Abstract
Infection from fluoroquinolone-resistant Enterobacteriaceae is an increasing health problem worldwide. In the present study, we developed a pyrosequencing-based high-throughput method for analyzing the nucleotide sequence of the quinolone resistance-determining regions (QRDRs) of gyrA and parC. By using this method, we successfully determined the QRDR sequences of 139 out of 140 clinical Escherichia coli isolates, 28% of which were nonsusceptible to ciprofloxacin. Sequence results obtained by the pyrosequencing method were in complete agreement with those obtained by the Sanger method. All fluoroquinolone-resistant isolates (n = 35; 25%) contained mutations leading to three or four amino acid substitutions in the QRDRs. In contrast, all isolates lacking a mutation in the QRDR (n = 81; 57%) were susceptible to ciprofloxacin, levofloxacin, and nalidixic acid. The qnr determinants, namely, the qnrA, qnrB, and qnrS genes, were not detected in the isolates, and the aac(6′)-Ib-cr gene was detected in 2 (1.4%) of the isolates. Multilocus sequence typing of 34 randomly selected isolates revealed that sequence type 131 (ST131) (n = 7; 20%) is the most prevalent lineage and is significantly resistant to quinolones (P < 0.01). The genetic background of quinolone-susceptible isolates seemed more diverse, and interestingly, neighboring STs of ST131 in the phylogenetic tree were all susceptible to ciprofloxacin. In conclusion, our investigation reveals the relationship between fluoroquinolone resistance caused by mutations of QRDRs and the population structure of clinical extraintestinal E. coli isolates. This high-throughput method for analyzing QRDR mutations by pyrosequencing is a powerful tool for epidemiological studies of fluoroquinolone resistance in bacteria.
INTRODUCTION
Fluoroquinolones are powerful broad-spectrum antimicrobial agents used for the treatment of a wide variety of community-acquired and nosocomial infections (1). However, following the introduction of fluoroquinolones in the 1980s, the population of fluoroquinolone-resistant bacteria has increased markedly over the years (2, 3). The Asia-Pacific region in particular is an area where fluoroquinolone resistance is endemic among clinical isolates of Escherichia coli (4, 5).
As quinolones inhibit bacterial DNA gyrase and topoisomerase IV, amino acid substitutions in the quinolone resistance-determining regions (QRDRs) of these enzymes might lead to changes that decrease the binding of quinolone (6). In fact, the accumulation of mutations in the QRDRs of gyrA and parC is recognized as the most common and important mechanism of quinolone resistance in Enterobacteriaceae (7). However, the relationship of quinolone resistance with the genetic background has not been clearly established.
The Clinical and Laboratory Standards Institute (CLSI) continues to reevaluate the breakpoints of fluoroquinolones for Enterobacteriaceae (8). Following previous reports (3, 9, 10), the CLSI recently released a new lowered breakpoint of ciprofloxacin for Salmonella enterica serotype Typhi and extraintestinal Salmonella spp. (susceptible at ≤0.06 μg/ml). Furthermore, revising the breakpoint of levofloxacin for this species is under discussion by a CLSI working group. Possible changes in the breakpoints for other Enterobacteriaceae were also discussed by this working group. Sometimes, however, the clinical response to an infection caused by an isolate considered “susceptible” according to the present CLSI breakpoint appears suboptimal (11, 12). As our current understanding of the genetic basis for the development of quinolone resistance is not sufficient, further understanding of the relationship between quinolone susceptibility and QRDR mutations in Enterobacteriaceae should provide some fundamental information for making rational decisions.
Pyrosequencing is a real-time sequence analysis method based on the detection of pyrophosphate that is released during the synthesis of DNA (13). Because pyrosequencing is less labor- and time-intensive than the conventional Sanger method for nucleotide sequence analysis, this method has already been used successfully to identify the resistance-conferring genes of several bacterial species (14–17). This method appears to be especially suitable as a tool for identifying “hot spot” mutations of QRDRs, namely, at amino acid positions 83 and 87 in GyrA and at positions 80 and 84 in ParC of E. coli. To the best of our knowledge, pyrosequencing has yet to be used as a tool for analyzing the QRDRs in E. coli.
In the present study, we used pyrosequencing to determine the QRDR mutations of gyrA and parC in clinical E. coli isolates obtained from a university hospital in Japan, a location where quinolone-resistant E. coli is endemic and plasmid-mediated quinolone resistance (PMQR) seems to be relatively uncommon (18). Additionally, we used multilocus sequence typing (MLST) for genotyping analysis and interpreted the results to delineate an evolutionary pathway of quinolone resistance.
MATERIALS AND METHODS
Bacterial strains.
We investigated 140 nonrepetitive consecutive clinical E. coli isolates, including 20 isolates from blood, 59 from sputum, and 61 from urine samples, all isolated in 2009 in the Toho University Omori Medical Center, which is a 972-bed university hospital located in Tokyo, Japan. All of the isolates were identified as E. coli by using the Vitek 2 system (bioMérieux, Lyon, France). E. coli ATCC 25922 was used as a control for MIC measurements.
Antimicrobial susceptibility test, detection of extended-spectrum beta-lactamase producers, and effect of efflux pump inhibitor.
