Abstract
Gram-positive bacteria naturally produce extracellular vesicles. However, little is known regarding the functions of Gram-positive bacterial extracellular vesicles, especially in the bacterial community. Here, we investigated the role of Staphylococcus aureus extracellular vesicles in interbacterial communication to cope with antibiotic stress. We found that S. aureus liberated BlaZ, a β-lactamase protein, via extracellular vesicles. These extracellular vesicles enabled other ampicillin-susceptible Gram-negative and Gram-positive bacteria to survive in the presence of ampicillin. However, S. aureus extracellular vesicles did not mediate the survival of tetracycline-, chloramphenicol-, or kanamycin-susceptible bacteria. Moreover, S. aureus extracellular vesicles did not contain the blaZ gene. In addition, the heat-treated S. aureus extracellular vesicles did not mediate the survival of ampicillin-susceptible bacteria. The β-lactamase activities of S. aureus soluble and extracellular vesicle-associated BlaZ were similar, but only the extracellular vesicle-associated BlaZ was resistant to protease digestion, which suggests that the enzymatic activity of BlaZ in extracellular vesicles is largely protected by the vesicle structure. Our observations provide evidence of the important role of S. aureus extracellular vesicles in antibiotic resistance, which allows the polymicrobial community to continue to evolve and prosper against antibiotics.
INTRODUCTION
Bacteria secrete diverse extracellular factors to communicate and to coordinate population behaviors. In addition to releasing soluble proteins and other molecules, many Gram-negative bacteria secrete extracellular vesicles (EVs), otherwise termed outer membrane vesicles or membrane vesicles (1–3). EVs are defined as spherical, bilayered proteolipids with an average diameter of 20 to 200 nm; they are composed of proteins, lipids, lipopolysaccharides, genetic materials, and other factors associated with virulence (4). These vesicles have been proposed to play diverse roles, including the transfer of proteins and genes between bacterial cells, delivery of toxins to host cells, cell-to-cell signaling, and biofilm formation (5–7).
Previous studies have reported that Gram-negative bacterial EVs contain β-lactamase, a periplasmic antibiotic-resistant protein (8–10). Moreover, Pseudomonas aeruginosa EVs containing β-lactamase were biologically active and degraded β-lactams in vitro (9). Thus, Gram-negative bacterial EVs act as transport vehicles to traffic antibiotic-resistant proteins from one bacterial cell to another (4). However, the underlying mechanisms of antibiotic resistance and the roles of EVs in multibacterial populations have not been elucidated in detail.
We recently reported for the first time that Gram-positive bacteria, including Staphylococcus aureus and Bacillus subtilis, naturally produce EVs during growth (11). This observation supports the notion that the secretion of EVs is an evolutionally conserved secretory pathway found in all bacterial species (11). However, in contrast to Gram-negative bacterial EVs, little is yet known about Gram-positive EVs. After the report on the secretion and proteome of S. aureus EVs (11), several groups, including ours examined the roles of Gram-positive bacterial EVs in the delivery of toxins, the induction of atopic dermatitis-like skin inflammation, the initiation of host cell death, and the induction of neutrophilic pulmonary inflammation (12–15). Although these studies support the important roles of Gram-positive bacterial EVs for intercellular communication, all have focused on functions in host-bacterium interactions. No studies have reported on the physiological roles of Gram-positive bacterial EVs in interbacterial communication. Our previous proteomic analysis showed that S. aureus ATCC 14458-derived EVs contained BlaZ, a β-lactamase protein (11). We therefore speculated that these S. aureus EVs may carry biologically active β-lactamase, which may work to protect the entire polymicrobial community.
In the present study, we showed that S. aureus packaged BlaZ into bacterium-free EVs. In addition, we investigated whether S. aureus EVs enabled other ampicillin-susceptible Gram-negative and Gram-positive bacteria to survive in the presence of ampicillin. S. aureus EVs were also tested whether they contained a β-lactamase-encoding gene, as well as whether they were resistant to heat or proteinase K. We further compared the β-lactamase activities of S. aureus soluble and EV-associated BlaZ.
MATERIALS AND METHODS
Bacteria and plasmids.
