Abstract
The genetic bases for antibiotic tolerance are obscure. Daptomycin (DAP) is a lipopeptide antibiotic with bactericidal activity against enterococci. Using time-kill assays, we provide evidence for the first time that a deletion of isoleucine in position 177 of LiaF, a member of the three-component regulatory system LiaFSR involved in the cell envelope response to antimicrobials, is directly responsible for a DAP-tolerant phenotype and is likely to negatively affect response to DAP therapy.
TEXT
Management of enterococcal infections is an important clinical challenge since these organisms exhibit intrinsic resistance to many antimicrobials. Furthermore, bactericidal therapy, paramount in treatment of severe infections, is compromised due to the fact that many strains are tolerant to common antienterococcal agents (i.e., are susceptible to the antibiotic but survive in the presence of the drug). During recent decades, the emergence and dissemination of multidrug-resistant (MDR) strains with resistance to ampicillin and vancomycin and/or with high-level resistance to aminoglycosides (1) have dramatically reduced the reliable bactericidal therapeutic options against MDR enterococci, further complicating the treatment of severe infections (2).
Daptomycin (DAP) is a cyclic lipopeptide antibiotic with activity against a wide range of Gram-positive organisms, including MDR enterococci. DAP is frequently used off-label for the management of enterococcal bacteremia as it retains bactericidal activity in vitro against enterococci in a dose-dependent fashion (3). Although the majority of clinical enterococcal isolates remain DAP susceptible (DAP-S), the development of resistance de novo and arising during therapy has been well described (4, 5, 6). The mechanism of DAP resistance is not fully understood. Nonetheless, mutations in liaFSR, encoding a three-component regulatory system that orchestrates the cell envelope response to stress, coupled with changes in genes encoding enzymes involved in phospholipid metabolism (cls and gdpD) have been linked with the development of DAP resistance in both Enterococcus faecalis and E. faecium (7). Using an allelic replacement strategy, we showed that introduction of a mutated liaF allele with a codon deletion resulting in an isoleucine deletion at position 177 of the predicted transmembrane protein LiaF produced a 3-fold increase in the DAP MIC of a DAP-S clinical isolate of vancomycin-resistant E. faecalis (7). Moreover, we subsequently provided evidence that a high proportion of DAP-S E. faecium clinical isolates with DAP MICs of between 3 and 4 μg/ml harbored mutations in at least one of the genes of the liaFSR system. Conversely, no changes in liaFSR were found in isolates with DAP MIC ≤ 2 μg/ml (8). These initial observations led us to postulate that mutations in the liaFSR system play a critical initial role in the development of DAP resistance in enterococci, producing a slight decrease in the susceptibility to the antibiotic (not sufficient to increase the MIC above the susceptibility breakpoint) but eventually leading to therapeutic failure. Indeed, several investigators have reported DAP clinical failure in patients with enterococcal bacteremia whose DAP MICs were in the higher range of susceptibility (3 to 4 μg/ml [the CLSI breakpoint is 4 μg/ml]) (9, 10). Because these isolates appear “susceptible” by standard testing, DAP monotherapy is usually administered, enhancing the possibilities of further selecting for resistance. Thus, identification of DAP-S enterococcal isolates that may be “predisposed” to DAP failure during therapy would be of paramount importance to improve outcomes in patients with severe enterococcal infections.
In order to evaluate the actual effect of the LiaF deletion on the bactericidal activity of DAP, we performed time-kill assays (see below) using the following previously reported vancomycin-resistant E. faecalis strains: (i) S613, a DAP-S isolate (MIC = 1 μg/ml) whose genome has been sequenced (available at the NCBI site http://www.ncbi.nlm.nih.gov/nuccore), recovered from the bloodstream of a patient before DAP therapy, (ii) R712, a DAP-resistant (DAP-R) derivative of S613 (MIC = 12 μg/ml) recovered from the patient's blood after DAP monotherapy, carrying the mutated liaF allele (with the ATT deletion in codon 177) and additional mutations in gdpD (encoding a glycerophosphodiester-phosphodiesterase) and cls (encoding a cardiolipin synthase enzyme) (http://www.ncbi.nlm.nih.gov/nuccore) leading to therapeutic failure (6), and (iii) a laboratory derivative of S613 in which the wild-type liaF allele was replaced with the mutated one from R712 (S613liaFΔIle177) (DAP MIC of 4 μg/ml) (7).
The bactericidal activity of DAP was evaluated with time-kill assays performed in triplicate with an initial bacterial inoculum of 107 CFU/ml from an overnight culture. Bacteria were grown in brain heart infusion (BHI) broth supplemented with calcium (50 mg/liter), and DAP was added at concentrations of 5× and 10× the MIC for each strain determined in BHI broth supplemented with calcium (the MICs obtained in BHI for S613, R712, and S613liaFΔIle177 were 2, 24, and 8 μg/ml, respectively). Quantitative bacterial counts were performed at 0, 6, and 24 h by plating appropriate dilutions on BHI agar plates. In order to control for antibiotic carryover, bacterial cell suspensions (1-ml samples) were centrifuged and the pelleted bacteria suspended in 0.9% saline solution before plating. Bactericidal activity was defined as a decrease of ≥3 log10 in bacterial counts at 24 h compared to the initial inoculum. The limit of detection of the assay was 200 CFU/ml, assuming maximum plating efficiency.
