Abstract
Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) for the identification of bacteria and fungi was recently introduced in microbiology laboratories. This technology could greatly improve the clinical management of patients and guidance for chemotherapy. In this study, we used a commercial MALDI Sepsityper extraction method to evaluate the performance of two commercial MALDI-TOF MS systems, the Vitek MS IVD (bioMérieux) and the Microflex LT Biotyper (Bruker Daltonics) for direct bacterial identification in positive blood cultures. In 181 monomicrobial cultures, both systems generated genus to species level identifications for >90% of the specimens (Biotyper, 177/181 [97.8%]; Vitek MS IVD, 167/181 [92.3%]). Overall, the Biotyper system generated significantly more accurate identifications than the Vitek MS IVD system (P = 0.016; 177 versus 167 out of 181 specimens). The Biotyper system identified the minority species among polymicrobial blood cultures. We also compared the performance of an in-house extraction method with that of the Sepsityper on both MALDI-TOF MS systems. The in-house method generated more correct identifications at the genus level than the Sepsityper (96.7% versus 93.5%) on the Biotyper system, whereas the two methods exhibited the same performance level (88.0% versus 88.0%) on the Vitek MS IVD system. Our study confirmed the practical advantages of MALDI-TOF MS, and our in-house extraction method reduced the reagent cost to $1 per specimen, with a shorter turnaround time of 3 h, which is highly cost-effective for a diagnostic microbiology service.
INTRODUCTION
Bloodstream infection is a condition associated with high morbidity and mortality (1, 2). The rapid identification of bloodstream pathogens is critical to clinical management and the choice of appropriate antibiotic treatments (3, 4). Currently, microbiologic diagnosis of bacteremia relies on subculture of positive blood culture broths on solid medium for an 18- to 24-h incubation period followed by biochemical tests or an automated preformed enzyme assay for identification of the bacteria (5). In general, laboratory diagnosis of common pathogens requires 18 to 48 h, while diagnosis of fastidious organisms requires longer incubation and identification procedures. Although the use of modern techniques such as fluorescence in situ hybridization and PCR can shorten the identification time (6), these operations require specialized equipment and technical expertise and the targeted pathogens are limited in a single run (7). Therefore, the introduction of a simple, rapid, broad-spectrum, and cost-effective system for the identification of blood culture pathogens is imperative.
Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) was developed in the 1980s, and this technique had been widely used for biomolecule analysis in the chemical industry for over 10 years. In recent years, MALDI-TOF MS was introduced for bacterial identification, which revolutionized the diagnostic microbiology service (8). Two manufacturers, bioMérieux (Marcy l'Etoile, France) and Bruker Daltonics (Bremen, Germany), are marketing efficient MALDI-TOF systems, the Vitek-MS and the Microflex LT, respectively, that allow the identification of bacteria and yeasts in a few minutes instead of the hours required by traditional methods. The practical use of MALDI-TOF MS for microorganism identification directly from positive blood culture specimens has been substantiated by a number of studies (9–13). Generally, studies have shown high identification rates for various types of organisms with the use of MALDI-TOF MS. However, various organism isolation methods from blood were used in those studies and therefore method standardization is necessary. The commercially available MALDI Sepsityper kit (Bruker Daltonics, Bremen, Germany) was recently developed to standardize the preparation of blood cultures prior to spectrometric analysis (14). However, the performance of the Sepsityper on MALDI-TOF platforms other than the Bruker system was not explored, and the high reagent cost also hindered the wide application of the Sepsityper in diagnostic settings.
This is the first study to compare the clinical performance and running costs of the two MALDI-TOF systems (Bruker Biotyper and bioMérieux Vitek-MS) on positive blood culture specimens. An in-house extraction method was also evaluated in this study, and its performance was compared with that of the commercial MALDI Sepsityper kit.
MATERIALS AND METHODS
The study was divided into two phases. The first phase was the evaluation of the bacterial identification performance of two MALDI-TOF systems, the Bruker Biotyper and the bioMérieux Vitek-MS, on blood culture specimens prepared with the commercially available MALDI Sepsityper extraction kit (Bruker Daltonics, Bremen, Germany). The second phase was the comparison of the performance of an in-house blood culture extraction method with that of the MALDI Sepsityper Kit.
Collection of blood cultures.
A total of 202 available blood culture broths that were Bactec positive as indicated by the Bactec system (Bactec Plus Aerobic, Bactec F Lytic Anaerobic, and Bactec Myco Lytic; Becton Dickinson, Franklin Lakes, NJ) were collected between March and July 2012. Samples collected from 8 a.m. to 5 p.m. were analyzed on the same day, while those broths that became positive after 5 p.m. were analyzed the next morning. Therefore, the incubation times of these blood culture broths after they became positive were longer than those for the others. Positive blood culture broths were also subjected to direct Gram staining and subculturing, followed by biochemical identification (Vitek II; bioMérieux, France) and/or 16S rRNA gene sequencing (15, 16). All of the 202 positive blood culture specimens were used for the comparison of the performance of the two MALDI-TOF systems, while 92 positive specimens were randomly selected for the comparison of the MALDI Sepsityper and the in-house blood culture extraction method.