S. aureus ATCC 14458, S. aureus RN6390, and Staphylococcus epidermidis ATCC 12228 were grown in nutrient liquid medium (0.5% peptone, 0.3% meat extracts [pH 7.0]) at 37°C. Escherichia coli DH5α, Salmonella enterica serovar Enteritidis (ATCC 4931), and E. coli O157:H7 FRIK2000 (ATCC BAA-1883) were cultured in lysogeny broth (LB) medium (1% tryptone, 0.5% yeast extract, 1% sodium chloride [pH 7.0]) or LB agar plate at 37°C. Plasmids pMSCV and pDsRed (Clontech, Palo Alto, CA) were used to transform E. coli DH5α.
Purification of bacterial EVs.
EVs were purified from S. aureus culture supernatant, as previously described (11), with some modifications. Briefly, S. aureus was grown in nutrient liquid medium up to A600 of 1.0 at 37°C with gentle shaking (150 rpm). After the removal of cells by centrifugation at 10,000 × g for 20 min, the culture supernatant was filtered through a 0.45-μm-pore-size filter, and the filtrate was concentrated by ultrafiltration (QuixStand; Amersham Biosciences, Piscataway, NJ) with a 100-kDa hollow fiber membrane (Amersham Biosciences). The retentate was again filtered through a 0.22-μm-pore-size filter to remove any remaining cells and cell debris. Thereafter, the resulting filtrate was subjected to ultracentrifugation at 150,000 × g for 3 h at 4°C. The resulting EVs were diluted in phosphate-buffered saline (PBS). The total protein concentration of the EVs was determined with Bradford assay (Bio-Rad Laboratories, Hercules, CA). EVs were also purified from E. coli DH5α transformed with pMSCV or pDsRed by the same procedure.
Sequencing of blaZ gene of S. aureus ATCC 14458.
Using S. aureus ATCC 14458 total RNA as a template, the DNA for blaZ gene was amplified by reverse transcription-PCR using the sense primer (5′-CATGCCATGGGCATGAAAGAGTTAAATGATTTAG-3′) and antisense primer (5′-ACGCGTCGACAAATTCCTTCATTACACT-3′). The 774-bp blaZ DNA fragment was purified from agarose gels, digested with restriction enzymes NcoI and SalI (New England BioLabs, Beverly, MA), and ligated into NcoI and SalI sites of pET-28a(+) plasmid (Novagen, Madison, WI). The nucleotide sequence of the cloned blaZ DNA was sequenced, and we confirmed the blaZ gene of S. aureus ATCC 14458 as type A (16).
Purification of native BlaZ from S. aureus.
BlaZ, a β-lactamase, was purified from S. aureus ATCC 14458, as previously described (17), with some modifications. Briefly, S. aureus was grown in nutrient broth containing 100 μg of ampicillin/ml up to an A600 of 1.5 at 37°C. Cells were harvested, washed once quickly with water, and then washed three times with buffer A (20 mM triethanolamine, 300 mM sodium phosphate, 200 mM sodium chloride [pH 7.0]). Each wash with buffer A was pooled, filtered through a 0.22-μm pore-size filter, and then ultrafiltrated through a Centriprep YM10 membrane (Amicon; Millipore, Bedford, MA). BlaZ was purified by a combination of Mono S cation-exchange column (Amersham Biosciences) and Superdex-75 10/300 GL prepacked column (Amersham Biosciences). We obtained 1.0 mg of BlaZ from 13.0 mg in total protein of BlaZ-containing extracts prepared by washing the S. aureus ATCC 14458 cells with buffer A. The purity of the isolated BlaZ was verified by using SDS-PAGE gels stained with Coomassie brilliant blue (CBB) and mass spectrometry (MS)-based proteomics.
MS analysis of the purified S. aureus BlaZ.