In order to place our results of the killing assay in the context of genetic and phenotypic changes associated with DAP resistance, five colonies from S613liaFΔIle177 were randomly selected at each time point (6 and 24 h). DAP susceptibility testing was performed on each colony in duplicate by Etest (bioMérieux, Marcy l'Etoile, France) using Mueller-Hinton agar plates following the manufacturer's recommendations. Additionally, the sequence analysis of liaF, liaS, liaR, cls, and gdpD (complete open reading frames) was performed using all of the colonies described above. The protocol for PCR amplification, DNA purification, sequencing, and analysis has been previously described (8). Apart from the Ile deletion in LiaF, we evaluated the presence of deletions previously described in R712 in the putative GdpD and Cls enzymes (deletion of Ile at position 170 and deletion of Lys at position 61, respectively) or novel mutations in the target genes. A mutation was defined as a nonsynonymous nucleotide change(s) that predicted a change in an amino acid that was not present in any other DAP-S isolates whose genomes are publicly available from NCBI.
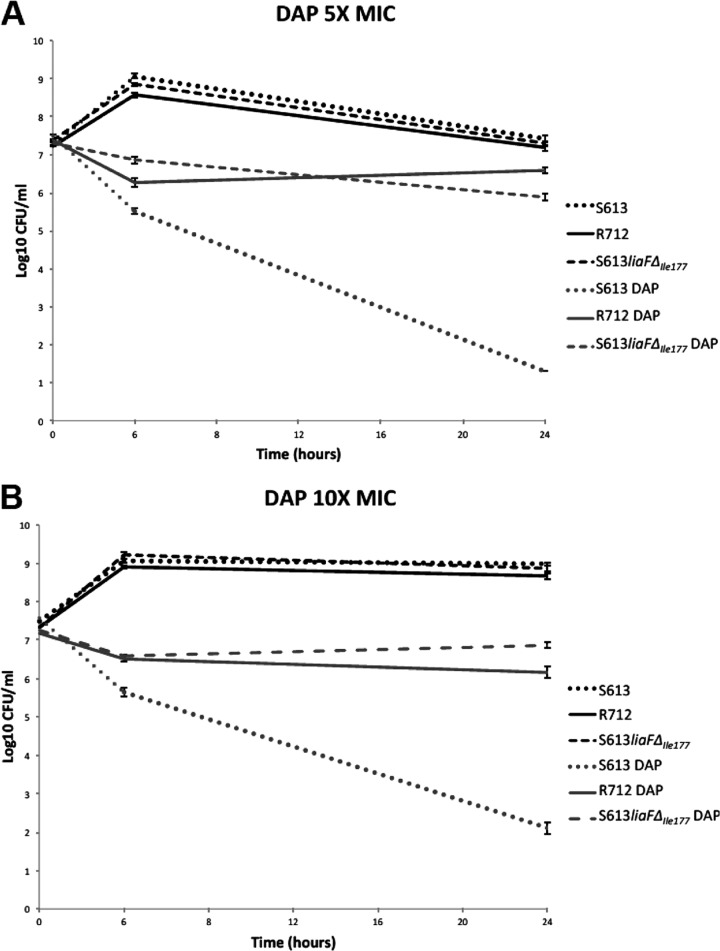
Figure 1 shows the time-kill curves for all the strains at different DAP concentrations. DAP bactericidal activity was evident for DAP-S S613, with a decrease of >3 log10 CFU/ml at concentrations of 5× and 10× the MIC. Conversely, DAP had no effect on the DAP-R R712 strain, with no killing effect observed at either concentration of the antibiotic. The most striking finding was that, for S613liaFΔIle177 (MIC = 4 μg/ml), DAP had no killing effect (no reduction in CFU count from the starting inoculum) at DAP concentrations of 5× or 10× the MIC, with the strain behaving in a fashion similar to that of R712 upon exposure to the antibiotic. Moreover, DAP MICs of all derivatives of S613liaFΔIle177 at 6 h and 24 h remained the same as those seen with the parental strain and were within the DAP susceptibility range (4 μg/ml). Apart from the ATT deletion in liaF, we found no other changes in cls or gdpD, suggesting that the liaF alteration was sufficient to affect the ability of DAP to kill S613liaFΔIle177.
Fig 1.
Time-kill assays with DAP at 5× the MIC (A) and 10× the MIC (B). The assays were performed using brain heart infusion broth supplemented with calcium to a final concentration of 50 μg/ml. CFU, colony-forming units.