MICs were determined by a broth microdilution method according to the CLSI testing standards (19). The MICs of cefepime, cefpodoxime, cefpodoxime-clavulanic acid, and meropenem were measured with frozen plates for antimicrobial susceptibility testing (Eiken Chemical Co., Ltd., Tokyo, Japan). The MICs of levofloxacin (Daiichi Sankyo Co., Ltd., Tokyo, Japan), ciprofloxacin (MP Biomedicals, LLC, Santa Ana, CA), and nalidixic acid (Sigma, St. Louis, MO) were measured separately. The results were interpreted according to the criteria recommended by the CLSI (8).
An extended-spectrum beta-lactamase (ESBL) producer was determined following the CLSI recommendations. Thus, when the MIC of cefpodoxime for an isolate was ≥4 μg/ml and clavulanic acid reduced the MIC by ≥2-fold, the isolate was considered to be an ESBL producer.
To assess the contribution of efflux pumps to quinolone resistance, MICs of ciprofloxacin and nalidixic acid were compared in the absence and presence of 20 μg/ml of Phe-Arg-β-naphthylamide (Sigma), an inhibitor of efflux pumps (20).
DNA extraction, PCR amplification, and Sanger DNA sequencing.
DNA templates for PCR amplification were prepared by resuspending fresh bacterial colonies in 500 μl of Tris-EDTA, heating the resuspended cells for 15 min at 100°C, and then centrifuging the mixture for 5 min at 10,000 rpm. Unless mentioned otherwise, all PCRs were performed using Ex Taq (TaKaRa Bio, Shiga, Japan). Nucleotide sequences of PCR-amplified DNAs of the region corresponding to amino acid positions from 81 to 87 in GyrA and from 78 to 84 in ParC were determined by the conventional Sanger method to confirm the reliability of the pyrosequencing results. Primers used in PCR amplification and nucleotide sequencing are listed in Table 1.
Table 1.
Primers used in the study
| Primer | Primer sequence (5′–3′) | PCR annealing temp (°C) | Reference |
|---|---|---|---|
| gyrA QRDR for pyrosequencing | |||
| gyrA-Pyro-F | CCTCTGGATTATGCGATGTCGGTCAT | 54 | This study |
| gyrA-Pyro-Rbiotina | TCAGCCCTTCAATGCTGATGTCT | This study | |
| gyrA-Pyro-S | TAATCGGTAAATACCATCCCCA | This study | |
| parC QRDR for pyrosequencing | |||
| parC-Pyro-F | ACTACTCCATGTACGTCATCATGGAC | 54 | This study |
| parC-Pyro-Rbiotina | AGCCACTTCGCGCAGGTTAT | This study | |
| parC-Pyro-S | TGGGTAAATACCATCCGCAC | This study | |
| parC-Pyro-S-alt | TGGGTAAATACCATCCGCAT | This study | |
| QRDR for Sanger sequencing | |||
| gyrA-QRDR-F | TCTGGATTATGCGATGTCGGTCAT | 54 | This study |
| gyrA-QRDR-R | TCAGCCCTTCAATGCTGATGTCT | This study | |
| parC-QRDR-F | ACTACTCCATGTACGTCATCATGGAC | 54 | This study |
| parC-QRDR-R | CGCCACTTCGCGCAGGTTAT | This study | |
| gyrB-QRDR-F | GCTGAGCGAATACCTGCTGG | 54 | This study |
| gyrB-QRDR-R | TCGGTCATGATGATGATGCTGTGAT | This study | |
| parE-QRDR-F | GCGGAAGATATCTGGGATCGCT | 54 | This study |
| parE-QRDR-R | CTGGCTCAGATCGTCGCTGT | This study | |
| qnr multiplex PCR detection | |||
| QnrAm-F | AGAGGATTTCTCACGCCAGG | 56 | 24 |
| QnrAm-R | TGCCAGGCACAGATCTTGAC | 24 | |
| QnrBm-F | GGMATHGAAATTCGCCACTG | 56 | 24 |
| QnrBm-R | TTTGCYGYYCGCCAGTCGAA | 24 | |
| QnrSm-F | GCAAGTTCATTGAACAGGGT | 56 | 24 |
| QnrSm-R | TCTAAACCGTCGAGTTCGGCG | 24 | |
| MLST | |||
| E. coli MLST adkF | ATTCTGCTTGGCGCTCCGGG | 58 | 21 |
| E. coli MLST adkR | CCGTCAACTTTCGCGTATTT | 21 | |
| E. coli MLST fumCF | TCACAGGTCGCCAGCGCTTC | 58 | 21 |
| E. coli MLST fumCR | GTACGCAGCGAAAAAGATTC | 21 | |
| E. coli MLST gyrBF | TCGGCGACACGGATGACGGC | 65 | 21 |
| E. coli MLST gyrBR | ATCAGGCCTTCACGCGCATC | 21 | |
| E. coli MLST icdF | ATGGAAAGTAAAGTAGTTGTTCCGGCACA | 58 | 21 |
| E. coli MLST icdR | GGACGCAGCAGGATCTGTT | 21 | |
| E. coli MLST mdhF | CCAGGCGCTTGCACTACTGTTAA | 58 | 21 |
| E. coli MLST mdhR | GCGATATCTTTCTTCAGCGTATC | 21 | |
| E. coli MLST purAF | CGCGCTGATGAAAGAGATGA | 65 | 21 |
| E. coli MLST purAR | CATACGGTAAGCCACGCAGA | 21 | |
| E. coli MLST recAF | CGGCAAACTCAACGTTCC | 58 | 21 |
| E. coli MLST recAR | CTGACGCTGCAGGTGAT | 21 | |
| aac(6′)-Ib-cr: T304C/A for pyrosequencing | |||
| T304C-Fbiotina | GGAGAGCCGATTGGGTATG | 58 | 16 |
| T304C-R | TAACTGGTCTATTCCGCGTACTC | 16 | |
| T304C-So | CGGTTTCTTCTTCCCAC | 16 | |
| aac(6′)-Ib-cr: G535T for pyrosequencing | |||
| G535T-Fbiotina | CGATCCGATGCTACGAGAAA | 58 | 16 |
| G535T-R | CATGTACACGGCTGGACCA | 16 | |
| G535T-So | TGTACACGGCTGGAC | 16 |
5′-biotinylated.
