°C in a Bactec MGIT960 instrument. Cultures that were flagged as positive were tested for contamination by placing one drop on a blood agar plate (NHLS, Cape Town, South Africa) and incubating at 37°C for 48 h. The presence of acid-fast bacilli (AFB) in positive liquid culture was confirmed by Ziehl-Neelsen staining and microscopy; only TTPs from AFB positive and noncontaminated cultures were used for analysis.
Xpert MTB/RIF assay.
An aliquot of digested sputum sample (1 to 2 ml) was frozen at −80°C at the time of culture processing. After thawing, the specimen was resuspended by vortexing, and 1 ml was used according to the instructions of the manufacturer. Briefly, the diluted sample was mixed with 2 ml of Xpert sample reagent, inverted 10 times, and incubated for 15 min at room temperature; inversion was repeated after the first 8 min. This mixture was then transferred into an Xpert cartridge and loaded into the GeneXpert instrument, which conducts all necessary steps automatically using GeneXpert Dx software (version 4.0; Cepheid). The software reports results as CTs, which represent the number of PCR cycles needed to reach a detection threshold; the CT of probe B was used for all analyses according to previously published data (6).
Analysis plan.
From the sputum samples available from the EBA study, we prospectively selected the two baseline samples collected on the days directly preceding the treatment initiation and those collected after 7 and 14 days of treatment, when we expected treatment effects to emerge. We analyzed the data in a stepwise fashion. First, we studied the baseline samples to gain an impression of the variation of the measurements without treatment effects. Second, individual values were plotted against one another and inspected for correlation. Next, we examined the correlation of individual treatment activities for all measurements. Finally, we quantified the treatment effects and the respective ability of the measurements to differentiate between treatment groups by comparing variation within the groups to the variation between the groups for the periods from 0 to 7 days and 0 to 14 days.
Statistical methods.
The group sample size of 15 patients and study duration of 14 days were predetermined by the parent EBA study. Correlations between the methods of detection were quantified with the Spearman rank sum test. Treatment activity was calculated as the arithmetic difference between the means of the baseline values (also termed day 0 values) and the values obtained after 7 or 14 days, first for each individual and then as an average to obtain a group result. The significance of treatment effects within groups from day 0 to 7 or day 0 to 14 was determined by Student's t test. Variance within groups and between groups was compared with F statistics (one-way analysis of variance [ANOVA]). The larger the F statistic is, the better the chance that a measurement can differentiate group effects. A P value of <0.05 was considered statistically significant.
RESULTS
Log CFU, TTP, and CT.
Out of a possible 360 values, 304 log CFU (84.4%), 341 TTP (94.7%), and 317 CT (88.1%) results were obtained. The missing values were due to contamination, no growth, no sample being received, or invalid results. There was no significant difference between groups regarding the proportions of available data or the magnitude of the mean baseline values. The day-to-day variance estimated using the baseline values within subjects was compared with the total baseline variance to give ratios of 0.1621/0.9853 (0.165) for log CFU, 748.7/3448.5 (0.220) for TTP, and 12.25/6.94 (0.567) for CT. While 16.5% and 22% are similar, the value of 56.7% for CT suggests a larger day-to-day variability and smaller between-subject variability for CT than for the culture-based measurements, log CFU count and TTP. Individual log CFU, TTP, and CT values were moderately correlated at baseline, day 7, and day 14 and are illustrated in Fig. 1.
Fig 1.
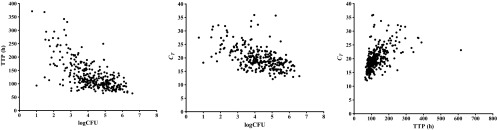
Individual log CFU, TTP, and CT values are shown for all patients and time points combined. Rho values were calculated by the Spearman rank sum test for log CFU versus TTP (rho = −0.613; P < 0.0001), log CFU versus CT (rho = −0.547; P < 0.0001), and TTP versus CT (rho = 0.632; P < 0.0001).
Treatment effects measured with log CFU, TTP, and CT.