The purified BlaZ (100 μg) was resolved in a digestion solution (6 M urea and 40 mM ammonium bicarbonate), reduced with 5 mM tris(2-carboxyethyl)phosphine hydrochloride, and subsequently alkylated with 25 mM iodoacetamide. The sample was digested in-solution with sequencing grade-modified trypsin (5 ng/μl; Promega, Madison, WI) for 16 h at 37°C. The tryptic peptides were separated using a homemade microcapillary C18 column and were then analyzed with an LTQ XL mass spectrometer equipped with a nano-ESI. The peptides were identified using SEQUEST version v.27 (Bioworks Browser Rev.3.1 SR1) (Thermo Fisher Scientific), matched to an S. aureus MW2 database (Uniprot Release 2011_06; 2,660 sequences).
Purification of polyclonal anti-BlaZ antibody.
Six-week-old female rabbits (Hyochang Science, Daegu, Republic of Korea) were immunized with the purified BlaZ (100 μg) emulsified in complete Freud adjuvant (Sigma, St. Louis, MO) for initial immunization and boosted on day 14 and 28 with the same amount of the purified BlaZ in incomplete Freund adjuvant (Sigma). On day 31, the rabbits were bled, and sera were recovered from the rabbit blood. The sera were dialyzed against 20 mM sodium phosphate (pH 7.0) overnight at 4°C and loaded on a HiTrap protein G column (Amersham Biosciences) in 20 mM sodium phosphate (pH 7.0). The adsorbed antibodies were eluted with 100 mM glycine-HCl (pH 2.7) and neutralized immediately with 1 M Tris-HCl (pH 9.0). The purified antibody was dialyzed against PBS overnight at 4°C.
SDS-PAGE and Western blotting.
Whole-cell lysates of S. aureus were prepared as described previously (18), with some modifications. Briefly, S. aureus cells were lysed with lysostaphin (20 μg/ml; Sigma) for 15 min at 37°C in Tris-EDTA buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]). The lysed cells were sonicated, and insoluble materials were removed by centrifugation at 8,000 × g for 10 min. Protein samples from whole-cell lysates, S. aureus EVs, and the purified BlaZ were analyzed with SDS-PAGE (12% resolving gel), as described previously (19). The proteins in the gel were transferred to a polyvinylidene difluoride membrane, and blocked with 3% skim milk (Difco, Detroit, MI) containing the control preimmune rabbit IgG. The blocked membrane was incubated with the lab-made polyclonal anti-BlaZ antibody and then incubated with goat anti-rabbit IgG-horseradish peroxidase (Santa Cruz Biotechnology, Santa Cruz, CA). The immunoreactive bands were visualized with a chemiluminescent substrate.
Measurement of bacterial survival.
EVs derived from S. aureus or E. coli were incubated with recipient bacteria in LB medium or LB agar plate containing antibiotics. Bacterial growth in the LB medium was measured by A600, whereas the growth on the LB agar plate was assessed by visible spot reactions of survived bacteria. Ampicillin, tetracycline, chloramphenicol, and kanamycin (Sigma) were used at 100, 10, 30, and 30 μg/ml, respectively.
PCR analysis.
S. aureus whole-cell lysates (1 μg) or EVs (1, 2, or 5 μg) were used as templates of PCR using the blaZ primer set, 5′-GCGATAAATAGTGCTATT-3′ and 5′-TCGTAAAAATGACTAAAAC-3′. The reactions were carried out in a volume of 50 μl, in which the initial Taq activation for 10 min at 94°C was followed by 30 cycles of denaturation for 1 min at 94°C, primer annealing for 1 min at 38°C, and extension for 1 min at 72°C.
Heat treatment and proteinase K digestion of S. aureus EVs and BlaZ.
The purified S. aureus EVs (1 μg) and purified BlaZ (0.02 μg) were subjected to heat treatment and proteinase K digestion. For heat treatment, S. aureus EVs and BlaZ were heated for 20 min at 100°C and cooled on ice. For proteinase K digestion, S. aureus EVs and BlaZ were incubated for 10 min at 37°C in the presence of proteinase K (100 μg/ml) (Invitrogen, Carlsbad, CA). After the digestion, phenylmethylsulfonyl fluoride (Sigma; a proteinase K inhibitor) was added to a final concentration of 1 mM.
Quantification of β-lactamase activity.