Pyrosequencing of QRDR.
The PCR primers used in the pyrosequencing method were designed using the PyroMark assay design software 2.0 (Qiagen, Hilden, Germany) on the basis of sequence information available for the QRDRs of the gyrA and parC genes (GenBank accession number U00096), with manual modifications to avoid possible template loops, self-annealing duplexes, or alternate annealing sites, and the sequences of these primers are listed in Table 1. The forward primers, labeled with “-F,” and the reverse primers, labeled with “-R,” were covalently coupled to biotin at the 5′ end to obtain a pyrosequencing template from the PCR product.
The PCR for pyrosequencing was performed in a reaction volume of 25 μl with amplification primers and a PyroMark PCR kit (Qiagen) and following the manufacturer's instructions. PCR began with a 15-min hot-start step at 95°C and was followed by 45 cycles of amplification consisting of 30 s at 95°C, 1 min at 54°C, and 30 s at 72°C.
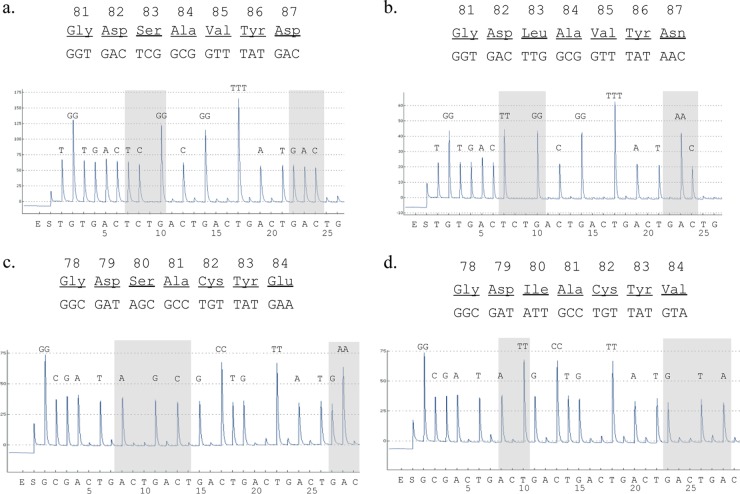
Pyrosequencing was carried out using a PyroMark Q24 system (Qiagen) based on a 4-enzyme solution system, according to the manufacturer's instructions. Briefly, PCR products were captured and separated by using streptavidin-Sepharose beads (GE health care, Pittsburgh, PA), and the resulting single-stranded DNA was used as a template for pyrosequencing. A sequence primer, labeled with “-S” at the end in Table 1, was annealed to a single-stranded PCR product. Single nucleotides were dispensed individually in a predetermined order to the reaction mixture: TGTGACT6(CTGA) for gyrA and GCGA8(CTGA) for parC (shown on the x axis in Fig. 1). The analysis range was the QRDRs of GyrA and ParC, corresponding to the amino acid positions from 81 to 87 in GyrA and from 78 to 84 in ParC. The signal strength, reflecting light emitted enzymatically from pyrophosphate, is proportional to the number of nucleotides incorporated in a single nucleotide flow. The pyrograms were analyzed using PyroMark Q24 software (Qiagen).
Fig 1.
Analysis of QRDRs in gyrA (a and b) and parC (c and d) by pyrosequencing in quinolone-susceptible (a and c) and quinolone-resistant (b and d) isolates. The shaded regions show polymorphisms at positions 83 and 87 in GyrA (a and b) and at positions 80 and 84 in ParC (c and d). Single nucleotides were dispensed individually in a predetermined order as shown on the x axis of the trace. The signal strength, reflecting light emitted enzymatically from pyrophosphate, is proportional to the number of nucleotides incorporated in a single nucleotide flow. The additions of enzymes and substrates to the reaction mix are indicated by E and S, respectively.
MLST.
Sequence types (STs) of 34 randomly selected E. coli isolates were determined according to the MLST scheme (21). These E. coli isolates included 14 from urine, 15 from sputum, and 5 from blood samples. Clustering of each sequence type (ST) was determined using the program eBURST version 3, and single-locus variants were used to define the clonal complexes (CCs) (22). Phylogenetic analysis was performed using the maximum-likelihood method by MEGA5 (23).
