Abstract
We assessed the performance of the Ceeram and Altona assays, the first two commercially available hepatitis E virus (HEV) RNA assays, using serial dilutions of 4 HEV-positive reference samples (genotypes 3a, 3c, 3e, and 3f). Both assays provided good analytical sensitivity and high reproducibility for detecting genotype 3 HEV RNA.
TEXT
Hepatitis E virus (HEV) is becoming increasingly important in industrialized countries (1, 2). Four main genotypes and several subtypes have been identified (3). Most infections in industrialized countries are due to zoonotic transmission, often of genotype 3 (HEV3); subtypes 3a and 3b are frequent in North America and Japan, and subtypes 3c, 3e, and 3f are more prevalent in Europe (3–5). HEV3 is an emerging concern for immunocompromised patients, as it can lead to chronic infection and cirrhosis (6–12).
As evaluations of anti-HEV IgM assays revealed appreciable variations in their performances (13, 14), it is important to diagnose HEV infections by detecting HEV RNA. Several in-house reverse transcription-PCRs (RT-PCRs) were recently evaluated, and their sensitivities were shown to differ greatly (15). We have also shown that genotype 3 diversity can influence the quantification of HEV RNA (16).
We have therefore assessed the performance of two newly available commercial HEV RNA assays, the Ceeram and Altona assays. We tested their ability to detect HEV RNA, particularly those subtypes of HEV3 that are most prevalent in industrialized countries.
We used the HEV RNA WHO international standard (WHO/BS/2011.2175), which is a HEV genotype 3a strain quantified at 250,000 IU/ml. Samples of HEV genotypes 3c, 3e, and 3f were collected from patients in France (17, 18). Each sample was diluted in HEV-negative plasma and quantified with a validated in-house RT-PCR protocol using a transcribed RNA as the quantification standard (1 IU/ml corresponds to 1.25 copies/ml) (16). HEV RNA was extracted from blood samples (140 μl) with the RNeasy minikit according to the manufacturer's instructions (Qiagen, Courtaboeuf, France). The HepatitisE@ceeramTools kit by Ceeram (La Chapelle sur Erdre, France) and the RealStar HEV RT-PCR kit, version 1.0, by Altona Diagnostics (Eurobio, Courtaboeuf, France) were used with the Light Cycler 480 instrument (Roche Diagnostics, Meylan, France) according to the manufacturers' instructions. The threshold cycle (CT) value of each sample was determined.
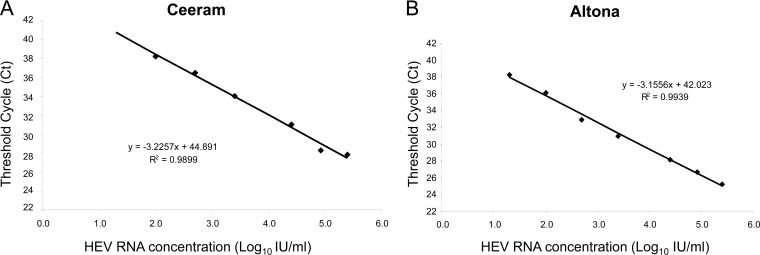
The linearity of both assays was assessed with serial dilutions of the WHO HEV reference standard. The Ceeram assay was linear from 100 to 250,000 IU/ml, and the Altona assay was linear from 20 to 250,000 IU/ml (Fig. 1). The standard curves gave amplification efficiencies of 2.08 for the Ceeram assay and 2.3 for the Altona assay. Reproducibility was estimated from the CT values for each dilution. The mean standard deviations were 0.7 CT (range: 0.4 to 1.6 CT) for the Ceeram RT-PCR and 0.4 CT (range: 0.1 to 1.4 CT) for the Altona RT-PCR.
Fig 1.
Standard curves generated using dilutions of the WHO reference standard. (A) Ceeram assay. (B) Altona assay. Data are means of three replicates for each standard dilution.