Individual treatment effects were strongly correlated between log CFU and TTP; log CFU and CT and TTP and CT were moderately correlated (Fig. 2). All three measurements showed an overall decrease from baseline, which was significant for TTP at day 7 and for all three measurements at day 14 (Table 1; Fig. 3). A significant difference between baseline and day 7 and/or day 14 was found in 4 groups with log CFU, in 5 groups with TTP, and in 2 groups with CT. The real test for discrimination between treatment effects at day 7 and day 14 is shown in Table 2. One-way ANOVA delivered the greatest F values for TTP, indicating that liquid culture had the most favorable ratio of within-group to between-group variation to discriminate between groups for treatment effects at day 7 and day 14. Log CFU from solid culture medium displayed acceptable discriminatory ability, while quantitative PCR, represented by Xpert CT, was relatively poor.
Fig 2.
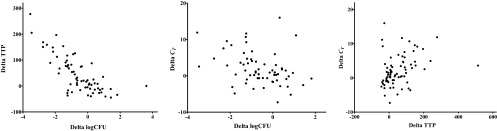
Individual activities of log CFU, TTP, and CT from day 0 to day 14 are plotted against each other. A strong correlation was found between log CFU and TTP (rho = −0.726; P < 0.0001), and a moderate correlation was found between log CFU and CT (rho = −0.385; P = 0.0017) and TTP and CT (rho = 0.400; P = 0.0004).
Table 1.
Treatment effects measured with log CFU, TTP, and CTa
| Measurement | Group | Baseline |
Days 0–7 |
Days 0–14 |
|||||
|---|---|---|---|---|---|---|---|---|---|
| n | Value | n | Change in value | P | n | Change in value | P | ||
| Log CFU | Group 1 | 14 | 5.51 (1.15) | 11 | −0.94 (1.76) | 0.096 | 12 | −0.97 (1.35) | 0.018 |
| Group 2 | 15 | 5.91 (0.77) | 14 | −0.32 (1.18) | 0.429 | 13 | −0.97 (0.72) | 0.009 | |
| Group 3 | 14 | 5.42 (0.97) | 13 | +0.47 (1.41) | 0.434 | 12 | +0.17 (1.41) | 0.994 | |
| Group 4 | 14 | 5.48 (0.91) | 12 | +0.19 (0.56) | 0.367 | 12 | +0.39 (0.76) | 0.385 | |
| Group 5 | 13 | 6.34 (0.79) | 12 | −1.17 (0.82) | 0.004 | 10 | −1.84 (1.03) | 0.0006 | |
| Group 6 | 13 | 5.67 (1.17) | 13 | +0.08 (0.47) | 0.860 | 12 | −0.28 (0.71) | 0.384 | |
| Total | 83 | 5.72 (1.00) | 75 | −0.26 (1.23) | 0.2112 | 71 | −0.55 (1.24) | 0.0006 | |
| TTP | Group 1 | 15 | 115.6 (39.4) | 15 | +83.7 (73.7) | 0.0005 | 15 | +91.5 (77.5) | <0.0001 |
| Group 2 | 15 | 100.0 (20.6) | 15 | +53.7 (40.6) | 0.0002 | 15 | +78.1 (54.5) | <0.0001 | |
| Group 3 | 15 | 120.4 (56.0) | 14 | +10.6 (62.2) | 0.607 | 12 | +6.68 (36.7) | 0.835 | |
| Group 4 | 15 | 125.7 (42.4) | 15 | −2.13 (23.2) | 0.897 | 15 | −11.3 (24.9) | 0.423 | |
| Group 5 | 15 | 124.3 (77.4) | 14 | +70.5 (50.9) | 0.073 | 13 | +151.1 (125.1) | 0.004 | |
| Group 6 | 15 | 110.1 (24.5) | 15 | −2.52 (21.9) | 0.770 | 14 | +13.2 (41.0) | 0.365 | |
| Total | 90 | 116.0 (47.6) | 88 | +35.5 (59.3) | 0.0002 | 84 | +54.8 (86.8) | <0.0001 | |
| CT | Group 1 | 14 | 19.1 (4.57) | 11 | +2.33 (2.80) | 0.331 | 14 | +3.87 (5.75) | 0.030 |
| Group 2 | 14 | 18.9 (3.81) | 12 | +1.06 (3.18) | 0.476 | 15 | +1.27 (3.32) | 0.311 | |
| Group 3 | 14 | 19.6 (3.54) | 10 | −0.92 (2.71) | 0.181 | 12 | +0.61 (3.86) | 0.478 | |
| Group 4 | 14 | 20.1 (4.45) | 14 | +0.77 (2.18) | 0.628 | 14 | +1.29 (3.60) | 0.524 | |
| Group 5 | 13 | 17.9 (3.27) | 12 | +0.91 (3.72) | 0.425 | 11 | +3.56 (2.63) | 0.021 | |
| Group 6 | 14 | 20.0 (3.34) | 14 | −0.87 (3.05) | 0.277 | 15 | +1.85 (5.58) | 0.350 | |
| Total | 83 | 19.3 (3.88) | 73 | +0.55 (3.07) | 0.6030 | 81 | +2.06 (4.37) | 0.0020 | |
Baseline values are means ± standard deviations. All P values were calculated using unpaired Student's t test. n, number of patients with measurements.
