Abstract
A novel commercial chromogenic technique, the βLACTA test (Bio-Rad, Marnes-la-Coquette, France), was evaluated to detect nonsusceptibility to ceftazidime in Pseudomonas aeruginosa isolates. Easily implemented in the routine microbiology laboratory, this rapid test was sensitive (95%) and specific (87%) and presented negative and positive predictive values of 99% and 100%, respectively.
TEXT
Pseudomonas aeruginosa is a nonfermenting Gram-negative bacillus commonly responsible for nosocomial infections that are associated with high mortality and morbidity (1). Moreover, it is known to display a predilection to infect immunocompromised patients and debilitated patients (2). The low permeability of its outer membrane, overexpression of various efflux pumps, and constitutive production of β-lactamases make P. aeruginosa intrinsically resistant to several β-lactams (2). According to most international therapy guidelines, first-line treatment options usually rely on ceftazidime (CAZ), piperacillin-tazobactam, or fluoroquinolones (3). Resistance mechanisms against broad-spectrum β-lactams in P. aeruginosa could result from reduced permeability through porin deficiency, increased active efflux, overproduction of constitutive cephalosporinase, or the acquisition of exogenous β-lactamases hydrolyzing various antipseudomonal β-lactams, including CAZ (4). Rapid identification of CAZ-nonsusceptible P. aeruginosa strains on primary culture may be useful to guide the decision on whether to use CAZ as empirical therapy for P. aeruginosa infections.
The βLACTA test (BLT) (Bio-Rad, Marnes-la-Coquette, France) is a qualitative colorimetric test based on the selective cleavage of a chromogenic substrate, HMRZ-86, which is structurally close to CAZ. HMRZ-86 is not hydrolyzed by narrow-spectrum β-lactamases, due to their poor affinity for this compound. On the other hand, this substrate is hydrolyzed by evolved β-lactamases, such as extended-spectrum-β-lactamases (ESBLs) and metallo-β-lactamases (MBLs), but also by stably derepressed chromosomal AmpC cephalosporinase (5). Hydrolysis of the β-lactam ring modifies the wavelength absorbed by the molecule, shifting the color of the compound from the initial yellow to orange to red to purple, depending on the degree of hydrolysis (6). BLT was primarily designed for the detection of Enterobacteriaceae strains with decreased susceptibility to extended-spectrum cephalosporins. This test appeared to be reliable for the detection of ESBL-producing Enterobacteriaceae (7, 8).
The aim of this study was to evaluate the ability of this newly developed test to rapidly discriminate directly from primary culture colonies between CAZ-susceptible and CAZ-nonsusceptible (intermediately of fully resistant) P. aeruginosa isolates.
A total of 164 P. aeruginosa isolates were tested in this study, including 100 consecutive nonduplicate strains isolated from routine clinical samples and 64 collection strains previously characterized for their β-lactamases at the molecular level by multiplex PCR and sequencing (9, 10). Routine clinical samples included mainly lower respiratory tract specimens (68%), but also urine specimens (10%), blood cultures (3%), and other specimens (19%). β-Lactamase-producing isolates from the collection included ESBLs (n = 22), MBLs (n = 24), producers of various oxacillinases (n = 9) and carbenicillinases (n = 5), and producers of both MBLs and ESBLs (n = 4) and are listed in Table 1. All P. aeruginosa strains were tested for susceptibility to 16 antimicrobials, including CAZ, by the disk diffusion method according to the CLSI recommendations (11). After 18 to 24 h of incubation at 35°C, CAZ inhibition zone diameters were recorded and the BLT was performed according to the manufacturer's instructions on fresh colonies grown on plates containing Trypticase soy agar (TSA) supplemented with 5% sheep blood (Becton, Dickinson, Le Pont de Chaix, France), which served as an antibiogram purity plate. Briefly, one drop each of two reagent solutions was added extemporaneously in a microtube. Isolated colonies picked up with a 1-μl loop were then suspended in the reaction mixture. Test results were recorded after up to 30 min of incubation at room temperature. Any colorimetric change from yellow to orange, red, or purple was considered a positive result, and the absence of a colorimetric change was considered a negative result. BLT was also evaluated under the same conditions for 20 representative strains (including 10 acquired β-lactamase-producing strains, 5 overexpressed β-lactamase-producing strains, and 5 wild-type susceptible strains), each grown on the following agar plates: TSA, MacConkey's agar (Becton, Dickinson, Le Pont de Chaix, France), cystine-lactose-electrolyte-deficient agar (bioMérieux, Marcy l'Etoile, France), and chocolate Haemophilus agar 2 (bioMérieux, Marcy l'Etoile, France).