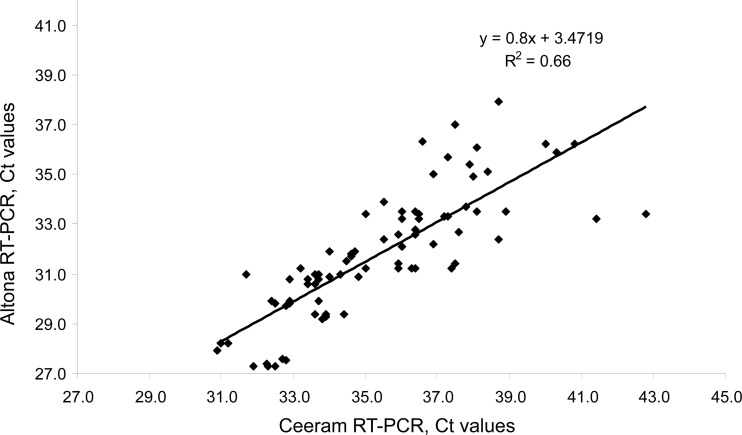
We assayed samples of strains 3a, 3c, 3e, and 3f to assess analytical sensitivity. The dilution concentrations were 2,500, 500, 100, and 20 IU/ml, and 6 replicates of each were assayed (Table 1). Both assays detected all the 2,500-IU/ml and 500-IU/ml samples. The Ceeram assay detected 21/24 100-IU/ml samples, while the Altona assay detected all 24. The Ceeram assay detected 8/24 of the lowest-concentration (20-IU/ml) samples, while the Altona assay detected 18/24 (P = 0.008) (Table 1). The poorer sensitivity of the Ceeram assay at this low HEV RNA concentration was independent of the particular genotype 3 subtype. The Ceeram assay gave higher CT values than the Altona assay (P = 0.003). The mean difference in the CT values was 3.4 CT. The differences were 2.9 CT for subtype 3a, 2.8 CT for genotype 3c, 5.2 CT for genotype 3e, and 2.7 CT for subtype 3f. The mean CT difference was greater for subtype 3e than for the other subtypes (P < 0.01). The Ceeram and the Altona RT-PCR results were correlated (ρ = 0.88, P < 0.001) (Fig. 2).
Table 1.
Data obtained with the Ceeram and Altona assays for the 4 reference strains
| Subtype | Sample concn (IU/ml) | Result for: |
CT difference (Ceeram − Altona) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ceeram |
Altona |
|||||||||
| No. of samples detected/no. tested |
CT |
No. of samples detected/no. tested |
CT |
|||||||
| Mean | SD | CVa (%) | Mean | SD | CV (%) | |||||
| 3a | 2,500 | 6/6 | 34.0 | 0.5 | 1.4 | 6/6 | 30.9 | 0.1 | 0.3 | 3.1 |
| 500 | 6/6 | 36.5 | 0.6 | 1.7 | 6/6 | 32.9 | 0.3 | 0.9 | 3.6 | |
| 100 | 5/6 | 38.2 | 1.1 | 3.0 | 6/6 | 36.0 | 1.4 | 3.8 | 2.1 | |
| 20 | 0/6 | 4/6 | 38.3 | 0.6 | 1.6 | |||||
| 3c | 2,500 | 6/6 | 32.9 | 0.5 | 1.4 | 6/6 | 29.8 | 0.1 | 0.3 | 3.0 |
| 500 | 6/6 | 34.6 | 0.3 | 0.9 | 6/6 | 31.7 | 0.3 | 0.9 | 2.9 | |
| 100 | 6/6 | 37.5 | 1.3 | 3.5 | 6/6 | 34.3 | 1.8 | 5.2 | 3.2 | |
| 20 | 1/6 | 38.4 | 6/6 | 36.3 | 1.0 | 2.7 | 2.1 | |||
| 3e | 2,500 | 6/6 | 32.4 | 0.3 | 1.0 | 6/6 | 27.4 | 0.1 | 0.5 | 5.0 |
| 500 | 6/6 | 33.9 | 0.3 | 0.8 | 6/6 | 29.4 | 0.1 | 0.3 | 4.6 | |
| 100 | 6/6 | 36.6 | 0.7 | 2.0 | 6/6 | 31.3 | 0.1 | 0.3 | 5.3 | |
| 20 | 5/6 | 39.2 | 3.0 | 7.7 | 6/6 | 33.3 | 0.5 | 1.4 | 5.9 | |
| 3f | 2,500 | 6/6 | 33 | 0.7 | 2.1 | 6/6 | 30.8 | 0.2 | 0.8 | 2.2 |
| 500 | 6/6 | 36.3 | 0.6 | 1.7 | 6/6 | 32.8 | 0.7 | 2 | 3.5 | |
| 100 | 4/6 | 37.9 | 1.6 | 4.3 | 6/6 | 35.9 | 0.7 | 2.1 | 2.0 | |
| 20 | 2/6 | 39.4 | 2/6 | 36.3 | 3.1 | |||||
| All samples | 77/96 | 0.9b | 2.42c | 90/96 | 0.5b | 1.5c | 3.4d | |||
CV, coefficient of variation.
Mean standard deviation.