Fig 3.
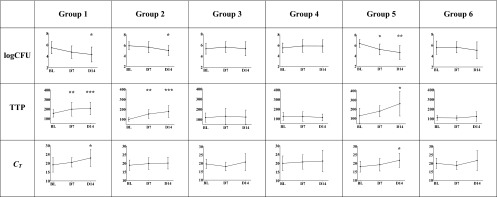
The mean change from baseline (SDs are shown as vertical bars) of log CFU, TTP, and CT after 7 and 14 days of treatment is shown for all six groups. Significantly different measurements for days 0 to 7 or days 0 to 14 were found for TTP in 2 and 3, for log CFU in 1 and 3, and for CT in 0 and 2 groups, respectively. Statistically significant differences between time points are indicated by asterisks (* = P < 0.05, ** = P < 0.001, *** = P < 0.0001, BL = baseline, D7 = day 7, D14 = day 14).
Table 2.
Discrimination between treatment effectsa
| Activity period | Log CFU |
TTP |
CT |
|||
|---|---|---|---|---|---|---|
| F | P | F | P | F | P | |
| Days 0 to 7 | 4.135 | 0.0024 | 9.012 | <0.0001 | 1.995 | 0.0910 |
| Days 0 to 14 | 7.277 | <0.0001 | 11.58 | <0.0001 | 1.203 | 0.3160 |
One-way ANOVA was used for treatment activities over 7 days and 14 days measured with log CFU, TTP, and CT. Greater F values represent a better ability to discriminate between groups for treatment effects (there were 6 treatment groups for each period).
DISCUSSION
This prospective study directly compared three different methods for the quantification of the mycobacterial load in sputum specimens collected 7 and 14 days after initiation of antituberculosis treatment. All three methods were able to demonstrate an average decrease of the mycobacterial sputum load. TTP in liquid culture detected treatment effects earlier and more frequently than the other methods, and it discriminated best between treatment groups. CFU counting on agar plates was comparable to TTP. Both culture-based methods had discriminatory power that was clearly superior to that of real-time PCR.
Why does PCR in the form of standardized Xpert CT perform so poorly in the detection of early treatment effects? This can be explained by the ability of Xpert to detect DNA from whole but metabolically inactive or dead bacteria or even free DNA causing false-positive signals. For establishing the diagnosis of tuberculosis, a high sensitivity is an advantage because the presence of mycobacterial DNA in sputum is associated with disease, and it does not matter whether the DNA detected comes from viable or dead bacteria. For monitoring of treatment effects, however, the distinction between DNA contained in viable mycobacteria and DNA originating from other sources becomes critical, because bacteria killed by antituberculosis treatment are subsequently expectorated in sputum, and it is known that mycobacterial DNA survives for an extended period (15). This does not encourage the hope that Xpert could emerge as a treatment-monitoring tool for the HIV-positive, smear-negative tuberculosis patients frequently encountered in countries such as South Africa. A modified protocol for Xpert intended to exclude DNA from nonviable mycobacteria from the reaction could remedy this problem, and research in this area is ongoing (16).
The analysis reported here is based on a single study of only 14 days' duration with only 4 time points assayed. We are confident, however, that the results are valid, since TTP and CT were similarly well correlated in a previous study at our laboratory with 74 sputum samples collected before treatment (rho = 0.539; present study, rho = 0.632) (13). Also, merged data from five different EBA studies revealed very similar correlations between 7-day treatment activities determined by TTP and log CFU (rho = 0.649; present study, rho = −0.673) (4).
In conclusion, our results demonstrate that culture-based methods remain the standard for measuring the decline of sputum bacterial load during the early phase of antituberculosis treatment. The Xpert's easy handling and quick results do not compensate for its low discriminatory power between groups with different treatments. Measures that reduce contamination with DNA from nonviable bacteria might transform the value of this technique.