Table 1.
βLACTA test results and CAZ 30-μg disk inhibition diameters of acquired β-lactamase-producing P. aeruginosa strains
| Resistance mechanism(s) | Resistance determinant(s) | CAZ inhibition zone diam (mm) | BLT result |
|---|---|---|---|
| ESBLs | BEL-1 | 17 | Purple |
| BEL-1 | 12 | Purple | |
| BEL-1 | 21 | Purple | |
| BEL typea | 6 | Purple | |
| BEL typea | 13 | Purple | |
| BEL typea | 14 | Purple | |
| GES-1 | 6 | Orange | |
| GES-5 | 10 | Orange | |
| GES-5 | 10 | Red | |
| GES-18 + CARB-1 | 17 | Orange | |
| GES typea | 13 | Red | |
| PER-1 | 6 | Purple | |
| PER-1 | 6 | Purple | |
| PER typea + OXA G2 | 6 | Purple | |
| PER typea | 6 | Purple | |
| PER typea | 6 | Purple | |
| PER typea | 6 | Purple | |
| VEB-1a | 6 | Purple | |
| VEB-1b | 6 | Purple | |
| VEB-1b | 6 | Purple | |
| VEB-1b + OXA-10 | 6 | Purple | |
| VEB-1 | 6 | Purple | |
| MBLs | IMP-7 | 6 | Purple |
| IMP-7 | 6 | Purple | |
| IMP-13 | 6 | Purple | |
| IMP-13 | 6 | Purple | |
| IMP-13 | 6 | Purple | |
| VIM-2 | 6 | Orange | |
| VIM-2 | 6 | Orange | |
| VIM-2 | 12 | Orange | |
| VIM-2 | 9 | Orange | |
| VIM-2 | 13 | Orange | |
| VIM-2 | 10 | Orange | |
| VIM-4 | 18 | Yellow | |
| VIM-4 | 6 | Orange | |
| VIM-4 | 25 | Orange | |
| VIM-4 | 7 | Red | |
| VIM-4 + OXA-35 | 12 | Orange | |
| VIM typea | 9 | Orange | |
| VIM typea | 12 | Yellow | |
| VIM typea | 8 | Orange | |
| VIM typea | 6 | Yellow | |
| VIM typea | 10 | Orange | |
| VIM typea | 6 | Red | |
| VIM typea | 10 | Orange | |
| VIM typea | 6 | Red | |
| Carbenicillinases | CARB typea | 21 | Orange |
| CARB typea | 25 | Orange | |
| CARB typea | 22 | Orange | |
| CARB typea | 23 | Orange | |
| CARB typea | 25 | Orange | |
| Oxacillinases | OXA-1 | 16 | Orange |
| OXA-2 | 21 | Orange | |
| OXA-2 | 20 | Red | |
| OXA-9 | 6 | Orange | |
| OXA-10 | 25 | Purple | |
| OXA-G10 | 24 | Purple | |
| OXA-18 + OXA-20 | 10 | Purple | |
| OXA-18 + OXA-20 | 6 | Purple | |
| OXA-198 | 27 | Orange | |
| MBLs with ESBLs | VIM-2 + BEL-1 | 6 | Purple |
| VIM-2 + BEL-2 | 8 | Purple | |
| VIM typea + BEL-typea | 6 | Purple | |
| VIM typea + BEL-typea | 6 | Purple |
Unsequenced gene.
