Abstract
Recent studies have shown that respiratory isolates from pulmonary disease patients and household water/biofilm isolates of Mycobacterium avium could be matched by DNA fingerprinting. To determine if this is true for Mycobacterium intracellulare, household water sources for 36 patients with Mycobacterium avium complex (MAC) lung disease were evaluated. MAC household water isolates from three published studies that included 37 additional MAC respiratory disease patients were also evaluated. Species identification was done initially using nonsequencing methods with confirmation by internal transcribed spacer (ITS) and/or partial 16S rRNA gene sequencing. M. intracellulare was identified by nonsequencing methods in 54 respiratory cultures and 41 household water/biofilm samples. By ITS sequencing, 49 (90.7%) respiratory isolates were M. intracellulare and 4 (7.4%) were Mycobacterium chimaera. In contrast, 30 (73%) household water samples were M. chimaera, 8 (20%) were other MAC X species (i.e., isolates positive with a MAC probe but negative with species-specific M. avium and M. intracellulare probes), and 3 (7%) were M. avium; none were M. intracellulare. In comparison, M. avium was recovered from 141 water/biofilm samples. These results indicate that M. intracellulare lung disease in the United States is acquired from environmental sources other than household water. Nonsequencing methods for identification of nontuberculous mycobacteria (including those of the MAC) might fail to distinguish closely related species (such as M. intracellulare and M. chimaera). This is the first report of M. chimaera recovery from household water. The study underscores the importance of taxonomy and distinguishing the many species and subspecies of the MAC.
INTRODUCTION
Previous studies have suggested household water (especially from bathroom showers) as a source of the Mycobacterium avium complex (MAC), which causes chronic lung disease (1, 2, 3, 4, 5). Both M. avium and M. intracellulare have been recovered from sputum, sinus, and household or potable water samples in multiple countries, including the United States and Japan (1, 4–8). In the majority of these studies, either hybridization probe methods (AccuProbe; Hologic Gen-Probe, Inc., San Diego, CA) or a multiplex 16S rRNA gene PCR was used for identification (1, 5, 9, 10). Isolates positive with the MAC probe but negative with the species-specific M. avium and M. intracellulare probes are collectively referred to as MAC X species.
Currently, the M. avium complex includes four M. avium subspecies (M. avium subsp. avium, M. avium subsp. hominissuis, M. avium subsp. silvaticum, and M. avium subsp. paratuberculosis) (11, 12) and eight species (M. avium, M. intracellulare, M. marseillense [13], M. timonense [13], M. bouchedurhonense [13], M. colombiense [14], M. vulneris [15], and M. chimaera [16]). The last of these species (M. chimaera) was first reported in 2004 by Tortoli et al., who described 12 isolates of a slowly growing species closely related to M. intracellulare and recovered from respiratory samples in Italy, including some from patients with definite lung infection (16). The new species differed by only 1 bp at position 450 in the entire 16S rRNA gene sequence of M. intracellulare but had a very different 16S to 23S internal transcribed spacer (ITS) region sequence (16). Tortoli and coauthors named this new species M. chimaera, as it included characteristics of several species and masqueraded as M. intracellulare (16).
We recently completed a study of variable-number tandem-repeat (VNTR) typing of clinical isolates of M. intracellulare using ITS sequencing to confirm the species designation (7). We then began household water biofilm studies for some of our patients and performed species identification and VNTR genotyping for MAC isolates as needed.
Preliminary studies suggested that respiratory isolates of M. intracellulare indeed belonged to that species based on ITS sequencing (7) but that biofilm isolates thought to be M. intracellulare were actually M. chimaera (R. J. Wallace, Jr., unpublished observation). We then expanded the household study to reassess water and biofilm isolates from three additional published studies (1, 2, 6) and one additional unpublished respiratory study in an attempt to validate this initial finding (17).
MATERIALS AND METHODS
Patients and isolates.
Patients with MAC nodular lung disease who were part of long-term studies at the University of Texas Health Science Center at Tyler (UTHSCT) and who had been diagnosed with a recent episode of active MAC lung disease in the past 6 months were recruited for household water biofilm studies. Sputum isolates of the MAC recovered from these patients during the previous 5 years were available. The patient and household water biofilm studies were done using a protocol approved by the human subjects committee of the UTHSCT.
