Abstract
A singleplex PCR assay using a single primer pair targeting the putative sugar transporter gene was developed here to distinguish Cryptococcus neoformans var. grubii, Cryptococcus neoformans var. neoformans, and Cryptococcus gattii according to the distinct size of the amplicon. The interspecies and intravarietal hybrids were also characterized on the basis of distinct combined profiles of amplicons. This PCR assay is a rapid, simple, and reliable approach suitable for laboratory diagnoses and large-scale epidemiologic studies.
TEXT
The Cryptococcus neoformans/Cryptococcus gattii species complex is the causative agent of cryptococcosis and has been classified into two sibling species, namely, C. neoformans and C. gattii. C. neoformans is an opportunistic pathogen and causes infections mainly in immunocompromised individuals. This species contains two varieties, Cryptococcus neoformans var. grubii and C. neoformans var. neoformans, which correspond to serotypes A and D, respectively. Contrary to C. neoformans, C. gattii is a primary pathogen and often causes infections in immunocompetent individuals. Recent surveys indicated that this pathogen is more widely distributed than previously thought, especially since the C. gattii outbreak occurred in the Pacific Northwest (1). Phylogenetic analyses and molecular typing studies have identified eight major genotypes within the C. neoformans/C. gattii species complex (2). The relationship of the major genotypes to species, varieties, and serotypes are as follows: VNI and VNII, C. neoformans var. grubii, serotype A; VNIII, AD hybrid, serotype AD; VNIV, C. neoformans var. neoformans, serotype D; and VGI, VGII, VGIII, and VGIV, C. gattii, serotypes B and C (2, 3). Additionally, a geographically restricted genotype, VNB, belonging to C. neoformans var. grubii from Botswana has been reported (4).
Of note, differences in epidemiology, pathogenicity, biology, clinical features, and drug susceptibility have been reported to be associated with species or varieties in the C. neoformans/C. gattii species complex (2, 5, 6). Besides the intravarietal AD hybrid, a few novel interspecies hybrid isolates between C. neoformans and C. gattii have been characterized, and specific pathogenic attributes may be related to the strains (7). Thus, a rapid method for identification of the species, varieties, and hybrids within the pathogen will play an important role in laboratory diagnosis and epidemiologic investigations.
Several molecular approaches, such as multiplex PCR, PCR-restriction fragment length polymorphism (RFLP) analysis, hyperbranched rolling circle amplification (HRCA), loop-mediated isothermal DNA amplification, and matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS), have been developed to identify species, varieties, and hybrids (2, 8–12). Overall, these methods, while reliable, usually require expensive instruments or complicated methods. Singleplex PCR was reported to differentiate the closely related species of Candida spp. according to the distinct sizes of amplicons, which are based mainly on intron loss or differences in intron size of the group I intron or the nuclear pre-mRNA intron (13–15). Introns are widely used as phylogenetic markers in evolutionary analysis. It is reported that intron loss significantly predominates over gain in the evolution of the Cryptococcus clade, and short introns (∼60 bp) were found to predominate in the genome of the C. neoformans/C. gattii species complex (16, 17). However, molecular typing based on difference of introns for this clinically important pathogen has never been reported up to now. Here, the intron loss and intron size differences were analyzed for differentiation of the species complex.