β-Lactamase activity was measured spectrophotometrically using penicillin G (Sigma) as previously described (20). Briefly, the reaction mixture containing S. aureus EVs or BlaZ with 1 μM penicillin G was incubated in 20 mM sodium phosphate buffer (pH 7.0) at 25°C. The decrease in absorbance of the reaction mixture at 240 nm was recorded for 15 min. The reactions were linear for 15 min. The enzymatic activity of BlaZ was calculated from the initial rate of penicillin G hydrolysis: change of absorbance units/min (ΔA240/min).
Statistical analysis.
All experiments were performed in triplicates. The data were presented as mean values with standard deviation. Statistical significance was evaluated using Student t test and differences were considered significant at a P of <0.05.
RESULTS AND DISCUSSION
Identification of BlaZ in S. aureus EVs.
Our previous proteomic analysis showed that S. aureus ATCC 14458 EVs contained BlaZ, a β-lactamase protein (11). By DNA sequencing the blaZ gene, we confirmed that S. aureus ATCC 14458 expressed type A BlaZ (16). We further validated the presence of BlaZ in EVs with Western blotting. We first isolated the native type A BlaZ from S. aureus ATCC 14458 cells by the combination of cation-exchange chromatography and gel filtration. The purity of the isolated BlaZ (15 μg) was confirmed by SDS-PAGE with CBB stain: we observed only one protein band (data not shown). Furthermore, nano-LC-MS/MS analysis revealed that the purified protein was BlaZ and not a contaminant of any other proteins. Western blotting showed that polyclonal anti-BlaZ antibody specifically detected the isolated BlaZ (0.02 μg) and the BlaZ in S. aureus ATCC 14458 whole-cell lysates (5 μg) and in EVs (1 μg) (Fig. 1A). Taken together, these results indicate that S. aureus ATCC 14458 EVs carry BlaZ, a β-lactamase protein.
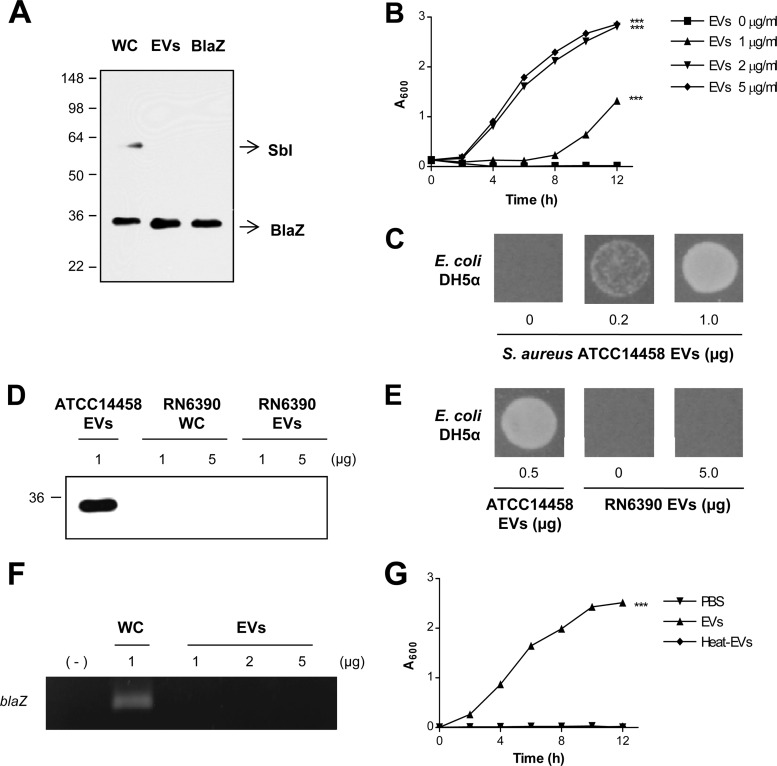
Fig 1.