Environmental household water and biofilm isolates identified as M. intracellulare were also obtained from one previously published study of mycobacterial sinusitis (6) and two previously published studies of pulmonary disease (1, 2), as well as an additional unpublished study of pulmonary disease comparing household biofilm and clinical MAC isolates (17) (Table 1). Cultures for each of these four studies were prepared in the same laboratory (Virginia Polytechnic Institute and University, Blacksburg, VA) with recovered MAC isolates that had been frozen until the time of the current study. Each study was approved by the human subjects committee for the involved institution.
Table 1.
Demographics of U.S. household water studies for the MAC
| Study no. | Type of disease | No. of patients | No. of households | No. of culture sites | Culture source |
Location(s) | Reference(s) | |
|---|---|---|---|---|---|---|---|---|
| Water | Biofilm | |||||||
| 1 | Sinus | 8 | 8 | 80 | + | + | New York City, San Francisco, CA, and Florida | 6 |
| 2 | Pulmonary | 26 | 26 | 224 | − | + | Philadelphia, PA | L. Lande, unpublished data; 17 |
| 3 | Pulmonary | 28 | 30 | 379 | + | + | 14 states | 1 |
| 4 | Pulmonary | 10 | 15 | 66 | − | + | Texas and Louisiana | R. J. Wallace, Jr., unpublished data |
| 5 | Pulmonary | 1 | 1 | 3 | + | + | New York City | 2 |
| Total | 73 | 80 | 752 | |||||
Control strains for PCR and sequencing included M. intracellulare strain ATCC 13950T and M. chimaera strain DSM 44623T.
Household water sampling.
The five studies utilized water samples and/or swab cultures of bathroom and kitchen faucet filters and pipes and showerhead filters and showerhead pipes, as well as samples from any other potential sites (e.g., air filters, hot tub filters, and bathtub inlet pipes, etc., when available) (Table 1). Water samples of 500 ml were concentrated by centrifugation (500 × g for 20 min), and then the pellet was resuspended in 1 ml of sterile tap water. Swabs were added to 2 ml of sterile tap water. Samples were decontaminated using the method of Thomson et al. (18) and spread onto Middlebrook 7H10 agar plates that were incubated for 3 weeks and then screened for acid-fast bacilli (AFB) (1, 2, 19).
Nonsequencing identification of M. intracellulare and M. avium.
Sputum isolates were identified as members of the MAC for all studies using a commercial DNA probe (AccuProbe). Sputum or water/biofilm isolates of MAC were initially identified as M. avium or M. intracellulare by nonsequencing methods using an M. avium or M. intracellulare DNA probe (AccuProbe) (10), a multiplex 16S rRNA gene PCR (9), or an hsp65 PCR of a 441-bp sequence followed by digestion with BstEII or HaeIII as described by Telenti et al. (20).
DNA sequencing for species identification of M. intracellulare and M. avium.
Sequencing of the 280-bp 16S to 23S internal transcribed spacer (ITS) region of each isolate of M. intracellulare (based on the preliminary identification) and the two reference strains was performed as described previously (11, 12, 16, 21). This method readily distinguishes the usual M. intracellulare sequence (Min-A) (7, 21) from the M. chimaera sequence MAC-A (16, 21). Sequences were compared to the Min-A sequence for M. intracellulare type strain ATCC 13950T (GenBank sequence no. AB026691) and the MAC-A sequence for M. chimaera type strain DSM 44623T (GenBank sequence no. EF521902).
Partial 16S rRNA gene sequencing of the first 500 bp of isolates identified as M. chimaera by the ITS region was also performed (7). This sequence contains the single base pair difference between M. intracellulare and M. chimaera (16).
Thirty-five M. avium isolates identified by nonsequencing methods also underwent ITS region sequencing. No nontuberculous mycobacteria (NTM) species other than M. avium was identified, which demonstrates that species confusion similar to that seen with M. intracellulare and M. chimaera is not seen with M. avium.
RESULTS
Patients.
A total of 73 patients with MAC respiratory disease were studied (Table 1); 65 met the American Thoracic Society (ATS) criteria for MAC lung disease (22). Isolates from eight patients with mycobacterial sinusitis and from their household water were also included for study (6). The patients were from multiple states and two large cities in the United States (Table 1).
Isolates and nonsequencing species identification.
One or more sputum culture isolates were available for 53 of the patients with MAC lung disease. Multiple sputum isolates were available from the 10 UTHSCT patients, many of whom had had prior episodes of MAC lung disease caused by species other than M. intracellulare or had mixed infections at the time of the water studies (study group 4; Tables 1 to 3). For the other lung disease patients, only a single respiratory culture was available (study groups 1 to 3 and 5; Tables 1 to 3), as was also the case for the patients with sinusitis.