The BLT yielded a positive test for 18 out of 19 clinical isolates that were found to be CAZ nonsusceptible by disk diffusion, while results were negative for all 81 CAZ-susceptible strains collected from the routine clinical samples. Eighteen of the 19 CAZ-nonsusceptible strains displayed a β-lactam resistance phenotype typical of chromosomal AmpC cephalosporinase overproduction (high-level resistance to ticarcillin, piperacillin, piperacillin-tazobactam, and ceftazidime and susceptibility to cefepime and aztreonam) associated or not with an outer membrane permeability defect (decreased susceptibility to imipenem). The only CAZ-nonsusceptible strain that was unrecognized by BLT was phenotypically compatible with a high-level active efflux-producing strain (high-level resistance to ticarcillin, piperacillin, ceftazidime, cefepime, aztreonam, and meropenem and intermediate resistance to piperacillin and piperacillin-tazobactam). Multiplex PCR targeting minor ESBL (including VEB, PER, BEL, and GES types), carbapenemase (including VIM, IMP, NDM, OXA-48, and KPC types), extended-spectrum penicillinase (including OXA-10, OXA-18, OXA-20, OXA-1, OXA-30, OXA-2, OXA-9, and OXA-198 types), and carbenicillinase (CARB-1 to -6) bla coding genes (9, 10) remained negative for this strain. Final positive colorimetric changes (orange, red, or purple) were variable and are detailed in Fig. 1. Based on these results, the negative and positive predictive values (NPV and PPV, respectively) of BLT were found to be 99% and 100%, respectively.
Fig 1.
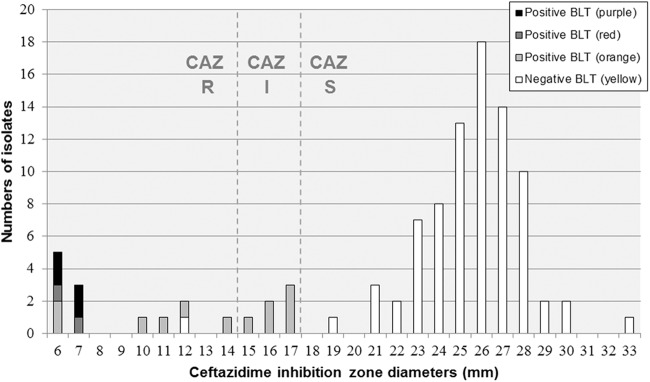
BLT results and CAZ 30-μg disk inhibition zone distribution of Pseudomonas aeruginosa isolates collected from routine clinical samples. Final positive colorimetric changes to purple, red, and orange are represented by black bars, dark gray bars, and light gray bars, respectively. Final negative colorimetric results are represented by white bars. R, fully resistant; I, intermediately resistant; S, susceptible.
In the collection of P. aeruginosa strains characterized for their β-lactamases, 61/64 were BLT positive (Table 1). Interestingly, HMRZ-86 hydrolyzing activity was detected in 12 P. aeruginosa isolates that were phenotypically CAZ susceptible (BEL-1 [n = 1]), VIM-4 [n = 1], CARB type [n = 5], OXA-2 [n = 2], OXA-10 [n = 2], and OXA-198 [n = 1]). On the other hand, 2 CAZ-nonsusceptible VIM-producing P. aeruginosa strains were found negative by the chromogenic test (Table 1). Among these isolates, colorimetric modification appeared to vary according to β-lactamase type. For the majority of the VIM-producing strains, colorimetric variation appeared slower, shifting from yellow to orange, and the BLT failed to detect 3 of the 19 VIM-producing isolates (including one CAZ-susceptible isolate). The overall BLT sensitivity and specificity, calculated for the 164 isolates on the basis of the CAZ-susceptible or -nonsusceptible phenotype, were found to be 95% and 87%, respectively. No colorimetric differences were observed in the results for each of the 20 representative strains tested from different agar plates.
On the whole, the BLT proved very accurate for the detection of CAZ hydrolysis associated with CAZ nonsusceptibility, against both routine P. aeruginosa isolates and those from this collection. In addition to its high NVP, PPV, sensitivity, and specificity, the BLT was easy to use and rapid to perform (about 30 min) with colonies grown on any agar plate types, and interpretation of results was simple.
In nosocomial infections with Pseudomonas aeruginosa, multidrug resistance (including resistance to CAZ) usually results from previous antibiotic selective pressure leading to chromosomal AmpC cephalosporinase and/or efflux pump overexpression (12) or from the acquisition of various transferable β-lactamases. According to our results, a negative BLT result accurately predicts in vitro susceptibility of P. aeruginosa to CAZ and would allow the continuation or initiation of treatment with this agent in case of infection. Since, the BLT result can be obtained 24 h earlier than the CAZ susceptibility results obtained by conventional methods, its use could avoid the prescription of broad-spectrum agents such as carbapenems for empirical treatment in infected patients. However, the lower sensitivity in detecting VIM-type-producing isolates has to be considered, especially during potential hospital epidemic outbreaks and in geographical regions characterized by high prevalence (13). A decrease in turnaround time for obtaining laboratory susceptibility results could have a significant beneficial impact on direct and indirect costs inherent in these infections and may also possibly curb the rise of microbial resistance. Previously, Bouza et al. evaluated prospectively the impact of direct Etest susceptibility testing on respiratory samples from patients with suspected ventilator-associated pneumonia (14). Reporting these early results was associated with substantial clinical benefits, including fewer days of fever, decreased antibiotic use and duration of mechanical ventilation, less Clostridium difficile-associated diarrhea, and lower cost of antimicrobial agents.