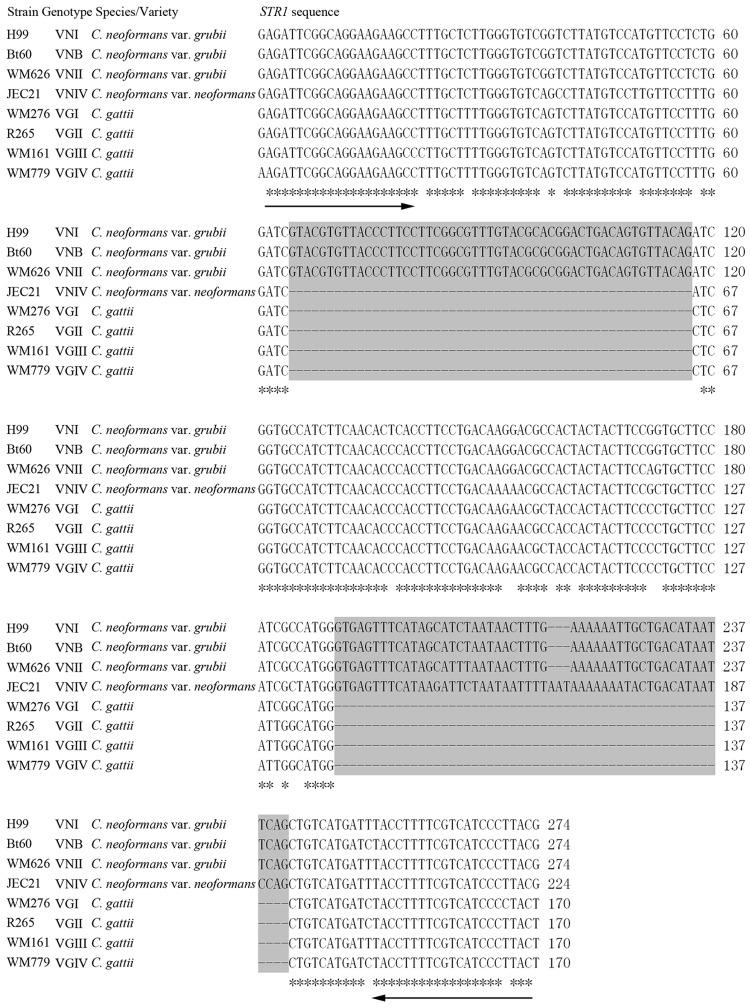
The genomic sequence alignment tool LAGAN (18) was used to analyze the intron loss and intron size differences among members of the species complex. The genome sequence and gene structure of C. neoformans var. grubii strain H99 and C. gattii strain R265 (available at http://www.broadinstitute.org/), and C. neoformans var. neoformans strain JEC21 and C. gattii strain WM276 (available at http://www.ncbi.nlm.nih.gov/genome/) were aligned and analyzed by this tool. Similarly, loss of short introns rather than intron size differences within longer introns between the species or varieties was detected in our study. Ultimately, two adjacent introns within the putative sugar transporter (STR1) gene were selected due to their different types of intron loss among the species and varieties. Sequence alignments of orthologs among the major genotypes were conducted using ClustalW2 and demonstrated that differences in size were the result of intron loss (Fig. 1).
Fig 1.
Multiple sequence alignment of fragments of the STR1 gene orthologs in type strains. Primers STR1F and STR1R are underlined. Two adjacent introns within the gene fragment are marked by a gray background. Gaps representing intron loss are indicated by dashes.
Primers targeted to the STR1 gene, STR1F (5′ GAGATTCGGCAGGAAGAAGC 3′) and STR1R (5′ CGTAAGGGATGACGAAAAGGTA 3′), were designed based on consensus nucleotide sequences of the exon region of the reference strains H99 (VNI), WM626 (VNII), Bt60 (VNB), JEC21 (VNIV), WM276 (VGI), R265 (VGII), WM161 (VGIII), WM779 (VGIV). PCR was performed in a final volume of 50 μl containing 50 ng DNA, 1× PCR buffer with 1.5 mM MgCl2, 0.2 mM (each) dATP, dCTP, dGTP, and dTTP, a 0.2 μM concentration of each primer, and 1.5 U of Taq polymerase. PCR was conducted in a Bio-Rad thermal cycler at 94°C for 5 min for initial denaturation, followed by 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s with a final extension step at 72°C for 6 min. PCR products were separated on a 1.6% (wt/vol) agarose gel at 100 V for 1 h.