Table 3.
Species in household water cultures (one culture per site) initially identified as M. intracellulare and then retested by ITS sequencing compared to the number of isolates of M. avium recovered from the same samples
| Study no. | No. of isolates identified by nonsequencing methodsa |
No. (%) of isolates identified by ITS sequencing |
Reference(s) | ||||
|---|---|---|---|---|---|---|---|
| M. avium | M. intracellulare | M. avium | MAC X | M. chimaera | M. intracellulare | ||
| 1 | 20 | 4 | 3 | 0 | 1 | 0 | 6 |
| 2 | 64 | 8 | 0 | 3 | 5 | 0 | L. Lande, unpublished data; 17 |
| 3 | 36 | 21b | 0 | 5 | 16 | 0 | 1 |
| 4 | 18 | 8 | 0 | 0 | 8 | 0 | R. J. Wallace, Jr., unpublished data |
| 5 | 3 | 0 | 0 | 0 | 0 | 0 | 2 |
| Total | 141 | 41 | 3 (7.3) | 8 (19.5) | 30 (73.2) | 0 | |
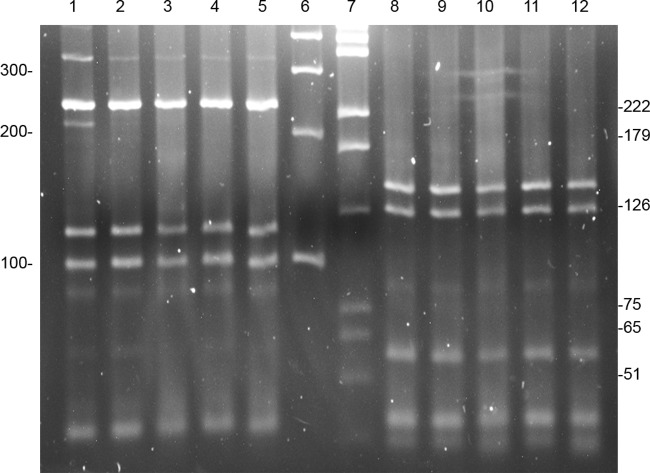
A comparison of the results from the commercial DNA M. intracellulare probe (AccuProbe) for isolates of M. intracellulare and M. chimaera showed that both species gave strongly positive results. A comparison of these two species using the multiplex 16S rRNA gene PCR (9) is shown in Fig. 1, and a comparison of the hsp65 PCR restriction enzyme analysis (PRA) (20) is shown in Fig. 2. None of these three nonsequencing methods distinguished M. intracellulare from M. chimaera.
Fig 1.
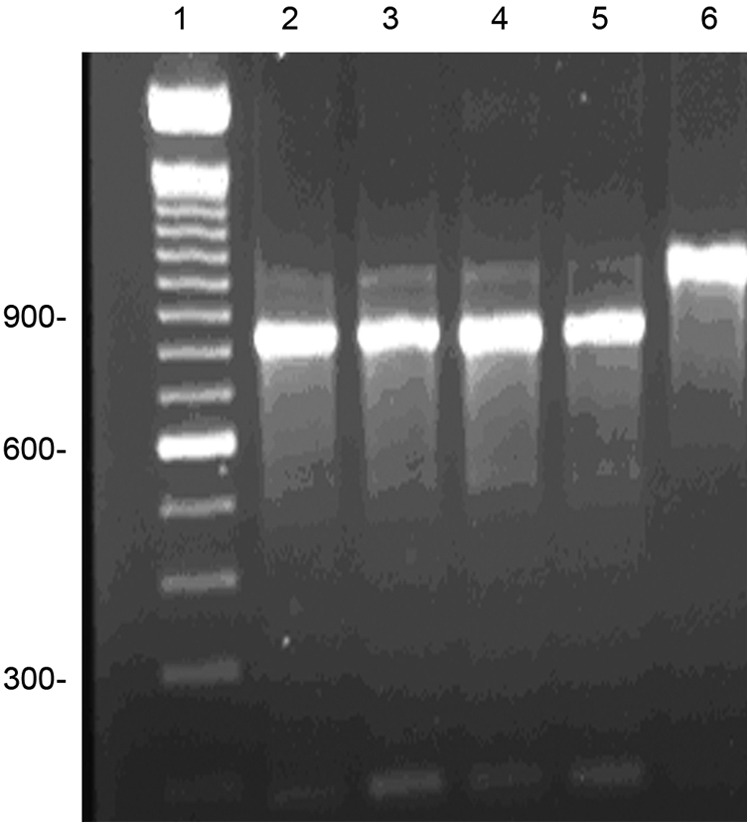
16S rRNA gene multiplex PCR comparing M. intracellulare and M. chimaera (9). The two species are indistinguishable by this method. Lane 1, 100-bp ladder; lane 2, M. chimaera type strain DSM 44623T; lane 3, M. chimaera isolate from household water sample; lane 4, M. intracellulare type strain ATCC 13950T; lane 5, M. intracellulare patient isolate; lane 6, M. marseillense isolate from household water sample. Molecular weight markers are shown to the left.