Blood culture extraction.
In order to extract and purify the bacterial protein from blood samples, we used the MALDI Sepsityper kit (Bruker Daltonics, Bremen, Germany) to extract all of the 202 samples. The in-house extraction method was used to extract protein from 92 samples randomly selected from the specimen pool. The extraction protocols of the two methods are described briefly.
MALDI Sepsityper extraction method.
For each blood culture bottle specimen, 1 ml of blood was transferred to a 1.5-ml capped tube (Eppendorf, Germany). Then, 200 μl of solution 1 (provided in the kit) was added to the 1 ml blood, followed by a centrifugation at 13,000 rpm for 1 min. The supernatant was discarded, and the pellet was resuspended with 1 ml of solution 2 (provided in the kit), with centrifugation again at 13,000 rpm for 1 min. The supernatant was discarded, and 300 μl of high-pressure liquid chromatography (HPLC)-grade distilled water (Fluka) was added to resuspend the pellet. Another 900 μl of HPLC-grade ethanol (Sigma-Aldrich) was added, and the mixture was centrifuged at 13,000 rpm for 2 min. Subsequently, the supernatant was carefully removed by pipetting, and 20 to 50 μl of formic acid was added depending on the size of the pellet. An equal volume of pure acetonitrile (Fluka) was then added, followed by centrifugation at 13,000 rpm for 2 min. The supernatant was then spotted onto target slides for MALDI-TOF analysis.
In-house extraction method.
Our in-house extraction method was a modified protocol published previously (17). The published protocol was modified by adding an ethanol-formic acid extraction after the washing steps. Briefly, 1 ml of the positive blood sample was added to 200 μl of 5% saponin solution (Sigma-Aldrich). The lysate was then vortexed thoroughly. After a 5-min room temperature incubation, the tube was centrifuged at 13,000 rpm for 1 min, and the supernatant was discarded. The pellet was further washed with 1 ml distilled water and the supernatant was discarded after centrifugation at 13,000 rpm for 1 min. A total of 100 μl distilled water was used to resuspend the pellet. Another 300 μl of ethanol was added, and the mixture was centrifuged at 13,000 rpm for 2 min. Subsequently, the supernatant was carefully removed by pipetting and resuspended with 50 μl formic acid. An equal volume of acetonitrile (Fluka) was then added, followed by centrifugation 13,000 rpm for 2 min. The supernatant was then spotted onto target slides for MALDI-TOF analysis.
MALDI-TOF analysis. (i) Bruker Microflex LT with Biotyper 3.0 system.
One microliter of each purified blood culture extract was transferred to an individual spot on the Bruker 96-spot reusable stainless steel target plate. Each spot was covered with 1 μl alpha-cyano-4-hydroxycinnamic acid (HCCA) matrix (Bruker Daltonics, Germany). The target plate was then read and analyzed by the Bruker Microflex LT system. A protein profile of each specimen with m/z values of 3,000 to 15,000 was generated based on a minimum of 240 laser shot measurements. The profiles were further analyzed by using the Biotyper 3.0 software (Bruker Daltonics, Germany) under the blood culture mode, which queried a reference database of 4,500 isolates and returned the top 10 identification matches along with confidence scores ranging from 0.0 to 3.0. We concluded the results by using the top scoring identification. Scores of ≥2.0 were considered high-confidence identification to the species level, while scores of 1.6 to 1.99 were considered intermediate-confidence identification to the genus level only. Scores of <1.6 were considered unacceptable identification according to manufacturer's recommendation.
(ii) bioMérieux Vitek MS IVD system.
One microliter of each purified blood culture extract was transferred to an individual spot on the 48-well Vitek MS-DS disposable target slide. Each spot was covered with 1 μl ready-to-use Vitek MS alpha-cyano-4-hydroxycinnamic acid (HCCA) matrix (bioMérieux, France). The target plate was then read and analyzed by the Vitek MS IVD system. The protein profile of each specimen with an m/z of 3,000 to 15,000 was generated based on 100 measurements. The profiles were further matched with the Vitek MS reference CE-IVD certified database, which included >20,000 spectra, and returned the best identification match along with confidence percentages from 0% to 99.9%. Spot results of 90 to 98% confidence were considered high at the genus level, while results of >98% confidence were considered high at the species level. Spots results of <90% confidence were considered unacceptable identification. For specimens showing more than one identification result under the same genus, we concluded that result to be a genus level match only. For specimens showing more than one identification result that included some results with mismatched genus or family identifications, we concluded that result to be an unacceptable identification.
Statistical methods.
Pearson's chi-square test was used to compare the results obtained by the two systems with the same specimens.
RESULTS
Routine identification.