Identification of PMQR genes.
qnrA, qnrB, and qnrS genes were detected using a multiplex PCR method and the primers listed in Table 1 (24). The aac(6′)-Ib-cr gene, which differed from aac(6′)-Ib by two single-nucleotide polymorphisms, namely, T304C/A and G535T, was detected by using a pyrosequencing method and the primers listed in Table 1 (16).
Statistical analysis.
A two-sided Fisher's exact test was used for analyzing categorical data. Differences at P values of <0.05 were considered statistically significant.
RESULTS
Prevalence of fluoroquinolone-resistant and ESBL-producing clinical E. coli isolates.
Table 2 summarizes the distribution of the MICs of each antimicrobial agent tested against the 140 E. coli isolates used in this study. According to the breakpoints reported by the CLSI in 2012, 35 (25%) of the isolates were resistant (MIC ≥ 4 μg/ml) and 40 (28%) of the isolates were nonsusceptible (MIC ≥ 2 μg/ml) to ciprofloxacin (8). Significantly, more isolates from sputum samples were resistant to ciprofloxacin than those from urine and blood samples (42% from sputum versus 13% and 10% from urine and blood, respectively; P < 0.01). Fifty-nine isolates (42.1%) were resistant to nalidixic acid (MIC ≥ 32 μg/ml).
Table 2.
Drug susceptibility of clinical Escherichia coli isolates (n = 140) used in the study
| Antimicrobial | No. of isolates at the indicated MIC (μg/ml)a |
Range (μg/ml) | MIC50 (μg/ml) | MIC90 (μg/ml) | Resistant isolatesb |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ≤0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | >128 | No. | Rate (%) | ||||
| Ciprofloxacin | 92 | 8 | 5 | 3 | 5 | 15 | 7 | 5h | ≤0.5 to >64 | ≤0.5 | 32 | 35 | 25 | ||
| Levofloxacin | 99 | 5 | 1 | 5 | 30e | ≤1 to >8 | ≤1 | >8 | 35 | 25 | |||||
| Nalidixic acid | 3 | 26 | 39 | 9 | 4 | 1 | 3 | 2 | 53 | 0.5 to >128 | 4 | >128 | 59 | 42 | |
| Cefepime | 122c | 3 | 3 | 1 | 5 | 6g | ≤0.5 to >32 | ≤1 | 4 | 11 | 7.9 | ||||
| Cefpodoxime | 109 | 9 | 1 | 1 | 2 | 18f | ≤0.5 to >16 | ≤0.5 | >16 | 20 | 14 | ||||
| Cefpodoxime-clavulanic acid | 126 | 7 | 5 | 2 | ≤0.5 to 8 | ≤0.5 | 0.5 | NAi | NAi | ||||||
| Meropenem | 140d | ≤2 | ≤2 | ≤2 | 0 | 0 | |||||||||
Vertical lines are shown between resistant and nonresistant isolates.
Breakpoint adopted from CLSI recommendations (8).
This number represents isolates for which the MIC was ≤1 μg/ml.
This number represents isolates for which the MIC was ≤2 μg/ml.
This number represents isolates for which the MIC was >8 μg/ml.
This number represents isolates for which the MIC was >16 μg/ml.
This number represents isolates for which the MIC was >32 μg/ml.
This number represents isolates for which the MIC was >64 μg/ml.
NA, not applicable.
Eighteen isolates (12.8%), which included both fluoroquinolone-resistant and -susceptible isolates, were identified as ESBL producers. Among them, 12 (66%) were from sputum samples, while 1 and 5 were from blood and urine samples, respectively. All 11 cefepime-resistant isolates were ESBL producers. None of the isolates was meropenem resistant.
Accumulations of mutations in QRDRs of gyrA and parC in relation to quinolone resistance.
Using the sequencing primers gyrA-Pyro-S and parC-Pyro-S (Table 1), we obtained sequence information on the QRDRs of the gyrA and parC genes, respectively, in 139 E. coli isolates by pyrosequencing (Fig. 1). While the gyrA-Pyro-S primer also yielded sequence information on the gyrA QRDR of the remaining isolate, the parC-Pyro-S primer failed to provide any sequence information on the parC QRDR due to a silent mutation in the primer region. However, another sequencing primer with one base change (parC-Pyro-S-alt) (Table 1) yielded sequence information on the parC QRDR of the remaining isolate. The sequencing results by the pyrosequencing method were found to be in complete agreement with those obtained by the Sanger sequencing used as a conventional methodology throughout this study.
These results were concordant with the established theory that the accumulation of QRDR mutations increases the level of resistance to fluoroquinolones (25). The MIC levels of ciprofloxacin and levofloxacin demonstrated a trimodal distribution (Table 3; also see Table S1 in the supplemental material). Eighty-one out of the 140 E. coli isolates (58%) had no mutations in the QRDRs of gyrA and parC, and the MIC50 of ciprofloxacin for these isolates was ≤0.008 μg/ml. Pyrosequencing identified nine different combinations of amino acid substitutions in the QRDRs. The numbers of isolates with a single mutation (found only in the gyrA QRDR) and double mutations (one in gyrA and one in parC QRDR) were 22 and 2, respectively, and the MIC50 of ciprofloxacin for these isolates was 0.5 μg/ml. Thirty-five isolates (25%) contained either three mutations (4 isolates containing 2 amino acid substitutions in GyrA QRDR and 1 amino acid substitution in ParC QRDR) or four mutations (31 isolates containing 2 amino acid substitutions in GyrA QRDR and 2 amino acid substitutions in ParC QRDR), and the MIC50 of ciprofloxacin for these isolates was 32 μg/ml.