Molecular techniques, such as endpoint PCR (15, 16), real-time PCR (17–19), or microarray technology (20), have also been proposed as alternative methods for the prediction of bacterial resistance. However, the molecular-based technologies require specific equipment and trained personnel, and they do not allow the detection of novel unidentified genes. On the other hand, the BLT detects phenotypic hydrolysis of CAZ, resulting from the expression of either acquired or constitutively overexpressed β-lactamases. Furthermore, the BLT is a simple test that may be easily implemented in routine laboratories, although local epidemiological data should also be taken into account.
In conclusion, the BLT is a rapid, sensitive, and specific test that may be implemented in any laboratory worldwide. The therapeutic added value of this test should be evaluated in the clinical field, particularly in the setting of life-threatening infections caused by P. aeruginosa—for example, in intensive care unit patients. A negative BLT could guide physicians to prescribe CAZ in such patients, ensuring an early appropriate empirical therapy and also avoiding the unnecessary use of broad-spectrum antibiotics.
ACKNOWLEDGMENTS
βLACTA test kits were kindly provided by Bio-Rad Laboratories, Clinical Microbiology Division, Steenvoorde, France. We especially thank Caroline Dallenne and Manette Juvin for helpful support throughout the study.
This work was supported by EU grant FP7-HEALTH-2009-SINGLE-STAGE TEMPOtest-QC, project 241742 and INAMI-RIZIV/WIV-ISP-funded National Reference Center for Antibiotic Resistant Pseudomonas and Acinetobacter.
Footnotes
Published ahead of print 10 April 2013
REFERENCES
- 1. American Thoracic Society and Infectious Diseases Society of America 2005. Guidelines for the management of adults with hospital-acquired, ventilator-associated, and healthcare-associated pneumonia. Am. J. Respir. Crit. Care Med. 171:388–416 [DOI] [PubMed] [Google Scholar]
- 2. Mesaros N, Nordmann P, Plésiat P, Roussel-Delvallez M, Van Eldere J, Glupczynski Y, Van Laethem Y, Jacobs F, Lebecque P, Malfroot A, Tulkens PM, Van Bambeke F. 2007. Pseudomonas aeruginosa: resistance and therapeutic options at the turn of the new millennium. Clin. Microbiol. Infect. 13:560–578 [DOI] [PubMed] [Google Scholar]
- 3. Belgian/Luxembourg Working Party on Antimicrobial Therapy 2010. Bacterial infections: recommended antibacterials against selected bacteria, p 113 In Gilbert DN, Moellering RC, Eliopoulos GM, Chambers HF, Saag MS. (ed), The Sanford guide to antimicrobial therapy Belgian/Luxembourg version 2010–2011, 22nd ed Antimicrobial Therapy, Inc, Sperryville, VA [Google Scholar]
- 4. Nordmann P, Naas T. 2010. β-Lactams and Pseudomonas aeruginosa, p 157–174 In Courvalin P, Leclercq R, Rice LB. (ed), Antibiogram. ASM Press, Washington, DC [Google Scholar]
- 5. Hanaki H, Koide Y, Yamazaki H, Kubo R, Nakano T, Atsuda K, Sunakawa K. 2007. Substrate specificity of HMRZ-86 for b-lactamases, including extended-spectrum β-lactamases (ESBLs). J. Infect. Chemother. 13:390–395 [DOI] [PubMed] [Google Scholar]
- 6. Hanaki H, Yamazaki H, Harada H, Kubo R, Kobayashi T, Atsuda K, Sunakawa K. 2005. The synthesis of 7-substituted-3-dinitrostyryl cephalosporins and their ability for detecting extended spectrum β-lactamases (ESBLs). J. Antibiot. 58:69–73 [DOI] [PubMed] [Google Scholar]