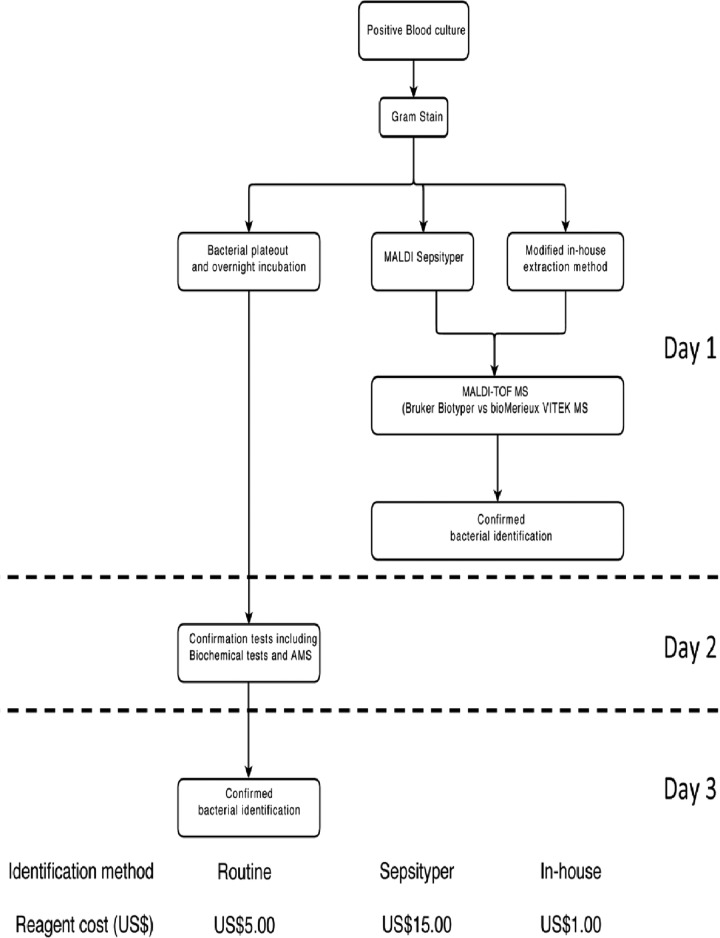
Among the 202 blood culture-positive specimens included in the study, 181 of them were identified as monomicrobial, of which 75 of them (41.4%) contained Gram-positive organisms and 106 of them (58.6%) contained Gram-negative organisms, through the use of our routine identification methods, including different selective media, biochemical tests, automated microbial systems (bioMérieux Vitek II and BD Phoenix), and 16S rRNA gene sequencing,. The remaining 21 blood cultures were identified to be polymicrobial. Identifications were generated by biochemical tests and automated microbial systems (Vitek II, bioMérieux, France) within 3 working days (Fig. 1), whereas 17 specimens containing coagulase-negative staphylococcus (CNS) or bacillus species required 16S rRNA gene sequencing for identification, which took another 3 working days to process.
Fig 1.
Workflow and cost comparison of routine identification and MALDI-TOF MS identification methods used on direct blood culture specimens. AMS, AutoMicrobic system.
MALDI-TOF showed high concordance to routine identification.
Of the 181 monomicrobial cultures, both MALDI-TOF systems (Biotyper [177/181, 97.8%]; Vitek MS IVD [167/181; 92.3%]) generated genus to species level identifications for >90% of specimens (Table 1). There were 3 Gram-positive (1 Bacillus circulans and 2 Bacillus horneckiae) and 1 Gram-negative (Brevundimonas diminuta) bacteria that were identified only by the 16S rRNA gene sequencing and not through the use of MALDI-TOF. In general, no genus level discordance was observed between the routine and the MALDI-TOF method. Nevertheless, there were 2 cases of Gram-positive bacteria showing discordant identifications at the species level; one was Streptococcus anginosus identified as Streptococcus constellatus by both MALDI-TOF systems and the other was Streptococcus bovis identified as Streptococcus gallolyticus by both MALDI-TOF systems. These discordant results might have been attributable to the small differences in the proteomic profiles, so that S. anginosus and S. constellatus both belonged to the S. anginosus group and S. bovis and S. gallolyticus both belonged to the S. bovis group (5). As S. bovis has been recently reclassified as S gallolyticus subsp. gallolyticus, the next version of the MS identification database may provide better discrimination power for this species.
Table 1.