The phenomenon of antibiotic tolerance in bacteria has been recognized for many years, but its genetic bases still remain obscure. Here we show, for the first time, a direct correlation between a mutation in a gene that encodes a transmembrane protein (LiaF), a member of a system predicted to modulate the cell envelope response to antimicrobials (11, 12), and antibiotic tolerance. Although we cannot exclude the possibility of the presence of additional mutations in other regions of the genome, the lack of changes in genes that have been previously associated with development of the DAP-R phenotype in the same strain strongly suggests that the liaF deletion was responsible for the tolerance phenotype. These findings, along with our previous observations in E. faecium (8) that a high proportion of the DAP-S isolates at the high end of the susceptible range (MICs of 3 to 4 μg/ml) harbor mutations in the LiaFSR system, continue to suggest that the enterococcal breakpoints for DAP may need to be revisited. The specific role of the liaFSR system in the development of tolerance and resistance to DAP is the object of our future studies.
ACKNOWLEDGMENTS
This work was supported by NIH grants R00 AI72961 and R01 AI093749 to C.A.A. from the National Institute of Allergy and Infectious Diseases (NIAID). B.E.M. was supported in part by NIH R01 AI047923.
C.A.A. has received lecture fees, research support, and consulting fees from Pfizer, Inc., and Cubist and research support from Forest Pharmaceuticals and Theravance and has served as speaker for Novartis. B.E.M. received grant support from Johnson & Johnson, Astellas, Palumed, Intercell, and Cubist and has served as consultant for Astellas (Theravance), Cubist, Targanta Therapeutics (now owned by The Medicines Company), Pfizer, The Medicines Company, Rib-X, and Durata Therapeutics. J.M.M, T.T.T., L.D., D.P., and J.R. declare that we have no conflicts of interest.
Footnotes
Published ahead of print 18 March 2013
REFERENCES
- 1. Galloway-Peña J, Roh JH, Latorre M, Qin X, Murray BE. 2012. Genomic and SNP analyses demonstrate a distant separation of the hospital and community-associated clades of Enterococcus faecium. PLoS One 7:e30187 doi:10.1371/journal.pone.0030187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arias CA, Contreras GA, Murray BE. 2010. Management of multidrug resistant enterococcal infections. Clin. Microbiol. Infect. 16:555–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hall AD, Steed ME, Arias CA, Murray BE, Rybak MJ. 2012. Evaluation of standard- and high-dose daptomycin versus linezolid against vancomycin-resistant Enterococcus isolates in an in vitro pharmacokinetic/pharmacodynamic model with simulated endocardial vegetations. Antimicrob. Agents Chemother. 56:3174–3180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kelesidis T, Humphries R, Uslan DZ, Pegues D. 2012. De novo daptomycin-nonsusceptible enterococcal infections. Emerg. Infect. Dis. 18:674–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lewis JS, II, Owens A, Cadena J, Sabol K, Patterson JE, Jorgensen JH. 2005. Emergence of daptomycin resistance in Enterococcus faecium during daptomycin therapy. Antimicrob. Agents Chemother. 49:1664–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Munoz-Price LS, Lolans K, Quinn JP. 2005. Emergence of resistance to daptomycin during treatment of vancomycin-resistant Enterococcus faecalis infection. Clin. Infect. Dis. 41:565–566 [DOI] [PubMed] [Google Scholar]
- 7. Arias CA, Panesso D, McGrath DM, Qin X, Mojica MF, Miller C, Diaz L, Tran TT, Rincon S, Barbu EM, Reyes J, Roh JH, Lobos E, Sodergren E, Pasqualini R, Arap W, Quinn JP, Shamoo Y, Murray BE, Weinstock GM. 2011. Genetic basis for in vivo daptomycin resistance in enterococci. N. Engl. J. Med. 365:892–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Munita JM, Panesso D, Diaz L, Tran TT, Reyes J, Wanger A, Murray BE, Arias CA. 2012. Correlation between mutations in liaFSR of Enterococcus faecium and MIC of daptomycin: revisiting daptomycin breakpoints. Antimicrob. Agents Chemother. 56:4354–4359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Arias CA, Torres HA, Singh KV, Panesso D, Moore J, Wanger A, Murray BE. 2007. Failure of daptomycin monotherapy for endocarditis caused by an Enterococcus faecium strain with vancomycin-resistant and vancomycin-susceptible subpopulations and evidence of in vivo loss of the vanA gene cluster. Clin. Infect. Dis. 45:1343–1346 [DOI] [PubMed] [Google Scholar]
- 10. Schwartz BS, Ngo PD, Guglielmo BJ. 2008. Daptomycin treatment failure for vancomycin-resistant Enterococcus faecium infective endocarditis: impact of protein binding? Ann. Pharmacother. 42:289–290 [DOI] [PubMed] [Google Scholar]
- 11. Eldholm V, Gutt B, Johnsborg O, Brückner R, Maurer P, Hakenbeck R, Mascher T, Håvarstein LS. 2010. The pneumococcal cell envelope stress-sensing system LiaFSR is activated by murein hydrolases and lipid II-interacting antibiotics. J. Bacteriol. 192:1761–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Suntharalingam P, Senadheera MD, Mair RW, Lévesque CM, Cvitkovitch DG. 2009. The LiaFSR system regulates the cell envelope stress response in Streptococcus mutans. J. Bacteriol. 191:2973–2984 [DOI] [PMC free article] [PubMed] [Google Scholar]