Table 3.
Relationship between quinolone resistance-determining region sequences and ciprofloxacin susceptibility of clinical Escherichia coli isolates
| Amino acid substitution |
No. of isolates | No. of isolates at a ciprofloxacin MIC (μg/ml) ofa: |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GyrA |
ParC |
≤0.008 | 0.015 | 0.03 | 0.06 | 0.13 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | >64 | |||
| Ser83 | Asp87 | Ser80 | Glu84 | ||||||||||||||||
| 81 | 51b (3) | 27 | 3 | ||||||||||||||||
| Leu | 20 | 2 | 2 (1) | 5 | 8 (1) | 3 | |||||||||||||
| Gly | 1 | 1 | |||||||||||||||||
| Asn | 1 | 1 | |||||||||||||||||
| Leu | Ile | 1 | 1 | ||||||||||||||||
| Leu | Gly | 1 | 1 | ||||||||||||||||
| Leu | Asn | Ile | 4 | 1 | 1 | 1 | 1 (1) | ||||||||||||
| Leu | Asn | Ile | Gly | 1 | 1 | ||||||||||||||
| Leu | Asn | Ile | Arg | 3 | 3 (3) | ||||||||||||||
| Leu | Asn | Ile | Val | 27 | 2 | 4 (1) | 15 (7) | 5b (1) | 1 | ||||||||||
| Total | 140 | 51 | 27 | 3 | 0 | 2 | 4 | 5 | 8 | 5 | 0 | 3 | 5 | 15 | 7 | 5 | |||
Numbers of ESBL-producing isolates are shown in parentheses.
One of the isolates harbored the aac(6′)-Ib-cr gene.
None of the isolates, however, revealed any other mutation in the QRDRs of gyrB or parE, or in the remaining regions of gyrA and parC, as determined by the Sanger sequencing method. The qnr determinants, namely, the qnrA, qnrB, and qnrS genes, were not detected in these isolates. However, the aac(6′)-Ib-cr gene was detected in 2 (1.4%) of the isolates, one of which harbored no QRDR mutation and the other which harbored four QRDR mutations. The efflux pump inhibitor Phe-Arg-β-naphthylamide affected the susceptibility of nalidixic acid but not that of ciprofloxacin or levofloxacin (data not shown).
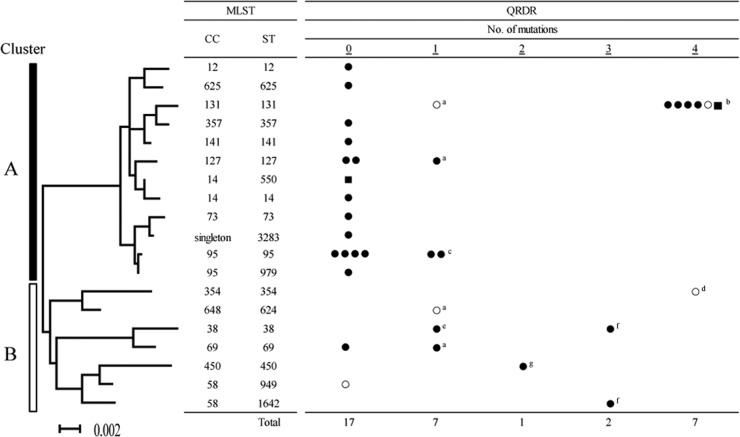
Diverse lineage of quinolone-susceptible isolates and predominance of ST131 among the quinolone-resistant isolates.
The 34 clinical E. coli isolates, selected in an unbiased manner, were assigned to 19 STs by MLST (Fig. 2). According to the results of phylogenetic analysis, we divided the isolates into two groups, clusters A and B. Quinolone-susceptible isolates seemed more diverse than quinolone-resistant isolates: seventeen isolates with no QRDR mutations belonged to 13 STs, 10 of which consisted of a single isolate. Similarly, seven isolates with a single QRDR mutation belonged to 6 different STs. In contrast, seven isolates with four QRDR mutations belonged to only 2 STs.
Fig 2.
Molecular phylogenetic analysis of 34 clinical E. coli isolates, including 9 ciprofloxacin-resistant isolates, according to the MLST scheme. The evolutionary history was estimated using the maximum-likelihood method, and the isolates were divided into one of two groups, namely, clusters A and B, according to the results of phylogenetic analysis. Isolates in cluster A, except ST131, had significantly fewer mutations in the QRDRs than the isolates in cluster B (P < 0.05). Underlined numbers indicate the numbers of mutations in the QRDRs. Closed squares, isolates harboring aac(6′)-Ib-cr; open circles, ESBL-producing isolates; closed circles, isolates not harboring aac(6′)-Ib-cr or producing ESBL. ST, sequence type; CC, clonal complex. a, S83L in GyrA; b, all isolates with S83L and D87N in GyrA and S80I and E84V in ParC; c, one isolate with S83L in GyrA and one with D87N in GyrA; d, S83L and D87N in GyrA and S80I and E84R in ParC; e, D87G in GyrA; f, S83L and D87N in GyrA and S80I in ParC; g, S83L in GyrA and S80I in ParC.