Identification results by Biotyper and Vitek-MS using recommended cutoff values (n = 181)
| Conventional bacterial identification (no. of strains) | No. (%) of strains identified with the Biotyper |
MALDI-TOF identification (no. of strains) | No. (%) of strains identified with the Vitek-MS |
MALDI-TOF identification (no. of strains) | ||||
|---|---|---|---|---|---|---|---|---|
| Species level | Genus level only | Not identified | Species level | Genus level only | Not identified | |||
| Gram-positive bacteria | ||||||||
| Enterococcus avium (1) | 1 | 0 | 0 | Enterococcus avium (1) | 1 | 0 | 0 | Enterococcus avium (1) |
| Enterococcus casseliflavus (1) | 1 | 0 | 0 | Enterococcus casseliflavus (1) | 1 | 0 | 0 | Enterococcus casseliflavus (1) |
| Enterococcus faecalis (5) | 5 | 0 | 0 | Enterococcus faecalis (5) | 5 | 0 | 0 | Enterococcus faecalis (5) |
| Enterococcus faecium (5) | 5 | 0 | 0 | Enterococcus faecium (5) | 5 | 0 | 0 | Enterococcus faecium (5) |
| Staphylococcus aureus (16) | 15 | 1 | 0 | Staphylococcus aureus (15) and Staphylococcus aureus (1)a | 14 | 0 | 2 | Staphylococcus aureus (14) and not identified (2) |
| Staphylococcus epidermidis (10) | 3 | 7 | 0 | Staphylococcus epidermidis (3) and Staphylococcus epidermidis (7)a | 6 | 0 | 4 | Staphylococcus epidermidis (6), not identified (3), and wrong identification (1) |
| Staphylococcus capitis (1) | 1 | 0 | 0 | Staphylococcus capitis (1) | 0 | 1 | 0 | Staphylococcus capitis (1)b and Staphylococcus caprae (1)b |
| Staphylococcus hominis (1) | 1 | 0 | 0 | Staphylococcus hominis (1) | 1 | 0 | 0 | Staphylococcus hominis (1) |
| Streptococcus agalactiae (4) | 4 | 0 | 0 | Streptococcus agalactiae (4) | 4 | 0 | 0 | Streptococcus agalactiae (4) |
| Streptococcus anginosus (1) | 0 | 1 | 0 | Streptococcus constellatus (1) | 1 | 0 | 0 | Streptococcus constellatus (1) |
| Streptococcus bovis (1) | 0 | 1 | 0 | Streptococcus gallolyticus (1) | 0 | 1 | 0 | Streptococcus gallolyticus (1) |
| Streptococcus pyogenes (2) | 2 | 0 | 0 | Streptococcus pyogenes (2) | 2 | 0 | 0 | Streptococcus pyogenes (2) |
| Streptococcus sanguinis (5) | 2 | 3 | 0 | Streptococcus sanguinis (2) and Streptococcus sanguinis (3)a | 5 | 0 | 0 | Streptococcus sanguinis (5) |
| Listeria monocytogenes (1) | 1 | 0 | 0 | Listeria monocytogenes (1) | 1 | 0 | 0 | Listeria monocytogenes (1) |
| Micrococcus luteus (1) | 1 | 0 | 0 | Micrococcus luteus (1) | 1 | 0 | 0 | Micrococcus luteus/lylae (1) |
| Propionibacterium acnes (4) | 3 | 1 | 0 | Propionibacterium acnes (3) and Propionibacterium acnes (1)a | 4 | 0 | 0 | Propionibacterium acnes (4) |
| Bacillus cereus/thuringiensis (9) | 7 | 2 | 0 | Bacillus cereus (7) and Bacillus weihenstephanensis (2)a | 0 | 9 | 0 | Bacillus mycoides (9)b, Bacillus cereus (9)b, and Bacillus thuringiensis (9)b |
| Bacillus circulans (1) | 0 | 0 | 1 | Not identified (1) | 0 | 0 | 1 | Not identified (1) |
| Bacillus flexus (1) | 1 | 0 | 0 | Bacillus flexus (1) | 0 | 1 | 0 | Bacillus megaterium (1) |
| Bacillus horneckiae (2) | 0 | 0 | 2 | Not identified (2) | 0 | 0 | 2 | Not identified (2) |
| Paenibacillus urinalis (1) | 1 | 0 | 0 | Paenibacillus urinalis (1) | 0 | 0 | 1 | Not identified (1) |
| Paenibacillus illinoisensis (1) | 0 | 1 | 0 | Paenibacillus illinoisensis (1)a | 0 | 1 | 0 | Paenibacillus pabuli (1)b and Paenibacillus spp. (1)b |
| Bacillus megaterium (1) | 0 | 1 | 0 | Bacillus flexus (1) | 0 | 0 | 1 | Not identified (1) |
| Subtotal (75) | 54 (72.0) | 18 (24.0) | 3 (4.0) | 51 (68.0) | 13 (17.3) | 11 (14.7) | ||
| Gram-negative bacteria | ||||||||
| Acinetobacter baumannii (6) | 4 | 2 | 0 | Acinetobacter baumannii (4) and, Acinetobacter baumannii (2)a | 6 | 0 | 0 | Acinetobacter baumannii (6) |
| Acinetobacter junii (1) | 1 | 0 | 0 | Acinetobacter junii (1) | 0 | 1 | 0 | Acinetobacter junii (1)a |
| Aeromonas hydrophila (1) | 0 | 1 | 0 | Aeromonas caviae (1) | 0 | 1 | 0 | Aeromonas sobria (1) |
| Brevundimonas diminuta (1) | 0 | 0 | 1 | Not identified (1) | 0 | 0 | 1 | Not identified (1) |