ACKNOWLEDGMENTS
We thank all patients for participation in this study and the staff from the Centre of Clinical Tuberculosis Research (CCTR) for technical assistance. This study was conducted as an activity of the Pan African Consortium for the Evaluation of Anti-tuberculosis Antibiotics (PanACEA). PanACEA is funded by EDCTP grants CT.2004.32011.001, IP.2007.32011.011, IP.2007.32011.012, and IP.2007.32011.013, the Bill and Melinda Gates Foundation, the UK Medical Research Council, and the German Ministry of Science and Technology (BmBF; grant 01KA0901). A specific Ph.D. stipend was given by PanACEA to X. A. Kayigire to foster local career development.
The Pan African Consortium for the Evaluation of Anti-tuberculosis Antibiotics (PanACEA) comprises the following individuals and institutions: Medical Centre of the University of Munich, Munich, Germany (Sonja Henne and Anna Maria Mekota, Norbert Heinrich, Andrea Rachow, Anke Kohlenberg, Elmar Saathoff, and Michael Hoelscher); University of St. Andrews, St Andrews, United Kingdom (Stephen Gillespie); Radboud University Nijmegen Medical Centre, Nijmegen, the Netherlands (Georgette Plemper van Balen, Marloes Weijers, Rob Aarnoutse, and Martin Boeree); University College of London, London, United Kingdom (Anna Bateson, Timothy McHugh, Kasha Singh, Robert Hunt, and Alimuddin Zumla); MRC Clinical Trials Unit, London, United Kingdom (Andrew Nunn and Patrick Phillips); University of Cape Town, Cape Town, South Africa (Rodney Dawson and Kim Narunsky); University of Stellenbosch, Cape Town, South Africa (Andreas Diacon, Jeannine du Bois, Amour Venter, and Sven Friedrich); University of the Witswatersrand, Johannesburg, South Africa (Ian Sanne, Karla Mellet, and Eefje de Jong); The Aurum Institute, Johannesburg, South Africa (Gavin Churchyard and Salome Charalambous); University of Zambia, Lusaka, Zambia (Peter Mwaba); NIMR-Mbeya Medical Research Centre, Mbeya, Tanzania (Nyanda Elias, Chacha Mangu, Gabriel Rojas-Ponce, Bariki Mtafya, and Leonard Maboko); Ifakara Health Institute-Bagamoyo Research and Training Centre, Bagamoyo, Tanzania (Klaus Reither and Levan Jugheli); Kilimanjaro Clinical Research Institute, Moshi, Tanzania (Noel Sam, Gibson Kibiki, Hadija Semvua, and Stellah Mpagama); Medical Research Unit—Albert Schweitzer Hospital, Lambarene, Gabon (Afsatou Traore and Ayola Akim Adegnika); Kenya Medical Research Institute, Nairobi, Kenya (Evans Amukoye); Makerere University, Kampala, Uganda (Alphonse Okwera).
No author has a conflict of interest to state.
Footnotes
Published ahead of print 17 April 2013
REFERENCES
- 1. Hong Kong Chest Service 1987. Five-year follow-up of a controlled trial of five 6-month regimens of chemotherapy for pulmonary tuberculosis. Am. Rev. Respir. Dis. 136:1339–1342 [DOI] [PubMed] [Google Scholar]
- 2. Jindani A, Aber VR, Edwards EA, Mitchison DA. 1980. The early bactericidal activity of drugs in patients with pulmonary tuberculosis. Am. Rev. Respir. Dis. 121:939–949 [DOI] [PubMed] [Google Scholar]
- 3. Pheiffer C, Carroll NM, Beyers N, Donald P, Duncan K, Uys P, Van Helden P. 2008. Time to detection of Mycobacterium tuberculosis in BACTEC systems as a viable alternative to colony counting. Int. J. Tuberc. Lung Dis. 12:792–798 [PubMed] [Google Scholar]
- 4. Diacon AH, Maritz JS, Venter A, Van Helden PD, Dawson R, Donald PR. 2012. Time to liquid culture positivity can substitute for colony counting on agar plates in early bactericidal activity studies of antituberculosis agents. Clin. Microbiol. Infect. 18(7):711–717 [DOI] [PubMed] [Google Scholar]