ST131 was outstanding with respect to both frequency and resistance. ST131 was the most common ST (n = 7; 21% of isolates) followed by ST95 (n = 6; 18%), ST127 (n = 3; 9%), ST38 (n = 2; 6%), and ST69 (n = 2; 6%). In addition, most of the highly quinolone-resistant isolates with four QRDR mutations (6/7) were the ST131 isolates, and ST131 isolates were more likely to exhibit ciprofloxacin resistance than were the isolates of other genotypes (P < 0.01). Moreover, the ST131 isolates included two ESBL producers and one isolate with the aac(6′)-Ib-cr gene. Interestingly, isolates of cluster A, except ST131, had significantly fewer QRDR mutations than isolates of cluster B, including ST38 and ST354 (P < 0.05) (Fig. 2). In an epidemiological investigation, seven ST131 isolates were recovered from patients in different wards, implying that nosocomial transmission was unlikely.
Besides the number of mutations in QRDR, the mutation content enabled us to further discriminate the isolates. For example, two ST95 isolates harbored different single mutations in the QRDRs. Similarly, two ST38 isolates had a mutation at position 87 in GyrA but did not share the same amino acid residue at that position. Although ST354 shared three QRDR mutations with ST131, the amino acid at position 84 in ParC differed from the amino acid at that position in these two STs.
DISCUSSION
We have successfully developed a method focusing on detecting hot-spot mutations of QRDRs in gyrA and parC using pyrosequencing as a real-time labor-saving sequencing technology. Although the read length is shorter than the read length for the Sanger method of DNA sequencing, the sequence information obtained by pyrosequencing is easy to interpret, and the target region in this study seems sufficient for estimating the level of quinolone resistance. As the measurement of the MICs of drugs with low concentration levels poses practical challenges for clinical microbiology laboratories, the availability of a uniform platform for genotyping seems to provide a considerable advantage.
Our data, obtained from 140 clinical E. coli isolates from a university hospital in Tokyo, Japan, in 2009, show a robust relationship between the quinolone resistance and the number of mutations in the QRDRs of gyrA and parC, as was reported previously (25). The prevalence of ciprofloxacin resistance (25%) was, however, higher than that previously reported in a Japanese study, where only 15% of E. coli isolates from urine samples were resistant to fluoroquinolone (26). Besides the difference of the study time, this discrepancy might be due to the fact that in this study, we also included sputum samples which were previously reported to contain more fluoroquinolone-resistant isolates than all other samples, including blood and urine (27, 28). In addition, the roles of PMQRs and efflux pumps in fluoroquinolone resistance were less apparent in our study than in previously reported studies (18, 29, 30), although the PCR detection in this study might not have covered all of the currently known PMQR determinants.
In the present study, at the currently recommended breakpoint concentration of fluoroquinolone (ciprofloxacin MIC ≥ 4 μg/ml), we were able to identify E. coli isolates harboring three or four QRDR mutations. In addition, a lower concentration of ciprofloxacin (MIC ≤ 0.06 μg/ml), which was the recommended breakpoint for S. Typhi and Salmonella spp., was useful for the detection of E. coli isolates with no QRDR mutations or with a single QRDR mutation.
Among the previous phylogenetic studies on clinical E. coli isolates, most applied different criteria in terms of isolate selection, drug resistance, and severity of infections (31, 32). This might lead to a biased representation of certain STs. Therefore, for MLST analysis in the present study, we analyzed the population structure of clinical E. coli strains by randomly selecting 34 isolates, a group which included both quinolone-susceptible and -resistant isolates.
ST131 has been reported as a globally disseminated clone with multidrug resistance, including resistance to cephalosporins and fluoroquinolones (33). Interestingly, fluoroquinolone resistance and ESBL production are often epidemiologically related in Enterobacteriaceae (4). In our data, most of the ST131 isolates showed high-level resistance to fluoroquinolones, including two ESBL producers and one isolate positive for the aac(6′)-Ib-cr gene. The other ESBL-producing and aac(6′)-Ib-cr gene-harboring isolates were distributed among 4 different STs, suggesting the role of horizontal gene transfer in these cases. Although the reason why the resistance of ST131 to fluoroquinolones was more extreme than that of the other isolates in the same cluster A is not clear, further investigations, such as a comparison of the whole genomes of a wider range of isolates, might reveal the actual pathway for the development of quinolone resistance in the clinical E. coli population.
According to the genotyping data, we hypothesize two possible pathways to the emergence of quinolone resistance among clinical E. coli isolates. The clonal spread of resistant strains such as ST131 might play a role, in part at least, although the epidemiological information did not provide robust evidence for nosocomial transmission. Alternatively, fluoroquinolone resistance might arise independently through target mutations of various strains. In fact, different gyrA mutations were observed in the same ST background, such as ST95 or ST38 isolates (Fig. 2). Further study is necessary to elucidate the true contribution of these two pathways.