S. aureus EVs carrying BlaZ mediated the survival of ampicillin-susceptible E. coli in the presence of ampicillin. (A) Western blotting of S. aureus ATCC 14458 whole-cell lysates (WC, 5 μg), EVs (1 μg), and the purified BlaZ (0.02 μg) detected with lab-made polyclonal anti-BlaZ antibodies. Molecular mass standards are indicated on the left (in kDa). (B) Dose-dependent effect of S. aureus ATCC 14458 EVs (0 to 5 μg/ml) on the growth of ampicillin-susceptible E. coli DH5α in LB-ampicillin (100 μg/ml) medium. ***, P < 0.001. (C) Dose-dependent effect of S. aureus ATCC 14458 EVs (0 to 1.0 μg) on the growth of ampicillin-susceptible E. coli DH5α on LB-ampicillin (100 μg/ml) agar plate. (D) Western blotting of S. aureus RN6390 whole-cell lysates (WC; 1 and 5 μg) and EVs (1 and 5 μg) detected with lab-made polyclonal anti-BlaZ antibodies. S. aureus ATCC 14458 EVs (1 μg) were used as a positive control. The molecular mass standard is indicated on the left (in kDa). (E) Effect of S. aureus RN6390 EVs (0 and 5 μg) on the growth of E. coli DH5α on LB-ampicillin (100 μg/ml) agar plate. S. aureus ATCC 14458 EVs (0.5 μg) were used as a positive control. (F) Determination of the presence of blaZ gene in S. aureus ATCC 14458 whole-cell lysates (WC, 1 μg) and EVs (1, 2, and 5 μg) by PCR. (G) Effect of intact and heat-treated S. aureus ATCC 14458 EVs (1 μg) on the growth of E. coli DH5α in LB-ampicillin (100 μg/ml) medium. ***, P < 0.001.
S. aureus EV-mediated survival of ampicillin-susceptible Gram-negative and Gram-positive bacteria in the presence of ampicillin.
To examine the role of S. aureus ATCC 14458 EV-associated BlaZ in antibiotic resistance, we incubated ampicillin-susceptible E. coli DH5α with the EVs in the presence of ampicillin. S. aureus ATCC 14458 EVs mediated the survival of E. coli DH5α in LB-ampicillin (100 μg/ml) medium in a dose-dependent manner (P < 0.001) (Fig. 1B). Application of S. aureus ATCC 14458 EVs directly to a lawn of E. coli DH5α in LB-ampicillin (100 μg/ml) agar plate produced a zone of survived bacteria (Fig. 1C). No colonies grew on LB and LB-ampicillin (100 μg/ml) agar plate with the isolated S. aureus ATCC 14458 EVs (data not shown), indicating that the purified EVs were not contaminated with any bacteria. S. aureus RN6390, which is susceptible to ampicillin, however, did not contain BlaZ in either whole-cell lysates or EVs (Fig. 1D). Consequently, S. aureus RN6390 EVs did not mediate survival of E. coli against ampicillin exposure (Fig. 1E). Similar to S. aureus ATCC 14458 EVs (11), transmission electron microscopy and dynamic light-scattering analyses showed that S. aureus RN6390 EVs were closed vesicular forms, and the diameter of purified EVs ranged from 20 to 100 nm, respectively. S. aureus ATCC 14458 EVs did not contain the blaZ gene (Fig. 1F). Moreover, heat-treated S. aureus EVs did not mediate the survival of E. coli DH5α in LB-ampicillin (100 μg/ml) medium (Fig. 1G). The isolated S. aureus BlaZ was heat labile because its enzymatic activity was lost by heat treatment (data not shown). These observations suggest that the major factor for EV-mediated ampicillin resistance should be heat-labile protein(s) or other factor(s).
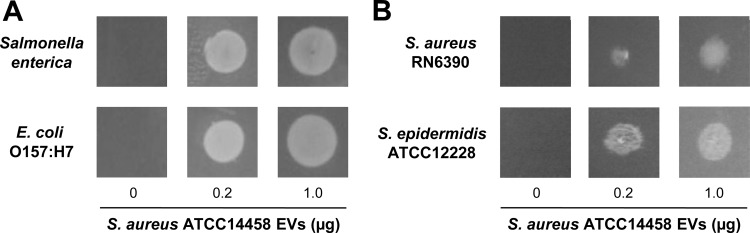
S. aureus ATCC 14458 EV-mediated bacterial survival was not confined to E. coli DH5α. S. aureus ATCC 14458 EVs mediated the survival of ampicillin-susceptible bacteria, including Gram-negative bacteria S. enterica serovar Enteritidis and E. coli O157:H7 (Fig. 2A), as well as Gram-positive bacteria S. aureus RN6390 and S. epidermidis ATCC 12228 (Fig. 2B), on LB-ampicillin (100 μg/ml) agar plate. These observations suggest that S. aureus ATCC 14458 EVs carrying BlaZ can mediate the survival of diverse ampicillin-susceptible Gram-negative and Gram-positive bacteria in the presence of ampicillin.