- 5. Hesseling AC, Walzl G, Enarson DA, Carroll NM, Duncan K, Lukey PT, Lombard C, Donald PR, Lawrence KA, Gie RP, Van Helden PD, Beyers N. 2010. Baseline sputum time to detection predicts month two culture conversion and relapse in non-HIV-infected patients. Int. J. Tuberc. Lung Dis. 14:560–570 [PubMed] [Google Scholar]
- 6. Blakemore R, Story E, Helb D, Kop J, Banada P, Owens MR, Chakravorty S, Jones M, Alland D. 2010. Evaluation of the analytical performance of the Xpert MTB/RIF assay. J. Clin. Microbiol. 48:2495–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Helb D, Jones M, Story E, Boehme C, Wallace E, Ho K, Kop J, Owens MR, Rodgers R, Banada P, Safi H, Blakemore R, Lan NT, Jones-Lopez EC, Levi M, Burday M, Ayakaka I, Mugerwa RD, McMillan B, Winn-Deen E, Christel L, Dailey P, Perkins MD, Persing DH, Alland D. 2010. Rapid detection of Mycobacterium tuberculosis and rifampin resistance by use of on-demand, near-patient technology. J. Clin. Microbiol. 48:229–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boehme CC, Nabeta P, Hillemann D, Nicol MP, Shenai S, Krapp F, Allen J, Tahirli R, Blakemore R, Rustomjee R, Milovic A, Jones M, O'Brien SM, Persing DH, Ruesch-Gerdes S, Gotuzzo E, Rodrigues C, Alland D, Perkins MD. 2010. Rapid molecular detection of tuberculosis and rifampin resistance. N. Engl. J. Med. 363:1005–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Van Zyl-Smit RN, Binder A, Meldau R, Mishra H, Semple PL, Theron G, Peter J, Whitelaw A, Sharma SK, Warren R, Bateman ED, Dheda K. 2011. Comparison of quantitative techniques including Xpert MTB/RIF to evaluate mycobacterial burden. PLoS One 6:e28815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rachow A, Zumla A, Heinrich N, Rojas-Ponce G, Mtafya B, Reither K, Ntinginya EN, O'Grady J, Huggett J, Dheda K, Boehme C, Perkins M, Saathoff E, Hoelscher M. 2011. Rapid and accurate detection of Mycobacterium tuberculosis in sputum samples by Cepheid Xpert MTB/RIF assay-A clinical validation study. PLoS One 6(6):e20458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Theron G, Peter J, Van Zyl-Smit R, Mishra H, Streicher E, Murray S, Dawson R, Whitelaw A, Hoelscher M, Sharma S, Pai M, Warren R, Dheda K. 2011. Evaluation of the Xpert MTB/RIF assay for the diagnosis of pulmonary tuberculosis in a high HIV prevalence setting. Am. J. Respir. Crit. Care Med. 184:132–140 [DOI] [PubMed] [Google Scholar]
- 12. Blakemore R, Nabeta P, Davidow AL, Vadwai V, Tahirli R, Munsamy V, Nicol M, Jones M, Persing DH, Hillemann D, Ruesch-Gerdes S, Leisegang F, Zamudio C, Rodrigues C, Boehme CC, Perkins MD, Alland D. 2011. A multisite assessment of the quantitative capabilities of the Xpert MTB/RIF assay. Am. J. Respir. Crit. Care Med. 184:1076–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Friedrich SO, Venter A, Kayigire XA, Dawson R, Donald PR, Diacon AH. 2011. Suitability of Xpert MTB/RIF and Genotype MTBDRplus for patient selection for a tuberculosis clinical trial. J. Clin. Microbiol. 49:2827–2831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. International Union Against Tuberculosis and Lung Dis 2000. Technical guide: Sputum examination for tuberculosis by direct microscopy in low income countries. 5th edition [Google Scholar]
- 15. Kennedy N, Gillespie SH, Saruni AO, Kisyombe G, McNerney R, Ngowi FI, Wilson S. 1994. Polymerase chain reaction for assessing treatment response in patients with pulmonary tuberculosis. J. Infect. Dis. 170:713–716 [DOI] [PubMed] [Google Scholar]
- 16. Miotto P, Bigoni S, Migliori GB, Matteelli A, Cirillo DM. 2012. Early tuberculosis treatment monitoring by Xpert(R) MTB/RIF. Eur. Respir. J. 39:1269–1271 [DOI] [PubMed] [Google Scholar]