Mean coefficient of variation.
Mean CT difference.
Fig 2.
Correlation between CT values obtained with the Ceeram and Altona assays for the 4 reference strains.
A recent evaluation of home brew HEV RNA assays using 10-fold serial dilutions of HEV reference samples (3a, 3b, 3f, and 4c) found an enormous difference in their sensitivities (100-fold to 1,000-fold) (15). We therefore estimated the analytical sensitivities of commercial assays by testing serial dilutions of genotype 3a, 3c, 3e, and 3f reference strains. Sensitivities were between 100 and 500 IU/ml for the Ceeram assay and between 20 and 100 IU/ml for the Altona assay. The Ceeram RT-PCR was less sensitive than the Altona RT-PCR when the HEV RNA concentration was low (20 IU/ml). Moreover, the mean difference between the CT values (3.4 CT) indicated that the Altona RT-PCR may be more sensitive than the Ceeram. But this sensitivity difference could be linked to differences in the recommended RNA input volumes: 5 μl for the Ceeram assay and 25 μl for the Altona assay.
The recent evaluation of 2 HEV RNA assays has demonstrated that it is essential to use an RT-PCR protocol based on open reading frame 3 (ORF3) in order to accurately quantify all the HEV genotype 3 subtypes, as this region is better conserved than most others (16). The 2 commercial assays tested include primers and a probe targeting this region. However, the mean difference between the CT values for genotype 3e may indicate that the Ceeram assay is less sensitive for this subtype. No data are yet available for HEV genotypes 1, 2, and 4, but these assays might be suitable for detecting them also, as ORF3 is highly conserved across HEV genotypes. These two points should be confirmed in further studies.
As transfusion-transmitted HEV3 infections have been reported in industrialized countries (19–22), sensitive HEV RNA assays may well be useful for screening blood products (23). Several studies have reported detecting HEV RNA in pooled plasma samples from European blood donors (24–27). Tests using the Altona assay found that 1.18% of plasma pools were positive in Germany (24). These two commercial assays now need to be compared for testing plasma pools.
The Ceeram and the Altona assays provide good analytical sensitivity with high reproducibility for detecting genotype 3 HEV RNA. They provide a useful complement to serological methods for detecting HEV infections.
ACKNOWLEDGMENTS
The National Reference Center for Hepatitis E is supported by a grant from the French Public Health authorities.
RT-PCR reagents were kindly provided by Ceeram and Eurobio.