Fig 2.
S. aureus EVs mediated the survival of ampicillin-susceptible Gram-negative and Gram-positive bacteria in the presence of ampicillin. (A) Dose-dependent effect of S. aureus ATCC 14458 EVs (0 to 1 μg) on the growth of S. enterica serovar Enteritidis and E. coli O157:H7 on LB-ampicillin (100 μg/ml) agar plate. (B) Dose-dependent effect of S. aureus ATCC 14458 EVs (0 to 1 μg) on the growth of S. aureus RN6390 and S. epidermidis ATCC 12228 on LB-ampicillin (100 μg/ml) agar plate.
S. aureus EV-mediated resistance against other antibiotics.
To investigate the role of S. aureus EVs in other antibiotic resistance, we examined the survival of E. coli DH5α on LB agar plate containing either tetracycline (10 μg/ml), chloramphenicol (30 μg/ml), or kanamycin (30 μg/ml). The proteins involved in the resistance to tetracycline, chloramphenicol, and kanamycin are tetracycline resistance protein, chloramphenicol acetyltransferase, and aminoglycoside phosphotransferase, respectively (21–23). Although S. aureus ATCC 14458 cells were resistant to tetracycline, chloramphenicol, and kanamycin (data not shown), their EVs did not mediate the survival of E. coli grown on LB agar plate containing these antibiotics (Fig. 3A). These results are consistent with previous proteomic data indicating that S. aureus EVs contained no resistance determinants other than BlaZ (11).
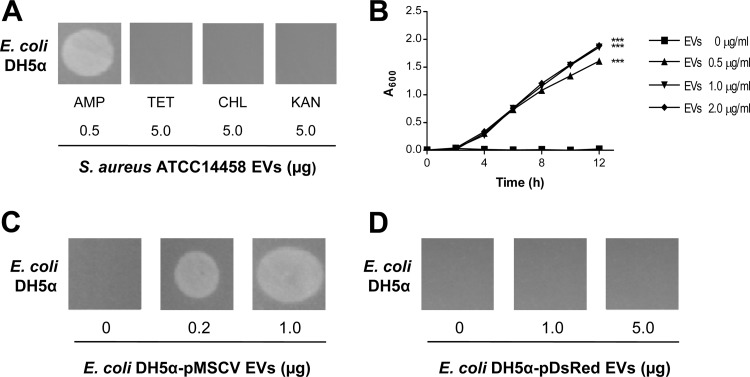
Fig 3.
S. aureus EVs did not mediate resistance to other antibiotics. (A) Effect of S. aureus ATCC 14458 EVs (5.0 μg) on the growth of E. coli DH5α on LB agar plate with tetracycline (TET, 10 μg/ml), chloramphenicol (CHL, 30 μg/ml), or kanamycin (KAN, 30 μg/ml). The effect of S. aureus ATCC 14458 EVs (0.5 μg) on the growth of E. coli DH5α on LB agar plate with ampicillin (AMP; 100 μg/ml) was used as a positive control. (B) Dose-dependent effect of EVs (0 to 2 μg/ml) derived from E. coli DH5α transformed with pMSCV (a plasmid with an ampicillin-resistant gene) on the growth of E. coli DH5α in LB-ampicillin (100 μg/ml) medium. ***, P < 0.001. (C) Dose-dependent effect of EVs (0 to 1 μg) derived from E. coli DH5α transformed with pMSCV (a plasmid with an ampicillin-resistant gene) on the growth of E. coli DH5α on LB-ampicillin (100 μg/ml) agar plate. (D) Dose-dependent effect of EVs (0 to 5 μg) derived from E. coli DH5α transformed with pDsRed (a plasmid with a kanamycin-resistant gene) on the growth of E. coli DH5α on LB-kanamycin (30 μg/ml) agar plate.