Fig 2.
PCR restriction enzyme analysis (PRA) of the 441-bp Telenti fragment of the hsp65 gene. The first five lanes include digests with BstEII (fragments of 235-115-100), and lanes 8 to 12 include the same isolates digested with HaeIII (fragments of 145-125-55). The digest patterns for M. marseillense, M. chimaera, and M. intracellulare are indistinguishable. Lanes 1 and 8, M. marseillense, household water; lanes 2 and 9, M. chimaera, household water; lanes 3 and 10, M. intracellulare, patient isolates; lanes 4 and 11, M. chimaera type strain DSM 44623T; lanes 5 and 12, M. intracellulare type strain ATCC 13950T; lane 6, 100-bp ladder (100, 200, 300, 400); lane 7, pGEM ladder (51, 65, 75, 126, 179, 222, 350). Molecular weight standard sizes are shown to the left and the right.
Sequencing for identification of M. intracellulare and M. avium.
All isolates of presumptive M. intracellulare using nonsequencing methods underwent ITS region sequencing. The isolates had 100% identity to Min-A (M. intracellulare) or MAC-A (M. chimaera) (as the two type strains), and several sequences belonged to other MAC X species (11, 16, 21). A comparison of the base pair differences in the ITS region sequences between M. intracellulare and M. chimaera is shown in Table 4. Unlike the 16S rRNA gene that differed by only 1 bp substitution, the ITS region sequences differed by 19 bp substitutions (Table 4).
Table 4.
Base pair differences in ITS region sequences between M. intracellulare (Min-A, -B, -C, and -D) and M. chimaera (MAC-A) and the MAC X species M. colombiense (MAC-U)
| ITS region sequencea | Base pair differences with ITS sequevar ofb: |
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 22 | 55 | 73 | 76 | 137 | 138 | 148 | 216 | 220 | 229 | 230 | 232 | 237 | 239 | 244 | 264 | 268 | 269 | 270 | 272 | |
| Min-A | T | C | G | C | G | A | A | G | A | A | G | G | T | C | G | C | G | G | T | T |
| Min-B | • | • | • | • | • | • | • | • | • | G | C | • | • | • | • | • | • | • | • | • |
| Min-C | • | • | • | • | • | • | • | • | • | G | T | • | • | • | • | • | • | • | • | • |
| Min-D | • | • | • | T | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • |
| MAC-A | C | T | A | • | A | G | Tc | A | G | G | T | Ac | C | T | A | T | A | A | C | C |
| MAC-U | • | T | A | • | • | • | • | A | G | A | T | • | C | T | A | • | • | • | • | • |
Note the similarity of the two MAC X species (M. chimaera and M. colombiense) and their dissimilarity with the four ITS sequences of M. intracellulare.
•, same base pairs as in Min-A.
Base pair difference unique among the M. avium complex (12).
The patient and water isolates of M. chimaera (identified by ITS sequencing) also underwent partial (initially 500 bp) 16S rRNA gene sequencing. This included the T→C base pair substitution at position 450 that distinguishes M. intracellulare from M. chimaera (16). All sequences were a 100% match to the GenBank sequence of M. chimaera submitted by Tortoli et al. (16) (accession no. NR_029003) and to the M. chimaera type strain sequenced as a control.
A total of 35 random M. avium isolates underwent ITS sequencing. All had Mav-A or Mav-B sequences typical of M. avium.
Respiratory isolates.
A total of 54 respiratory cultures were identified as M. intracellulare by nonsequencing methods. By ITS sequencing, 49 isolates (90.7%) had the Min-A sequence of M. intracellulare, and 4 isolates (7.4%) had the MAC-A sequence of M. chimaera (Table 2).