Footnotes
Published ahead of print 20 March 2013
REFERENCES
- 1. Hoofnagle JH, Nelson KE, Purcell RH. 2012. Hepatitis E. N. Engl. J. Med. 367:1237–1244 [DOI] [PubMed] [Google Scholar]
- 2. Kamar N, Bendall R, Legrand-Abravanel F, Xia NS, Ijaz S, Izopet J, Dalton HR. 2012. Hepatitis E. Lancet 379:2477–2488 [DOI] [PubMed] [Google Scholar]
- 3. Lu L, Li C, Hagedorn CH. 2006. Phylogenetic analysis of global hepatitis E virus sequences: genetic diversity, subtypes and zoonosis. Rev. Med. Virol. 16:5–36 [DOI] [PubMed] [Google Scholar]
- 4. Legrand-Abravanel F, Mansuy JM, Dubois M, Kamar N, Peron JM, Rostaing L, Izopet J. 2009. Hepatitis E virus genotype 3 diversity, France. Emerg. Infect. Dis. 15:110–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rutjes SA, Lodder WJ, Lodder-Verschoor F, van den Berg HH, Vennema H, Duizer E, Koopmans M, de Roda Husman AM. 2009. Sources of hepatitis E virus genotype 3 in The Netherlands. Emerg. Infect. Dis. 15:381–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Colson P, Kaba M, Moreau J, Brouqui P. 2009. Hepatitis E in an HIV-infected patient. J. Clin. Virol. 45:269–271 [DOI] [PubMed] [Google Scholar]
- 7. Dalton HR, Bendall RP, Keane FE, Tedder RS, Ijaz S. 2009. Persistent carriage of hepatitis E virus in patients with HIV infection. N. Engl. J. Med. 361:1025–1027 [DOI] [PubMed] [Google Scholar]
- 8. Haagsma EB, van den Berg AP, Porte RJ, Benne CA, Vennema H, Reimerink JH, Koopmans MP. 2008. Chronic hepatitis E virus infection in liver transplant recipients. Liver Transpl. 14:547–553 [DOI] [PubMed] [Google Scholar]
- 9. Kamar N, Selves J, Mansuy JM, Ouezzani L, Peron JM, Guitard J, Cointault O, Esposito L, Abravanel F, Danjoux M, Durand D, Vinel JP, Izopet J, Rostaing L. 2008. Hepatitis E virus and chronic hepatitis in organ-transplant recipients. N. Engl. J. Med. 358:811–817 [DOI] [PubMed] [Google Scholar]
- 10. Ollier L, Tieulie N, Sanderson F, Heudier P, Giordanengo V, Fuzibet JG, Nicand E. 2009. Chronic hepatitis after hepatitis E virus infection in a patient with non-Hodgkin lymphoma taking rituximab. Ann. Intern. Med. 150:430–431 [DOI] [PubMed] [Google Scholar]
- 11. Tavitian S, Peron JM, Huynh A, Mansuy JM, Ysebaert L, Huguet F, Vinel JP, Attal M, Izopet J, Recher C. 2010. Hepatitis E virus excretion can be prolonged in patients with hematological malignancies. J. Clin. Virol. 49:141–144 [DOI] [PubMed] [Google Scholar]
- 12. Kamar N, Mansuy JM, Cointault O, Selves J, Abravanel F, Danjoux M, Otal P, Esposito L, Durand D, Izopet J, Rostaing L. 2008. Hepatitis E virus-related cirrhosis in kidney- and kidney-pancreas-transplant recipients. Am. J. Transplant. 8:1744–1748 [DOI] [PubMed] [Google Scholar]
- 13. Drobeniuc J, Meng J, Reuter G, Greene-Montfort T, Khudyakova N, Dimitrova Z, Kamili S, Teo CG. 2010. Serologic assays specific to immunoglobulin M antibodies against hepatitis E virus: pangenotypic evaluation of performances. Clin. Infect. Dis. 51:e24–e27 [DOI] [PubMed] [Google Scholar]