In conclusion, in the present study, MLST analysis of these isolates provided their likely phylogeny with respect to drug resistance to fluoroquinolones. In addition, we successfully used pyrosequencing as a tool and unraveled the QRDR mutation pattern among the clinical E. coli isolates obtained from a university hospital in Japan. This methodology thus represents a powerful tool for epidemiological studies of fluoroquinolone-resistant isolates. Future investigations should be undertaken to examine wider ranges of isolates to elucidate their population structures and to understand the relationship between molecular evolution and the emergence of resistant clones in bacteria.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Toho project research grant no. 23-8 of the Toho University School of Medicine.
We thank Ryoko Shimada from Qiagen, Japan, for her technical support and for helpful discussions on pyrosequencing. We are also grateful to Kiyoshi Sugihara for his technical support.
Footnotes
Published ahead of print 20 March 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.03049-12.
REFERENCES
- 1. Hooper DC. 2000. Mechanisms of action and resistance of older and newer fluoroquinolones. Clin. Infect. Dis. 31(Suppl 2):S24–S28 [DOI] [PubMed] [Google Scholar]
- 2. Threlfall EJ, Cheasty T, Graham A, Rowe B. 1997. High-level resistance to ciprofloxacin in Escherichia coli. Lancet 349:403. [DOI] [PubMed] [Google Scholar]
- 3. Threlfall EJ, Ward LR, Skinner JA, Smith HR, Lacey S. 1999. Ciprofloxacin-resistant Salmonella typhi and treatment failure. Lancet 353:1590–1591 [DOI] [PubMed] [Google Scholar]
- 4. Ko WC, Hsueh PR. 2009. Increasing extended-spectrum beta-lactamase production and quinolone resistance among Gram-negative bacilli causing intra-abdominal infections in the Asia/Pacific region: data from the Smart Study 2002–2006. J. Infect. 59:95–103 [DOI] [PubMed] [Google Scholar]
- 5. Yamaguchi K, Ohno A, Ishii Y, Tateda K, Iwata M, Kanda M, Akizawa K, Shimizu C, Kon S, Nakamura K, Matsuda K, Tominaga M, Nakagawa T, Sugita A, Ito T, Kato J, Suwabe A, Yamahata K, Kawamura C, Tashiro H, Horiuchi H, Katayama Y, Kondou S, Misawa S, Murata M, Kobayashi Y, Okamoto H, Yamazaki K, Okada M, Haruki K, Kanno H, Aihara M, Maesaki S, Hashikita G, Miyajima E, Sumitomo M, Saito T, Yamane N, Kawashima C, Akiyama T, Ieiri T, Yamamoto Y, Okamoto Y, Okabe H, Moro K, Shigeta M, Yoshida H, Yamashita M, Hida Y, Takubo T, et al. 2009. In vitro susceptibilities to levofloxacin and various antibacterial agents of 12,919 clinical isolates obtained from 72 centers in 2007. Jpn. J. Antibiot. 62:346–370 (In Japanese.) [PubMed] [Google Scholar]
- 6. Hawkey PM. 2003. Mechanisms of quinolone action and microbial response. J. Antimicrob. Chemother. 51(Suppl 1):29–35 [DOI] [PubMed] [Google Scholar]
- 7. Hopkins KL, Davies RH, Threlfall EJ. 2005. Mechanisms of quinolone resistance in Escherichia coli and Salmonella: recent developments. Int. J. Antimicrob. Agents 25:358–373 [DOI] [PubMed] [Google Scholar]
- 8. Clinical and Laboratory Standards Institute 2012. Performance standards for antimicrobial susceptibility testing; 22nd informational supplement. CLSI M100-S22. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 9. Molbak K, Baggesen DL, Aarestrup FM, Ebbesen JM, Engberg J, Frydendahl K, Gerner-Smidt P, Petersen AM, Wegener HC. 1999. An outbreak of multidrug-resistant, quinolone-resistant Salmonella enterica serotype typhimurium DT104. N. Engl. J. Med. 341:1420–1425 [DOI] [PubMed] [Google Scholar]
- 10. Wain J, Hoa NT, Chinh NT, Vinh H, Everett MJ, Diep TS, Day NP, Solomon T, White NJ, Piddock LJ, Parry CM. 1997. Quinolone-resistant Salmonella typhi in Viet Nam: molecular basis of resistance and clinical response to treatment. Clin. Infect. Dis. 25:1404–1410 [DOI] [PubMed] [Google Scholar]
- 11. Defife R, Scheetz MH, Feinglass JM, Postelnick MJ, Scarsi KK. 2009. Effect of differences in MIC values on clinical outcomes in patients with bloodstream infections caused by gram-negative organisms treated with levofloxacin. Antimicrob. Agents Chemother. 53:1074–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zelenitsky SA, Ariano RE. 2010. Support for higher ciprofloxacin AUC24/MIC targets in treating Enterobacteriaceae bloodstream infection. J. Antimicrob. Chemother. 65:1725–1732 [DOI] [PubMed] [Google Scholar]
- 13. Ronaghi M. 2001. Pyrosequencing sheds light on DNA sequencing. Genome Res. 11:3–11 [DOI] [PubMed] [Google Scholar]