Table 2.
Respiratory or sinus cultures of MAC initially identified as M. intracellulare by nonsequencing methods and then retested by ITS sequencing
| Study no. | No. of M. intracellulare isolates identified by nonsequencing methods | No. (%) of isolates identified by ITS sequencing |
Reference(s) | |||
|---|---|---|---|---|---|---|
| M. avium | MAC X | M. chimaera | M. intracellulare | |||
| 1 | 1 | 1 | 0 | 0 | 0 | 6 |
| 2 | 3 | 0 | 0 | 0 | 3 | L. Lande, unpublished data; 17 |
| 3 | 10 | 0 | 0 | 1 | 9 | 1 |
| 4a | 40 | 0 | 0 | 3 | 37 | R. J. Wallace, Jr., unpublished data |
| 5 | 0 | NAb | NA | NA | NA | 2 |
| Totals | 54 | 1 | 0 | 4 (7.4) | 49 (90.7) | |
Multiple isolates were available for all patients in this study.
NA, not applicable.
Household water sampling.
A total of 752 individual sites were sampled for the 73 patients from 80 households (Table 1). A total of 41 household water/biofilm isolates identified as M. intracellulare by nonsequencing methods underwent ITS and/or 16S rRNA partial gene sequencing, which revealed that the majority of these isolates (30/41, 73%) were M. chimaera or other MAC X species (8/41, 19.5%). Based on the ITS region sequencing, no isolate of M. intracellulare was recovered from any of the 752 sampled sites (Table 3).
DISCUSSION
This study indicates that environmental sources other than household water are the source of M. intracellulare nodular-type lung disease in the United States. More than 700 water plumbing sites from 71 patients from the Southwest, the Northeast, and more than 10 additional states were cultured for MAC. This makes the failure to recover M. intracellulare from household water samples in the United States not likely to be a function of either geography or inadequate sampling. There is no evidence that the decontamination procedure or the growth medium inhibits the growth of M. intracellulare (18). ITS sequence analysis detects only the most abundant microbial species. Thus, while ITS sequencing results failed to detect M. intracellulare in water samples, quantitative PCR sequencing needs to be done to definitively rule out its absence.
M. intracellulare is a recognized cause of MAC lung disease in the United States. Because species identification within the complex is not (yet) considered clinically important, it is usually not done except as part of prospective studies. Multiple studies of MAC lung disease have been done over the past 20 years at the UTHSCT with patients primarily from Texas and Louisiana. Species identification has been done in many of these studies. They have shown that 70% to 90% of MAC isolates from cases of nodular bronchiectatic disease and upper lobe fibrocavitary disease are M. intracellulare (23–25). (Some isolates from these older studies might have been M. chimaera, as all used nonsequencing species identification.) Few studies of the species of the MAC causing chronic lung disease that meet the ATS diagnostic criteria have been done in other areas of the United States, but M. intracellulare is considered an important cause of lung disease in these areas as well.
M. intracellulare is a major cause of MAC lung disease in several other countries. A recent study from Queensland, Australia, by Thomson compared NTM isolates obtained in 1999 and 2005 (26). There was a striking increase in M. intracellulare lung infections, with 79/97 (81%) of MAC lung disease cases being due to M. intracellulare and 19% due to M. avium. The method of species identification was not provided (26). A recent study of 590 Korean patients with MAC lung disease from 2000 to 2009 reported that 267 (45%) of the patients had M. intracellulare infections (27).
In contrast to the studies from the United States, Queensland, Australia, and Korea, previous studies from Japan reported that M. avium was the predominant cause of MAC lung disease (4). M. avium has also been shown to be the predominant or exclusive MAC species in water samples of residential bathrooms. In a 2009 study, Nishiuchi et al. found that 32/33 residential bathroom water MAC isolates were M. avium, and only one isolate was reported as M. intracellulare (utilizing a PCR method for species identification) (5). Nishiuchi et al. did report M. intracellulare in an earlier study from 3/9 bathrooms using the 16S to 23S internal spacer region for species identification (4). Neither of these studies reported the recovery of M. chimaera or other MAC X species from the water samples (4, 5). Thus, the characterization of the variability of MAC species populating water supplies in different parts of the world clearly warrants further investigation with more accurate sequencing techniques.