- 14. Legrand-Abravanel F, Thevenet I, Mansuy JM, Saune K, Vischi F, Peron JM, Kamar N, Rostaing L, Izopet J. 2009. Good performance of immunoglobulin M assays in diagnosing genotype 3 hepatitis E virus infections. Clin. Vaccine Immunol. 16:772–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Baylis SA, Hanschmann KM, Blumel J, Nubling CM. 2011. Standardization of hepatitis E virus (HEV) nucleic acid amplification technique-based assays: an initial study to evaluate a panel of HEV strains and investigate laboratory performance. J. Clin. Microbiol. 49:1234–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Abravanel F, Sandres-Saune K, Lhomme S, Dubois M, Mansuy JM, Izopet J. 2012. Genotype 3 diversity and quantification of hepatitis E virus RNA. J. Clin. Microbiol. 50:897–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Legrand-Abravanel F, Kamar N, Sandres-Saune K, Lhomme S, Mansuy JM, Muscari F, Sallusto F, Rostaing L, Izopet J. 2011. Hepatitis E virus infection without reactivation in solid-organ transplant recipients, France. Emerg. Infect. Dis. 17:30–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mansuy JM, Abravanel F, Miedouge M, Mengelle C, Merviel C, Dubois M, Kamar N, Rostaing L, Alric L, Moreau J, Peron JM, Izopet J. 2009. Acute hepatitis E in south-west France over a 5-year period. J. Clin. Virol. 44:74–77 [DOI] [PubMed] [Google Scholar]
- 19. Colson P, Coze C, Gallian P, Henry M, De Micco P, Tamalet C. 2007. Transfusion-associated hepatitis E, France. Emerg. Infect. Dis. 13:648–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Haim-Boukobza S, Ferey MP, Vetillard AL, Jeblaoui A, Pelissier E, Pelletier G, Teillet L, Roque-Afonso AM. 2012. Transfusion-transmitted hepatitis E in a misleading context of autoimmunity and drug-induced toxicity. J. Hepatol. 57:1374–1378 [DOI] [PubMed] [Google Scholar]
- 21. Boxall E, Herborn A, Kochethu G, Pratt G, Adams D, Ijaz S, Teo CG. 2006. Transfusion-transmitted hepatitis E in a ‘nonhyperendemic’ country. Transfus Med. 16:79–83 [DOI] [PubMed] [Google Scholar]
- 22. Matsubayashi K, Kang JH, Sakata H, Takahashi K, Shindo M, Kato M, Sato S, Kato T, Nishimori H, Tsuji K, Maguchi H, Yoshida J, Maekubo H, Mishiro S, Ikeda H. 2008. A case of transfusion-transmitted hepatitis E caused by blood from a donor infected with hepatitis E virus via zoonotic food-borne route. Transfusion 48:1368–1375 [DOI] [PubMed] [Google Scholar]
- 23. Aggarwal R. 2013. Diagnosis of hepatitis E. Nat. Rev. Gastroenterol. Hepatol. 10:24–33 [DOI] [PubMed] [Google Scholar]
- 24. Corman VM, Drexler JF, Eckerle I, Roth WK, Drosten C, Eis-Hubinger AM. 2013. Zoonotic hepatitis E virus strains in German blood donors. Vox Sang. 104:179–180 [DOI] [PubMed] [Google Scholar]
- 25. Ijaz S, Szypulska R, Tettmar KI, Kitchen A, Tedder RS. 2012. Detection of hepatitis E virus RNA in plasma mini-pools from blood donors in England. Vox Sang. 102:272. [DOI] [PubMed] [Google Scholar]
- 26. Baylis SA, Gartner T, Nick S, Ovemyr J, Blumel J. 2012. Occurrence of hepatitis E virus RNA in plasma donations from Sweden, Germany and the United States. Vox Sang. 103:89–90 [DOI] [PubMed] [Google Scholar]
- 27. Baylis SA, Koc O, Nick S, Blumel J. 2012. Widespread distribution of hepatitis E virus in plasma fractionation pools. Vox Sang. 102:182–183 [DOI] [PubMed] [Google Scholar]