This PCR assay was applied to our strain collection for identification. Genomic DNA was prepared from each strain as described previously (19). A total of 255 isolates belonging to the C. neoformans/C. gattii species complex were reference strains and those characterized by using conventional methods and PCR fingerprinting and PCR-RFLP analysis of the GEF1 gene, as previously reported (2, 9). The strains included C. neoformans var. grubii (n = 166), C. neoformans var. neoformans (n = 14), C. gattii (n = 56), AD hybrid (n = 17), AB hybrid (n = 1), and BD hybrid (n = 1). Another 42 pathogenic yeast isolates, which were either reference strains or strains that had been characterized by sequencing of the internal transcribed spacer (ITS) region as previously described (20), were chosen to detect the specificity of the primer pair of STR1F-STR1R. Detailed information regarding the strains used here is presented in Table S1 in the supplemental material. In addition, amplified PCR products of the three hybrid strains WM628 (AD), CBS10496 (AB), and CBS10488 (BD) were cloned and sequenced to determine their hybrid identities. The sequences obtained in this study and those from GenBank were used for phylogenetic analysis performed by using MEGA version 4.0 software (available at http://www.megasoftware.net/). A dendrogram was produced by neighbor-joining analysis using sequences alignment with the Kimura 2-parameter method. Introns were excluded from the phylogenetic analysis. Statistical support for each clade was assessed using bootstrap analysis with 500 replicates (Fig. 2).
Fig 2.
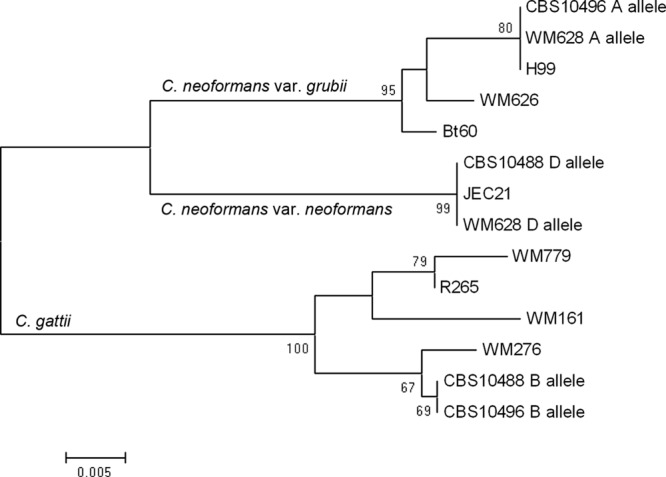
Neighbor-joining tree for type strains, including hybrid strains obtained by analysis of 170-bp fragments of the STR1 gene. Bootstrap values (500 replicates) are indicated for the main branches. Intron is excluded from the phylogenetic analysis.
No amplicon was produced when PCR was performed with the other pathogenic yeasts tested. As expected, the singleplex PCR amplified one fragment for C. neoformans var. grubii (274 bp), C. neoformans var. neoformans (224 bp), and C. gattii (170 bp) and two fragments for the AD hybrid (274 and 224 bp), AB hybrid (274 bp and 170 bp) and BD hybrid (224 and 170 bp) (Fig. 3). All amplicon sizes were confirmed by sequencing of the corresponding representative strains. By PCR analysis, 253 of 255 (99.2%) isolates of the C. neoformans/C. gattii species complex, including the two novel AB and BD hybrids, were correctly assigned to the relevant species, varieties, or hybrids. Nevertheless, 2 of 17 (11.8%) AD hybrid strains (IUM92-6198 and CBS132) produced only one amplicon corresponding to C. neoformans var. neoformans and C. neoformans var. grubii, respectively, by the PCR analysis, which was in correspondence with other reports using single or a few target genes (3, 10, 11). This result may be due to genomic instability in AD hybrid strains with loss of heterozygosity for the allele of STR1 gene, which may be resolved by molecular tools designed for use at the whole-genome level, such as amplified fragment length polymorphism (AFLP) and PCR fingerprinting (2, 7, 19). Phylogenetic analysis revealed that strains representing the three hybrid types had two distinct alleles clustered with the corresponding species, varieties, and genotypes.
Fig 3.