- 14. Gharizadeh B, Akhras M, Unemo M, Wretlind B, Nyren P, Pourmand N. 2005. Detection of gyrA mutations associated with ciprofloxacin resistance in Neisseria gonorrhoeae by rapid and reliable pre-programmed short DNA sequencing. Int. J. Antimicrob. Agents 26:486–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gorgani N, Ahlbrand S, Patterson A, Pourmand N. 2009. Detection of point mutations associated with antibiotic resistance in Pseudomonas aeruginosa. Int. J. Antimicrob. Agents 34:414–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guillard T, Duval V, Moret H, Brasme L, Vernet-Garnier V, de Champs C. 2010. Rapid detection of aac(6′)-Ib-cr quinolone resistance gene by pyrosequencing. J. Clin. Microbiol. 48:286–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hopkins KL, Arnold C, Threlfall EJ. 2007. Rapid detection of gyrA and parC mutations in quinolone-resistant Salmonella enterica using pyrosequencing technology. J. Microbiol. Methods 68:163–171 [DOI] [PubMed] [Google Scholar]
- 18. Ode T, Saito R, Kumita W, Sato K, Okugawa S, Moriya K, Koike K, Okamura N. 2009. Analysis of plasmid-mediated multidrug resistance in Escherichia coli and Klebsiella oxytoca isolates from clinical specimens in Japan. Int. J. Antimicrob. Agents 34:347–350 [DOI] [PubMed] [Google Scholar]
- 19. Clinical and Laboratory Standards Institute 2009. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically— approved standard, 8th ed. CLSI M07-A8. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 20. Saenz Y, Ruiz J, Zarazaga M, Teixido M, Torres C, Vila J. 2004. Effect of the efflux pump inhibitor Phe-Arg-β-naphthylamide on the MIC values of the quinolones, tetracycline and chloramphenicol, in Escherichia coli isolates of different origin. J. Antimicrob. Chemother. 53:544–545 [DOI] [PubMed] [Google Scholar]
- 21. Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MC, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60:1136–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cattoir V, Poirel L, Rotimi V, Soussy CJ, Nordmann P. 2007. Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. J. Antimicrob. Chemother. 60:394–397 [DOI] [PubMed] [Google Scholar]
- 25. Morgan-Linnell SK, Zechiedrich L. 2007. Contributions of the combined effects of topoisomerase mutations toward fluoroquinolone resistance in Escherichia coli. Antimicrob. Agents Chemother. 51:4205–4208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Takahashi A, Muratani T, Yasuda M, Takahashi S, Monden K, Ishikawa K, Kiyota H, Arakawa S, Matsumoto T, Shima H, Kurazono H, Yamamoto S. 2009. Genetic profiles of fluoroquinolone-resistant Escherichia coli isolates obtained from patients with cystitis: phylogeny, virulence factors, PAIusp subtypes, and mutation patterns. J. Clin. Microbiol. 47:791–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Boyd LB, Atmar RL, Randall GL, Hamill RJ, Steffen D, Zechiedrich L. 2008. Increased fluoroquinolone resistance with time in Escherichia coli from >17,000 patients at a large county hospital as a function of culture site, age, sex, and location. BMC Infect. Dis. 8:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Murillo Llanes J, Varon J, Velarde Felix JS, Gonzalez-Ibarra FP. 2012. Antimicrobial resistance of Escherichia coli in Mexico: how serious is the problem? J. Infect. Dev. Ctries. 6:126–131 [DOI] [PubMed] [Google Scholar]
- 29. Matsumura Y, Yamamoto M, Higuchi T, Komori T, Tsuboi F, Hayashi A, Sugimoto Y, Hotta G, Matsushima A, Nagao M, Takakura S, Ichiyama S. 2012. Prevalence of plasmid-mediated AmpC beta-lactamase-producing Escherichia coli and spread of the ST131 clone among extended-spectrum beta-lactamase-producing E. coli in Japan. Int. J. Antimicrob. Agents 40:158–162 [DOI] [PubMed] [Google Scholar]
- 30. Saga T, Akasaka T, Takase H, Tanaka M, Sato K, Kaku M. 2007. First detection of the plasmid-mediated quinolone resistance determinant qnrA in Enterobacteriaceae clinical isolates in Japan. Int. J. Antimicrob. Agents 29:738–739 [DOI] [PubMed] [Google Scholar]
- 31. Gibreel TM, Dodgson AR, Cheesbrough J, Fox AJ, Bolton FJ, Upton M. 2011. Population structure, virulence potential and antibiotic susceptibility of uropathogenic Escherichia coli from Northwest England. J. Antimicrob. Chemother. 67:346–356 [DOI] [PubMed] [Google Scholar]
- 32. Rogers BA, Sidjabat HE, Paterson DL. 2011. Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J. Antimicrob. Chemother. 66:1–14 [DOI] [PubMed] [Google Scholar]
- 33. Johnson JR, Menard M, Johnston B, Kuskowski MA, Nichol K, Zhanel GG. 2009. Epidemic clonal groups of Escherichia coli as a cause of antimicrobial-resistant urinary tract infections in Canada, 2002 to 2004. Antimicrob. Agents Chemother. 53:2733–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.