- 7. García-Castillo M, Morosini MI, Tato M, Curiao T, Gijón D, Valverde A, Ruiz Garbajosa P, Cantón R. 2012. Rapid detection of b-lactamase-hydrolyzing extended spectrum cephalosporins in Enterobacteriaceae using the new chromogenic βLACTA test. Abstr. 52nd Intersci. Conf. Antimicrob. Agents Chemother., abstr D-753 [Google Scholar]
- 8. Ben Soltana M, Dallenne C, Birgy A, Compain F, Vimont S, Verdet C, Favier C, Juvin M, Arlet G. 2012. Evaluation of a new chromogenic test (βLACTA) for rapid detection of third-generation cephalosporins non-susceptible Enterobacteriaceae. Abstr. 22nd Eur. Congr. Clin. Microbiol. Infect. Dis., abstr P-1729 [Google Scholar]
- 9. Glupczynski Y, Bogaerts P, Deplano A, Berhin C, Huang TD, Van Eldere J, Rodriguez-Villalobos H. 2010. Detection and characterization of class A extended-spectrum-beta-lactamase-producing Pseudomonas aeruginosa isolates in Belgian hospitals. J. Antimicrob. Chemother. 65:866–871 [DOI] [PubMed] [Google Scholar]
- 10. Bogaerts P, Huang TD, Rodriguez-Villalobos H, Bauraing C, Deplano A, Struelens MJ, Glupczynski Y. 2008. Nosocomial infections caused by multidrug-resistant Pseudomonas putida isolates producing VIM-2 and VIM-4 metallo-beta-lactamases. J. Antimicrob. Chemother. 61:749–751 [DOI] [PubMed] [Google Scholar]
- 11. Clinical and Laboratory Standards Institute 2012. Performance standards for antimicrobial susceptibility testing: 22nd informational supplement. M100-S22 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 12. Reinhardt A, Köhler T, Wood P, Rohner P, Dumas JL, Ricou B, van Delden C. 2007. Development and persistence of antimicrobial resistance in Pseudomonas aeruginosa: a longitudinal observation in mechanically ventilated patients. Antimicrob. Agents Chemother. 51:1341–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dortet L, Poirel L, Nordmann P. 2012. Rapid identification of carbapenemase types in Enterobacteriaceae and Pseudomonas spp. by using a biochemical test. Antimicrob. Agents Chemother. 56:6437–6440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bouza E, Torres MV, Radice C, Cercenado E, de Diego R, Sánchez-Carrillo C, Muñoz P. 2007. Direct E-test (AB Biodisk) of respiratory samples improves antimicrobial use in ventilator-associated pneumonia. Clin. Infect. Dis. 44:382–387 [DOI] [PubMed] [Google Scholar]
- 15. Dallenne C, Da Costa A, Decré D, Favier C, Arlet G. 2010. Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 65:490–495 [DOI] [PubMed] [Google Scholar]
- 16. Poirel L, Walsh TR, Cuvillier V, Nordmann P. 2011. Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 70:119–123 [DOI] [PubMed] [Google Scholar]
- 17. Monteiro J, Widen RH, Pignatari AC, Kubasek C, Silbert S. 2012. Rapid detection of carbapenemase genes by multiplex real-time PCR. J. Antimicrob. Chemother. 67:906–909 [DOI] [PubMed] [Google Scholar]
- 18. Naas T, Ergani A, Carrër A, Nordmann P. 2011. Real-time PCR for detection of NDM-1 carbapenemase genes from spiked stool samples. Antimicrob. Agents Chemother. 55:4038–4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Swayne RL, Ludlam HA, Shet VG, Woodford N, Curran MD. 2011. Real-time TaqMan PCR for rapid detection of genes encoding five types of non-metallo- (class A and D) carbapenemases in Enterobacteriaceae. Int. J. Antimicrob. Agents 38:35–38 [DOI] [PubMed] [Google Scholar]
- 20. Bogaerts P, Hujer AM, Naas T, de Castro RR, Endimiani A, Nordmann P, Glupczynski Y, Bonomo RA. 2011. Multicenter evaluation of a new DNA microarray for rapid detection of clinically relevant bla genes from beta-lactam-resistant Gram-negative bacteria. Antimicrob. Agents Chemother. 55:4457–4460 [DOI] [PMC free article] [PubMed] [Google Scholar]