| Citrobacter freundii (4) | 4 | 0 | 0 | Citrobacter freundii (4) | 3 | 1 | 0 | Citrobacter freundii (3) and Citrobacter youngae (1) |
| Escherichia coli (53) | 53 | 0 | 0 | Escherichia coli (53) | 53 | 0 | 0 | Escherichia coli (53) |
| Enterobacter aerogenes (4) | 4 | 0 | 0 | Enterobacter aerogenes (4) | 4 | 0 | 0 | Enterobacter aerogenes (4) |
| Enterobacter cloacae (1) | 0 | 1 | 0 | Enterobacter asburiae (1) | 0 | 1 | 0 | Enterobacter cloacae (1) and Enterobacter asburiae (1)b |
| Enterobacter kobei (2) | 1 | 1 | 0 | Enterobacter asburiae (1) and Enterobacter kobei (1) | 0 | 2 | 0 | Enterobacter cloacae (2) and Enterobacter asburiae (2)b |
| Klebsiella pneumoniae (16) | 16 | 0 | 0 | Klebsiella pneumonia (16) | 16 | 0 | 0 | Klebsiella pneumonia (16) |
| Proteus mirabilis (4) | 4 | 0 | 0 | Proteus mirabilis (4) | 4 | 0 | 0 | Proteus mirabilis (4) |
| Pseudomonas aeruginosa (3) | 3 | 0 | 0 | Pseudomonas aeruginosa (3) | 3 | 0 | 0 | Pseudomonas aeruginosa (3) |
| Pseudomonas putida (3) | 2 | 1 | 0 | Pseudomonas putida (2) and Pseudomonas putida (1)a | 3 | 0 | 0 | Pseudomonas putida (3) |
| Salmonella enteritidis (3) | 0 | 3 | 0 | Salmonella spp. (3)a | 0 | 3 | 0 | Salmonella spp. (3)a |
| Serratia marcescens (1) | 1 | 0 | 0 | Serratia marcescens (1) | 1 | 0 | 0 | Serratia marcescens (1) |
| Stenotrophomonas maltophilia (2) | 1 | 1 | 0 | Stenotrophomonas maltophilia (1) and Stenotrophomonas maltophilia (1)a | 2 | 0 | 0 | Stenotrophomonas maltophilia (2) |
| Vibrio vulnificus (1) | 0 | 1 | 0 | Vibrio vulnificus (1)a | 0 | 0 | 1 | Not identified (1) |
| Subtotal (106) | 94 (88.7) | 11 (10.4) | 1 (0.9) | 95 (89.6) | 9 (8.5) | 2 (1.9) | ||
| Total (181) | 148 (81.8) | 29 (16.0) | 4 (2.2) | 146 (80.7) | 22 (12.2) | 13 (7.2) | ||
Identifications at genus confidence level only (Bruker Score < 2.0 or Vitek-MS confidence < 90.0).
More than one identification generated at the same confidence level.
Our study cohort demonstrated that overall the Biotyper system (177/181) generated significantly more accurate identifications than the Vitek MS IVD system (167/181) (Pearson's chi-square; P = 0.016). The Biotyper system (84.4%; 103/122) demonstrated similar concordance to species level identifications of both Gram-positive (54/75 for Biotyper versus 51/75 for Vitek MS IVD) and Gram-negative (94/106 for Biotyper versus 95/106 for Vitek MS IVD) bacteria. However, the Gram-negative Vibrio vulnificus-containing specimen was identified only by the Biotyper system at the genus level and failed to be interpreted by the Vitek MS IVD system. On the other hand, for specimens with Bacillus cereus, the Vitek MS IVD system generated multiple Bacillus species identifications for all 8 of the Bacillus cereus-containing specimens, while the Biotyper system generated accurate species level identification for 6 of them. Also, both MALDI-TOF systems generated only genus level identifications of Salmonella enteritidis. Serogrouping should be followed up separately from the MALDI-TOF result in order to distinguish this organism from the highly pathogenic Salmonella typhi.
For Gram-positive bacterial identification, Staphylococcus epidermidis (7/10 at the genus level only) and Streptococcus sanguinis (3/5 at the genus level only) were the two species that were demonstrated to present difficulties in species level identification with the Biotyper system.
Polymicrobial cultures.
Twenty-one blood cultures in our cohort were identified by the routine methods to be composed of 2 bacterial species. The Vitek MS IVD system identified only the majority species composition in each of the 21 mixed cultures, while the Biotyper system identified both of the two species with >1.6 confidence scores in 5 out of the 21 mixed cultures through the Biotyper option “Top 10 matched pattern choices” (Table 2). For the other 16 mixed cultures, the Biotyper system identified only the major composition of the mixed cultures.
Table 2.