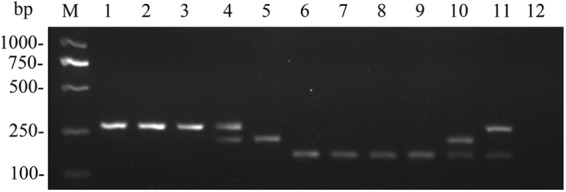
Agarose gel electrophoresis of PCR products. Species, variety and major genotype are depicted in brackets. Lanes: M, DL2000 ladder; 1, WM148 (C. neoformans var. grubii VNI); 2, Bt60 (C. neoformans var. grubii VNB); 3, WM626 (C. neoformans var. grubii VNII); 4, WM628 (C. neoformans AD hybrid VNIII); 5, WM629 (C. neoformans var. neoformans VNIV); 6, WM179 (C. gattii VGI); 7, WM178 (C. gattii VGII); 8, WM161 (C. gattii VGIII); 9, WM779 (C. gattii VGIV); 10, CBS10488 (C. neoformans×C. gattii BD hybrid); 11, CBS10496 (C. neoformans × C. gattii AB hybrid); 12, negative control.
Our study demonstrates that differences in intron loss in protein-coding genes may be used as a reliable genetic marker for molecular typing of pathogenic fungi. Thus, we report here a simple PCR assay with a single primer pair, which performed well in discrimination between members of the C. neoformans/C. gattii species complex. The simple method and the rapidity and reliability of this technique make it a useful tool for laboratory diagnoses and large-scale epidemiologic studies.
Nucleotide sequence accession numbers.
The sequences of the STR1 gene fragment of reference strains Bt60, WM626, WM161, and WM779 were deposited under the accession numbers KC429573 to KC429576.
Supplementary Material
ACKNOWLEDGMENTS
We thank Joseph Heitman, John R. Perfect, Anastasia P. Litvintseva, Wiley A. Schell, and Anna Floyd (Duke University Medical Center, Durham, NC); Wieland Meyer and Dee Carter (University of Sydney, Sydney, Australia); C. De Vroey (Prince Leopold Institut of Tropical Medicine, Belgium); Maria Anna Viviani (Istituto di Igiene e Medicina Preventiva, Milan, Italy); Kyung J. Kwon-Chung (National Institutes of Health, Bethesda, MD); James W. Kronstad (University of British Columbia, Vancouver, BC, Canada); Teun Boekhout and Ferry Hagen (Centraalbureau voor Schimmelcultures–Fungal Biodiversity Centre, Utrecht, the Netherlands); Jianping Xu (McMaster University, Hamilton, Canada); James Swezey (ARS Culture Collection, United States); Orazio Romeo (University of Messina, Italy); and Claudete R. Paula (University of São Paulo, São Paulo, Brazil) for the strains.
This work was supported by grants from the National Natural Science Foundation of China (no. 31000549) and Shanghai Key Laboratory of Molecular Medical Mycology Fund (no. 20110001).
Footnotes
Published ahead of print 27 March 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.00064-13.