To further confirm the role of bacterial EVs in antibiotic resistance, we isolated EVs from E. coli DH5α transformed with the ampicillin-resistant plasmid pMSCV and the kanamycin-resistant plasmid pDsRed, which encode TEM-1 β-lactamase (24) and aminoglycoside phosphotransferase (25), respectively. E. coli DH5α-pMSCV EVs mediated the survival of ampicillin-susceptible E. coli DH5α on LB-ampicillin (100 μg/ml) agar plate in a dose-dependent manner (P < 0.001) (Fig. 3B and C). However, E. coli DH5α-pDsRed EVs did not mediate the survival of kanamycin-susceptible E. coli DH5α on LB-kanamycin (30 μg/ml) agar plate (Fig. 3D).
Enzymatic activities of S. aureus soluble and EV-associated BlaZ.
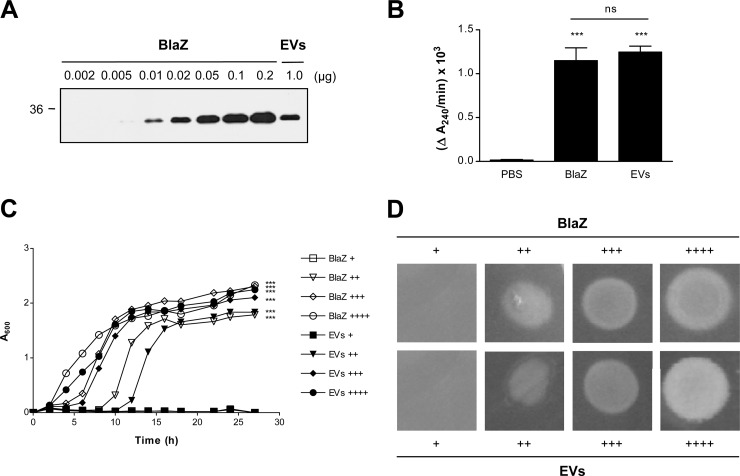
We next quantified the amount of BlaZ in S. aureus EVs by Western blotting (26). Three independent experiments showed that 1.0 μg of S. aureus EVs in total protein contained 0.02 ± 0.001 μg of BlaZ (Fig. 4A). We next determined the β-lactamase activity (ΔA240/min) by measuring the initial hydrolysis rate of penicillin (20). No significant difference (P > 0.05) was observed in the β-lactamase activity between soluble and EV-associated BlaZ (Fig. 4B): the β-lactamase activities of soluble BlaZ (0.02 μg/ml) and S. aureus EVs (1 μg/ml in total protein concentration) were 1.15 × 10−3 and 1.25 × 10−3 ΔA240/min, respectively. Moreover, soluble and EV-associated BlaZ similarly increased the survival of ampicillin-susceptible E. coli DH5α both in LB-ampicillin (100 μg/ml) medium (Fig. 4C) and in LB-ampicillin (100 μg/ml) agar plate (Fig. 4D). These results suggest that the enzymatic activity of BlaZ present in S. aureus EVs is similar to that of soluble BlaZ.
Fig 4.
Enzymatic activities of S. aureus soluble and EV-associated BlaZ were similar. (A) Western blotting of the purified S. aureus ATCC 14458 soluble BlaZ (0.002 to 0.2 μg) and EVs (1.0 μg) detected with lab-made polyclonal anti-BlaZ antibodies. The molecular mass standard is indicated on the left (in kDa). Note that 1.0 μg of S. aureus EVs contained 0.02 ± 0.001 μg of BlaZ. (B) β-Lactamase activities of the purified S. aureus ATCC 14458 soluble BlaZ (0.02 μg/ml) and EVs (1.0 μg/ml). The activities were the initial rates of hydrolysis of penicillin G (1 μM) for 15 min and expressed in change of absorbance units/min (ΔA240/min). ***, P < 0.001; ns, not significant. (C and D) Comparison of the dose-dependent effects of S. aureus ATCC 14458 soluble BlaZ and EVs on the growth of ampicillin-susceptible E. coli DH5α in the presence of ampicillin. The concentrations of soluble BlaZ and EVs in LB-ampicillin (100 μg/ml) medium (μg/ml) were as follows in panel C: +, 0.002/0.1; ++, 0.004/0.2; +++, 0.01/0.5; and ++++, 0.02/1.0. The amounts of soluble BlaZ and EVs on LB-ampicillin (100 μg/ml) agar plate (μg) were as follows in panel D: +, 0.0004/0.02; ++, 0.001/0.05; +++, 0.002/0.1; and ++++, 0.01/0.5. ***, P < 0.001.