Specimens with polymicrobial identifications generated by the Biotyper MALDI-TOF system
| Mixed culture identified by routine methods | Biotyper |
Vitek MS IVD (confidence %) | |
|---|---|---|---|
| Top match (score) | Other species in top 10 matches list with scores of >1.7 (score) | ||
| Escherichia coli + Klebsiella pneumoniae | Escherichia coli (2.33) | Klebsiella pneumoniae (2.07) | Escherichia coli (99.9) |
| Escherichia coli + Klebsiella pneumoniae | Escherichia coli (2.32) | Klebsiella pneumoniae (2.20) | Escherichia coli (99.9) |
| Escherichia coli + Klebsiella pneumoniae | Klebsiella pneumoniae (2.27) | Escherichia coli (2.13) | Klebsiella pneumoniae (99.9) |
| Klebsiella pneumoniae + Enterococcus faecalis | Klebsiella pneumoniae (2.23) | Enterococcus faecalis (1.91) | Klebsiella pneumoniae (99.9) |
| Acinetobacter baumannii + Staphylococcus epidermidis | Acinetobacter baumannii (1.97) | Staphylococcus epidermidis (1.76) | Acinetobacter baumannii complex (99.9) |
Comparison between MALDI Sepsityper and in-house extraction method.
The in-house sample extraction method was performed on a total of 92 monomicrobial specimens in order to compare the performance of the in-house and MALDI Sepsityper methods on the two MALDI-TOF systems (see Table S1 in the supplemental material).
Used with the Biotyper system, our in-house extraction method correctly identified 96.7% (89/92) and 80.4% (74/92) of the organisms to the genus and species levels, respectively. Used with the MALDI Sepsityper kit, the in-house method generated correct genus identification in 93.5% (86/92) of specimens and correct species identification in 81.5% (75/92). The misidentified rates (confidence score, <1.6) of the in-house and the MALDI Sepsityper extraction methods were 3.3% (3/92) and 6.5% (6/92), respectively.
On the other hand, the in-house extraction method used with the Vitek MS IVD system identified only 88.0% (81/92) and 79.3% (73/92) of the organisms to the genus and species levels, respectively. Correct genus identification was generated in 88.0% (81/92) of specimens, and species level identification was reached in 81.5% (75/92) of them by using the MALDI Sepsityper method. The identification-missing rate of the Vitek MS IVD system was 12.0% (11/92) with both the in-house and the MALDI Sepsityper methods.
DISCUSSION
Bacteremia causes serious conditions in patients, and rapid identification of the bacterial species in direct blood cultures is critical for early diagnosis and treatment. Currently, the routine diagnostic method requires 18 to 24 h for preliminary bacterial identification. With the introduction of MALDI-TOF MS, the diagnostic time might be largely reduced, which would have a positive impact on patient care. The high concordance of MALDI-TOF MS and routine diagnostic methods for the identification of bacterial cultures has also been confirmed by many studies (18–20). Despite these advantages, there have been few studies performed to evaluate the performance of MALDI-TOF MS for rapid identification of microorganisms from direct blood cultures of bacteremia patients. Direct comparison of the two commercial systems has not been available because the commercial blood culture extraction kit is relatively expensive for routine laboratory diagnosis, and therefore the introduction of an in-house extraction was necessary. Therefore, in our study we aimed to identify the best option for implementing MALDI-TOF MS in a clinical diagnostic laboratory. To our knowledge, this is first published article to evaluate the two MALDI-TOF systems and compare an in-house extraction method with a commercial kit for use on positive blood cultures.
In the first phase of this study, we demonstrated that for monobacterial direct blood culture specimens, both of the MALDI-TOF MS systems for clinical microbiological diagnosis generated accurate identification at rates of >80% at the species level and >90% at the genus level. A higher percentage of correct identifications to the species level was obtained using the Bruker Microflex LT with the Biotyper version 3.0 system (P = 0.016). The Bruker Biotyper system was found to generate more accurate identification than the bioMérieux Vitek-MS system for Gram-positive bacteria at both the species and genus levels, especially for Bacillus species identification. On the other hand, for Gram-negative bacteria the two systems demonstrated similar levels of identification performance at the species and genus levels.
In comparison to the rates of species level identification (84 to 95%) generated by the two MALDI-TOF systems, as reported for previous studies focusing on bacterial culture isolates (21, 22), our study demonstrated that the generation of high-quality identification of direct blood culture specimens was more difficult with both the Biotyper (81.8%) and Vitek MS (80.7%) systems. The poor performance in generating species level identifications for Bacillus species by both systems may be due to the spore-forming nature of bacilli, because the formic acid and CHCA matrix could not efficiently break down the cell walls of the spores.
Many studies have shown that patient outcome could be improved with the earlier use of appropriate antibiotics. Vlek et al. recently demonstrated that direct performance of MALDI-TOF MS on positive blood culture broths significantly reduced time to organism identification by 28.8 h compared with identification by conventional methods (4). Rapid identification to the genus level would be very useful to guide clinical management, such as the rapid differentiation of Gram-negative bacteria to the genus level (for example, Acinetobacter versus Klebsiella). Importantly, rapid and precise results were associated with an 11.3% increase in the proportion of patients receiving appropriate antibiotic treatment within 1 day of culture positivity (4). The clinical impact is likely to be greatest for the more virulent organisms (such as Staphylococcus aureus, Listeria monocytogenes, and Vibrio vulnificus) and for organisms with predictable resistance to cephalosporins (Enterobacter spp.) and carbapenems (Stenotrophomonas maltophilia and Flavobacterium meningosepticum) (23–28).