REFERENCES
- 1. MacDougall L, Kidd SE, Galanis E, Mak S, Leslie MJ, Cieslak PR, Kronstad JW, Morshed MG, Bartlett KH. 2007. Spread of Cryptococcus gattii in British Columbia, Canada, and detection in the Pacific Northwest, U. S. A. Emerg. Infect. Dis. 13:42–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Meyer W, Castaneda A, Jackson S, Huynh M, Castaneda E. 2003. Molecular typing of IberoAmerican Cryptococcus neoformans isolates. Emerg. Infect. Dis. 9:189–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Feng X, Yao Z, Ren D, Liao W, Wu J. 2008. Genotype and mating type analysis of Cryptococcus neoformans and Cryptococcus gattii isolates from China that mainly originated from non-HIV-infected patients. FEMS Yeast Res. 8:930–938 [DOI] [PubMed] [Google Scholar]
- 4. Litvintseva AP, Thakur R, Vilgalys R, Mitchell TG. 2006. Multilocus sequence typing reveals three genetic subpopulations of Cryptococcus neoformans var. grubii (serotype A), including a unique population in Botswana. Genetics 172:2223–2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Speed B, Dunt D. 1995. Clinical and host differences between infections with the two varieties of Cryptococcus neoformans. Clin. Infect. Dis. 21:28–34 [DOI] [PubMed] [Google Scholar]
- 6. Trilles L, Fernandez-Torres B, Lazera MS, Wanke B, Guarro J. 2004. In vitro antifungal susceptibility of Cryptococcus gattii. J. Clin. Microbiol. 42:4815–4817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bovers M, Hagen F, Kuramae EE, Hoogveld HL, Dromer F, St-Germain G, Boekhout T. 2008. AIDS patient death caused by novel Cryptococcus neoformans × C. gattii hybrid. Emerg. Infect. Dis. 14:1105–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Leal AL, Faganello J, Bassanesi MC, Vainstein MH. 2008. Cryptococcus species identification by multiplex PCR. Med. Mycol. 46:377–383 [DOI] [PubMed] [Google Scholar]
- 9. Feng X, Yao Z, Ren D, Liao W. 2008. Simultaneous identification of molecular and mating types within the Cryptococcus species complex by PCR-RFLP analysis. J. Med. Microbiol. 57:1481–1490 [DOI] [PubMed] [Google Scholar]
- 10. Kaocharoen S, Wang B, Tsui KM, Trilles L, Kong F, Meyer W. 2008. Hyperbranched rolling circle amplification as a rapid and sensitive method for species identification within the Cryptococcus species complex. Electrophoresis 29:3183–3191 [DOI] [PubMed] [Google Scholar]
- 11. Lucas S, Da LMM, Flores O, Meyer W, Spencer-Martins I, Inacio J. 2010. Differentiation of Cryptococcus neoformans varieties and Cryptococcus gattii using CAP59-based loop-mediated isothermal DNA amplification. Clin. Microbiol. Infect. 16:711–714 [DOI] [PubMed] [Google Scholar]
- 12. McTaggart LR, Lei E, Richardson SE, Hoang L, Fothergill A, Zhang SX. 2011. Rapid identification of Cryptococcus neoformans and Cryptococcus gattii by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 49:3050–3053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Romeo O, Racco C, Criseo G. 2006. Amplification of the hyphal wall protein 1 gene to distinguish Candida albicans from Candida dubliniensis. J. Clin. Microbiol. 44:2590–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tamura M, Watanabe K, Mikami Y, Yazawa K, Nishimura K. 2001. Molecular characterization of new clinical isolates of Candida albicans and C. dubliniensis in Japan: analysis reveals a new genotype of C. albicans with group I intron. J. Clin. Microbiol. 39:4309–4315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Enache-Angoulvant A, Guitard J, Grenouillet F, Martin T, Durrens P, Fairhead C, Hennequin C. 2011. Rapid discrimination between Candida glabrata, Candida nivariensis, and Candida bracarensis by use of a singleplex PCR. J. Clin. Microbiol. 49:3375–3379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sharpton TJ, Neafsey DE, Galagan JE, Taylor JW. 2008. Mechanisms of intron gain and loss in Cryptococcus. Genome Biol. 9:R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hughes SS, Buckley CO, Neafsey DE. 2008. Complex selection on intron size in Cryptococcus neoformans. Mol. Biol. Evol. 25:247–253 [DOI] [PubMed] [Google Scholar]
- 18. Brudno M, Do CB, Cooper GM, Kim MF, Davydov E, Green ED, Sidow A, Batzoglou S. 2003. LAGAN and Multi-LAGAN: efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 13:721–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lin X, Litvintseva AP, Nielsen K, Patel S, Floyd A, Mitchell TG, Heitman J. 2007. alpha AD alpha hybrids of Cryptococcus neoformans: evidence of same-sex mating in nature and hybrid fitness. PLoS Genet. 3:1975–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W. 2012. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. U. S. A. 109:6241–6246 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.