In 2009, Feazel et al. reported the use of 16S rRNA gene sequencing to determine the microbial composition of 45 shower sites collected from nine cities in the United States (3). Nontuberculous mycobacteria (NTM) were identified in approximately one-third of the samples, with the predominant species being M. gordonae and M. avium. M. avium was found in approximately 20% of all samples. Only two sampled sites, both in New York City, yielded clones of M. intracellulare, albeit in low numbers (3). In addition, the region of the 16S rRNA gene sequenced in this study excluded the base pair substitution at position 450 that distinguishes M. chimaera from M. intracellulare; therefore, these clones might have represented M. chimaera (3). The percentage of sites positive for NTM using 16S rRNA gene sequencing was similar to that with culture techniques (1, 6). Together, these studies indicate that M. intracellulare is absent from U.S. household water systems, in contrast to the prevalence of M. avium (3).
As noted previously, the individual species in the M. avium complex are rarely reported clinically in the United States. When reporting clinical studies, most U.S. investigators use the nonsequencing methods that include a hybridization DNA probe assay, a 16S rRNA gene multiplex PCR, or restriction fragment length polymorphism (RFLP) analysis of the PCR product of amplification of the hsp65 gene (1, 5, 9, 10, 20). This study points out the inadequacies of nonsequencing identification methods for closely related NTM species such as M. intracellulare and M. chimaera (16). Figures 1 and 2 and the results obtained here with the AccuProbe (Hologic Gen-Probe, Inc.) confirm the findings reported in the initial study of M. chimaera by Tortoli et al. (16).
Few environmental studies have been done for the MAC other than tests of household or hospital water (28), and these studies were done before sequencing was readily available. Soil would seem the most logical alternative source, and one study of aerosols of potting soil found M. intracellulare (identified by 16S rRNA PCR [9] and hsp65 PCR amplification and endonuclease restriction fragment patterns [20, 29]). Those methods would not have distinguished M. chimaera from M. intracellulare. More environmental studies of potential sources of M. intracellulare responsible for nodular and cavitary lung disease are clearly needed and will require attention to the distinction between M. intracellulare and M. chimaera.
The ITS sequence of M. chimaera from a respiratory sample in the United States was first reported by Frothingham and Wilson in a 1994 study of the use of ITS to produce subgroups within the complex (21). Patient details were not provided. We are aware of only one report (published in 2009) of M. chimaera as a respiratory pathogen in the United States. The authors described a patient whose infection occurred in the setting of chronic obstructive lung disease. The DNA was sequenced from pathological lung samples, but the organism was not recovered by culture (30). There have been no additional reports of M. chimaera in the United States that we were able to be identified; furthermore, the current study is the first description of M. chimaera from environmental sources. The studies mentioned above suggest that household water might be the reservoir for M. chimaera lung disease. More studies are needed to determine the relative importance of M. chimaera as a pathogen and the relationship of household water/biofilm and sputum isolates and to determine whether previous case definitions for NTM lung disease utilized for other species of the MAC will identify disease due to this new species (16).
More broadly, the data support the notion that lumping mycobacteria into groups and/or complexes, here the Mycobacterium avium complex (now including four M. avium subspecies and at least eight other species), will obscure unique characteristics. Those characteristics include their ecology, epidemiology, virulence (M. chimaera might have reduced virulence) (31), and even susceptibility to antibiotics.
ACKNOWLEDGMENT
We acknowledge the financial support of the Amon G. Carter Foundation.