For polymicrobial blood cultures, we demonstrated that the Bruker Biotyper was the only system that generated polymicrobial identifications in 5 out of the 21 mixed-culture specimens (23.8%). According to this observation, at this stage neither system was ready for direct use with polymicrobial blood cultures. When we mixed a blood culture with 2 or more aerobic bacteria, we were able to subculture 1 ml of the blood culture broth on blood agar, incubate this mixture in a CO2 chamber at 35°C for 2 to 3 h, and then pick hazy colonies for MALDI-TOF MS. However, this method worked only on aerobic bacteria since the colonies of most of the anaerobic bacteria in the first 2 to 3 h of incubation were too small for MALDI-TOF MS identification.
In the second phase of the study, we used 92 specimens to evaluate the performance of our modified in-house extraction method against that of the commercial MALDI Sepsityper kit (Bruker Daltonics, Germany). On both MALDI-TOF MS systems, the analytical performance exhibited by the modified in-house method was similar to that of the MALDI Sepsityper kit. In addition, the cost per sample was only $1 for the in-house method versus $15 for the commercial kit. For the routine identification method using the automated microbial system, the reagent cost would go up to $5 per sample (Fig. 1). Also, the processing time of the in-house method was around 2 h, and the in-house method was technically easy to handle.
In this study, we evaluated an in-house blood culture extraction protocol on two MALDI-TOF systems for bacterial identification. In terms of running cost and analytical performance, the application of an in-house method and the Bruker Daltonics Microflex LT Biotyper version 3.0 were shown to be the best options for a diagnostic microbiology setting.
Supplementary Material
Footnotes
Published ahead of print 20 March 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.03259-12.
REFERENCES
- 1. Ho PL, Chan WM, Tsang KW, Wong SS, Young K. 2002. Bacteremia caused by Escherichia coli producing extended-spectrum beta-lactamase: a case-control study of risk factors and outcomes. Scand. J. Infect. Dis. 34:567–573 [DOI] [PubMed] [Google Scholar]
- 2. Ho PL, Que TL, Ng TK, Chiu SS, Yung RW, Tsang KW. 2006. Clinical outcomes of bacteremic pneumococcal infections in an area with high resistance. Eur. J. Clin. Microbiol. Infect. Dis. 25:323–327 [DOI] [PubMed] [Google Scholar]
- 3. Seifert H. 2009. The clinical importance of microbiological findings in the diagnosis and management of bloodstream infections. Clin. Infect. Dis. 48(Suppl 4):S238–S245 [DOI] [PubMed] [Google Scholar]
- 4. Vlek AL, Bonten MJ, Boel CH. 2012. Direct matrix-assisted laser desorption ionization time-of-flight mass spectrometry improves appropriateness of antibiotic treatment of bacteremia. PLoS One 7:e32589 doi:10.1371/journal.pone.0032589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Versalovic J, Carroll KC, Funke G, Jorfensen JH, Landry ML, Warnock DW. (ed). 2011. Manual of clinical microbiology, 10th ed ASM Press, Washington, DC [Google Scholar]
- 6. Peters RP, Savelkoul PH, Simoons-Smit AM, Danner SA, Vandenbroucke-Grauls CM, van Agtmael MA. 2006. Faster identification of pathogens in positive blood cultures by fluorescence in situ hybridization in routine practice. J. Clin. Microbiol. 44:119–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Karumaa S, Karpanoja P, Sarkkinen H. 2012. PCR identification of bacteria in blood culture does not fit the daily workflow of a routine microbiology laboratory. J. Clin. Microbiol. 50:1031–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tan KE, Ellis BC, Lee R, Stamper PD, Zhang SX, Carroll KC. 2012. Prospective evaluation of a matrix-assisted laser desorption ionization–time of flight mass spectrometry system in a hospital clinical microbiology laboratory for identification of bacteria and yeasts: a bench-by-bench study for assessing the impact on time to identification and cost-effectiveness. J. Clin. Microbiol. 50:3301–3308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Klein S, Zimmermann S, Kohler C, Mischnik A, Alle W, Bode KA. 2012. Integration of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry in blood culture diagnostics: a fast and effective approach. J. Med. Microbiol. 61:323–331 [DOI] [PubMed] [Google Scholar]
- 10. Spanu T, Posteraro B, Fiori B, D'Inzeo T, Campoli S, Ruggeri A, Tumbarello M, Canu G, Trecarichi EM, Parisi G, Tronci M, Sanguinetti M, Fadda G. 2012. Direct maldi-tof mass spectrometry assay of blood culture broths for rapid identification of Candida species causing bloodstream infections: an observational study in two large microbiology laboratories. J. Clin. Microbiol. 50:176–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stevenson LG, Drake SK, Murray PR. 2010. Rapid identification of bacteria in positive blood culture broths by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 48:444–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wuppenhorst N, Consoir C, Lorch D, Schneider C. 2012. Direct identification of bacteria from charcoal-containing blood culture bottles using matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry. Eur. J. Clin. Microbiol. Infect. Dis. 31:2843–2850 [DOI] [PubMed] [Google Scholar]
- 13. Yan Y, He Y, Maier T, Quinn C, Shi G, Li H, Stratton CW, Kostrzewa M, Tang YW. 2011. Improved identification of yeast species directly from positive blood culture media by combining Sepsityper specimen processing and Microflex analysis with the matrix-assisted laser desorption ionization Biotyper system. J. Clin. Microbiol. 49:2528–2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kok J, Thomas LC, Olma T, Chen SC, Iredell JR. 2011. Identification of bacteria in blood culture broths using matrix-assisted laser desorption-ionization Sepsityper and time of flight mass spectrometry. PLoS One 6:e23285 doi:10.1371/journal.pone.0023285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Janda JM, Abbott SL. 2007. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J. Clin. Microbiol. 45:2761–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Woo PC, Teng JL, Yeung JM, Tse H, Lau SK, Yuen KY. 2011. Automated identification of medically important bacteria by 16S rRNA gene sequencing using a novel comprehensive database, 16SpathDB. J. Clin. Microbiol. 49:1799–1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Martiny D, Dediste A, Vandenberg O. 2012. Comparison of an in-house method and the commercial Sepsityper kit for bacterial identification directly from positive blood culture broths by matrix-assisted laser desorption-ionisation time-of-flight mass spectrometry. Eur. J. Clin. Microbiol. Infect. Dis. 31:2269–2281 [DOI] [PubMed] [Google Scholar]
- 18. Bizzini A, Greub G. 2010. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry, a revolution in clinical microbial identification. Clin. Microbiol. Infect. 16:1614–1619 [DOI] [PubMed] [Google Scholar]
- 19. Mellmann A, Cloud J, Maier T, Keckevoet U, Ramminger I, Iwen P, Dunn J, Hall G, Wilson D, Lasala P, Kostrzewa M, Harmsen D. 2008. Evaluation of matrix-assisted laser desorption ionization–time-of-flight mass spectrometry in comparison to 16S rRNA gene sequencing for species identification of nonfermenting bacteria. J. Clin. Microbiol. 46:1946–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Saffert RT, Cunningham SA, Ihde SM, Jobe KE, Mandrekar J, Patel R. 2011. Comparison of Bruker Biotyper matrix-assisted laser desorption ionization–time of flight mass spectrometer to BD Phoenix automated microbiology system for identification of Gram-negative bacilli. J. Clin. Microbiol. 49:887–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Martiny D, Busson L, Wybo I, El Haj RA, Dediste A, Vandenberg O. 2012. Comparison of the Microflex LT and Vitek MS systems for routine identification of bacteria by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 50:1313–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Seng P, Drancourt M, Gouriet F, La Scola B, Fournier PE, Rolain JM, Raoult D. 2009. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Infect. Dis. 49:543–551 [DOI] [PubMed] [Google Scholar]
- 23. Ho PL, Johnson DR, Yue AW, Tsang DN, Que TL, Beall B, Kaplan EL. 2003. Epidemiologic analysis of invasive and noninvasive group a streptococcal isolates in Hong Kong. J. Clin. Microbiol. 41:937–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ho PL, Shek RH, Chow KH, Duan RS, Mak GC, Lai EL, Yam WC, Tsang KW, Lai WM. 2005. Detection and characterization of extended-spectrum beta-lactamases among bloodstream isolates of Enterobacter spp. in Hong Kong, 2000-2002. J. Antimicrob. Chemother. 55:326–332 [DOI] [PubMed] [Google Scholar]
- 25. Kang CI, Kim SH, Park WB, Lee KD, Kim HB, Kim EC, Oh MD, Choe KW. 2005. Bloodstream infections caused by antibiotic-resistant Gram-negative bacilli: risk factors for mortality and impact of inappropriate initial antimicrobial therapy on outcome. Antimicrob. Agents Chemother. 49:760–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kim SH, Park WB, Lee CS, Kang CI, Bang JW, Kim HB, Kim NJ, Kim EC, Oh MD, Choe KW. 2006. Outcome of inappropriate empirical antibiotic therapy in patients with Staphylococcus aureus bacteraemia: analytical strategy using propensity scores. Clin. Microbiol. Infect. 12:13–21 [DOI] [PubMed] [Google Scholar]
- 27. Lee CC, Lee CH, Chuang MC, Hong MY, Hsu HC, Ko WC. 2012. Impact of inappropriate empirical antibiotic therapy on outcome of bacteremic adults visiting the ED. Am. J. Emerg. Med. 30:1447–1456 [DOI] [PubMed] [Google Scholar]
- 28. Tang WM, Ho PL, Fung KK, Yuen KY, Leong JC. 2001. Necrotising fasciitis of a limb. J. Bone Joint Surg. Brit. 83:709–714 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.