Footnotes
Published ahead of print 27 March 2013
REFERENCES
- 1. Falkinham JO., III 2011. Nontuberculous mycobacteria from household plumbing of patients with nontuberculous mycobacteria disease. Emerg. Infect. Dis. 17:419–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Falkinham JO, III, Iseman MD, de Haas P, van Soolingen D. 2008. Mycobacterium avium in a shower linked to pulmonary disease. J. Water Health 6:209–213 [DOI] [PubMed] [Google Scholar]
- 3. Feazel LM, Baumgartner LK, Peterson KL, Frank DN, Harris JK, Pace NR. 2009. Opportunistic pathogens enriched in showerhead biofilms. Proc. Natl. Acad. Sci. U. S. A. 106:16393–16399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nishiuchi Y, Maekura R, Kitada S, Taaru A, Taguri T, Kira Y, Hiraga T, Hirotani A, Yoshimura K, Miki M, Ito M. 2007. The recovery of Mycobacterium avium-intracellulare complex (MAC) from the residential bathrooms of patients with pulmonary MAC. Clin. Infect. Dis. 45:347–351 [DOI] [PubMed] [Google Scholar]
- 5. Nishiuchi Y, Tamaru A, Kitada S, Taguri T, Matsumoto S, Tateishi Y, Yoshimura M, Ozeki Y, Matsumura N, Ogura H, Mackura R. 2009. Mycobacterium avium complex organisms predominantly colonize in the bathtub inlets of patients' bathrooms. Jpn. J. Infect. Dis. 62:182–186 [PubMed] [Google Scholar]
- 6. Tichenor WS, Thurlow J, McNulty S, Brown-Elliott BA, Wallace RJ, Jr, Falkinham JO., III 2012. Nontuberculous mycobacteria in household plumbing as possible cause of chronic rhinosinusitis. Emerg. Infect. Dis. 18:1612–1617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Iakhiaeva E, McNulty S, Brown-Elliott BA, Falkinham JO, III, Williams MD, Vasireddy R, Wilson RW, Turenne C, Wallace RJ., Jr 2013. MIRU-VNTR of Mycobacterium intracellulare for strain comparison with establishment of a PCR data base. J. Clin. Microbiol. 51:409–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Whiley H, Keegan A, Giglio S, Bentham R. 2012. Mycobacterium avium complex—the role of potable water in disease transmission. J. Appl. Microbiol. 113:223–232 [DOI] [PubMed] [Google Scholar]
- 9. Wilton S, Cousins D. 1992. Detection and identification of multiple mycobacterial pathogens by DNA amplification in a single tube. PCR Methods Appl. 1:269–273 [DOI] [PubMed] [Google Scholar]
- 10. Kiehn TE, Edwards FF. 1987. Rapid identification using a specific DNA probe of Mycobacterium avium complex from patients with acquired immunodeficiency syndrome. J. Clin. Microbiol. 25:1551–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Turenne CY, Semret M, Cousins DV, Collins DM, Behr MA. 2006. Sequencing of hsp65 distinguishes among subsets of the Mycobacterium avium complex. J. Clin. Microbiol. 44:433–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mijs W, de Haas P, Rossau R, Van der Laan T, Rigouts L, Portaels F, van Soolingen D. 2002. Molecular evidence to support a proposal to reserve the designation Mycobacterium avium subsp. avium for bird-type isolates and “M. avium subsp. hominissuis” for the human/porcine type of M. avium. Int. J. Syst. Evol. Microbiol. 52:1505–1518 [DOI] [PubMed] [Google Scholar]
- 13. Iskandar BS, Cayrou C, Raoult D, Drancourt M. 2009. Mycobacterium marseillense sp. nov., Mycobacterium timonense sp. nov. and Mycobacterium bouchedurhonense sp. nov., members of the Mycobacterium avium complex. Int. J. Syst. Evol. Microbiol. 59:2803–2808 [DOI] [PubMed] [Google Scholar]
- 14. Murcia MI, Tortoli E, Menendez MC, Palenque E, Garcia MJ. 2006. Mycobacterium colombiense sp. nov., a novel member of the Mycobacterium avium complex and description of MAC-X as a new ITS genetic variant. Int. J. Syst. Evol. Microbiol. 56:2049–2054 [DOI] [PubMed] [Google Scholar]
- 15. van Ingen J, Boeree MJ, Kösters K, Wieland A, Tortoli E, Dekhuijzen PNR, van Soolingen D. 2009. Proposal to elevate Mycobacterium avium complex ITS sequevars MAC-Q. to Mycobacterium vulneris sp. nov. Int. J. Syst. Evol. Microbiol. 59:2277–2282 [DOI] [PubMed] [Google Scholar]
- 16. Tortoli E, Rindi L, Garcia MJ, Chiaradonna P, Dei R, Garzelli C, Kroppenstedt RM, Lari N, Mattei R, Mariottini A, Mazzarelli G, Murcia MI, Nanetti A, Piccoli P, Scarparo C. 2004. Proposal to elevate the genetic variant MAC-A, included in the Mycobacterium avium complex, to species rank as Mycobacterium chimaera sp. nov. Int. J. Syst. Evol. Microbiol. 54:1277–1285 15280303 [DOI] [PubMed] [Google Scholar]
- 17. Lande L, Peterson DD, Sawicki J, Kwait R, Williams MD, Iakhiaeva E, Wallace RJ, Jr, Falkinham JO., III 2013. Municipal water supply as a major source for pulmonary Mycobacterium avium lung disease: a comparison of household and respiratory isolates. Abstr. Am. Thor. Soc. Int. Conf., Philadelphia, PA [Google Scholar]
- 18. Thomson R, Carter R, Gilpin C, Coulter C, Hargreaves M. 2008. Comparison of methods for processing drinking water samples for the isolation of Mycobacterium avium and Mycobacterium intracellulare. Appl. Environ. Microbiol. 74:3094–3098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Falkinham JO, III, Norton CD, LeChevallier MW. 2001. Factors influencing numbers of Mycobacterium avium and Mycobacterium intracellulare and other mycobacteria in drinking water systems. Appl. Environ. Microbiol. 67:1225–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Telenti A, Marchesi F, Balz M, Bally F, Böttger EC, Bodmer T. 1993. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J. Clin. Microbiol. 31:175–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Frothingham R, Wilson KH. 1994. Molecular phylogeny of the Mycobacterium avium complex demonstrates clinically meaningful divisions. J. Infect. Dis. 169:305–312 [DOI] [PubMed] [Google Scholar]
- 22. Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, Holland SM, Horsburgh R, Huitt G, Iademarco MF, Iseman M, Olivier K, Ruoss S, von Reyn CF, Wallace RJ, Jr, Winthrop K. 2007. An official ATS/IDSA statement: diagnosis, treatment and prevention of nontuberculous mycobacterial diseases. Am. J. Respir. Crit. Care Med. 175:367–416 [DOI] [PubMed] [Google Scholar]
- 23. Wallace RJ, Jr, Zhang Y, Brown BA, Dawson D, Murphy DT, Wilson R, Griffith DE. 1998. Polyclonal Mycobacterium avium complex infections in patients with nodular bronchiectasis. Am. J. Respir. Crit. Care Med. 158:1235–1244 [DOI] [PubMed] [Google Scholar]
- 24. Griffith DE, Brown-Elliott BA, Langsjoen B, Zhang Y, Pan X, Girard W, Nelson K, Caccitolo J, Alvarez J, Shepherd S, Wilson R, Graviss EA, Wallace RJ., Jr 2006. Clinical and molecular analysis of macrolide resistance in Mycobacterium avium complex lung disease. Am. J. Respir. Crit. Care Med. 174:928–934 [DOI] [PubMed] [Google Scholar]
- 25. Wallace RJ, Jr, Zhang Y, Brown-Elliott BA, Yakrus MA, Wilson RW, Mann L, Couch L, Girard WM, Griffith DE. 2002. Repeat positive cultures in Mycobacterium intracellulare lung disease after macrolide therapy represent new infections in patients with nodular bronchiectasis. J. Infect. Dis. 186:266–273 [DOI] [PubMed] [Google Scholar]
- 26. Thomson RM. 2010. Changing epidemiology of pulmonary nontuberculous mycobacteria infections. Emerg. Infect. Dis. 16:1576–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Koh WJ, Jeong B-H, Jeon K, Lee NY, Lee KS, Woo SY, Shin SJ, Kwon OJ. 2012. Clinical significance of differentiation between Mycobacterium avium and Mycobacterium intracellulare in M. avium complex lung disease. Chest 142:1482–1488 [DOI] [PubMed] [Google Scholar]
- 28. Smole SC, McAleese F, Ngampasutadol J, von Reyn CF, Arbeit RD. 2002. Clinical and epidemiological correlates of genotypes within the Mycobacterium avium complex defined by restriction and sequence analysis of hsp65. J. Clin. Microbiol. 40:3374–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. De Groote MA, Pace NR, Fulton K, Falkinham JO., III 2006. Relationship between Mycobacterium isolates from patients with pulmonary infection and potting soils. Appl. Environ. Microbiol. 72:7602–7606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bills ND, Hinrichs SH, Aden TA, Wickert RS, Iwen PC. 2009. Molecular identification of Mycobacterium chimaera as a cause of infection in a patient with chronic obstructive pulmonary disease. Diagn. Microbiol. Infect. Dis. 63:292–295 [DOI] [PubMed] [Google Scholar]
- 31. Schweickert B, Goldenberg O, Richter E, Göbel UB, Petrich A, Buchholz P, Moter A. 2008. Occurrence and clinical relevance of Mycobacterium chimaera sp. nov., Germany. Emerg. Infect. Dis. 14:1443–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]