Several studies have reported that EV-associated proteins exhibit greater activity than soluble forms. For instance, Helicobacter pylori EV-associated toxin (VacA) caused significantly greater vacuolating activity than the soluble form (27), and enteropathogenic E. coli EV-associated toxin (ClyA) mediated higher cytotoxic activity in host cells than the soluble form (10). Moreover, in mammalian EVs, known as exosomes and microvesicles, vesicle-associated proteins such as adhesion molecule and tumor antigen exert greater immune modulating activity than the soluble forms of the proteins (28, 29). However, such is not always the case, particularly for proteins with enzymatic activity. In our study, the enzymatic activity of S. aureus EV-associated β-lactamase was similar to that of soluble β-lactamase. Consistent with this result, enzymes such as murine hydrolase exhibit similar activities inside EVs as in the soluble form (30, 31). This difference may be caused by different protein locations in the EVs; unlike surface-exposed proteins, proteins present inside the EVs may not exhibit greater activity. Examining the differences in protein activities based on their locations in EVs will be a valuable aspect.
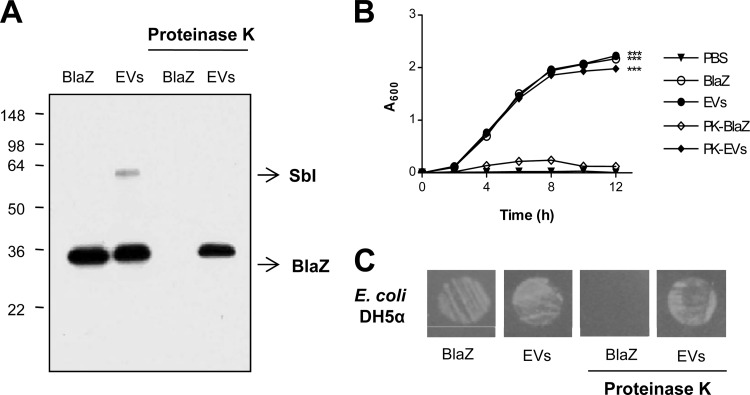
Proteinase K resistance of S. aureus EV-associated BlaZ.
To determine the location of BlaZ in S. aureus EVs, soluble BlaZ and EVs were subjected to proteolysis by proteinase K. Soluble BlaZ was digested by proteinase K treatment, while the EV-associated BlaZ was resistant to proteinase K, as confirmed by Western blotting (Fig. 5A). SbI present on the surface of S. aureus EVs (11), however, was digested by proteinase K. These results indicate that BlaZ is mainly located inside S. aureus EVs. Further, proteinase K-digested soluble BlaZ could not mediate the survival of ampicillin-susceptible E. coli DH5α in the presence of ampicillin (Fig. 5B and C). However, intact S. aureus EVs and proteinase K-digested S. aureus EVs mediated the survival of ampicillin-susceptible E. coli DH5α similarly in LB-ampicillin (100 μg/ml) medium (P < 0.001) (Fig. 5B) and on an LB-ampicillin (100 μg/ml) agar plate (Fig. 5C). Collectively, these results indicate that BlaZ in S. aureus EVs might be resistant to proteolysis by proteinase K. Thus, S. aureus EVs might mediate the survival of susceptible bacteria against β-lactam exposure even under protease-rich pathophysiological conditions (32).
Fig 5.
S. aureus EV-associated BlaZ was resistant to proteinase K treatment. (A) Western blotting of the purified S. aureus ATCC 14458 soluble BlaZ and EVs, which were digested with proteinase K, detected with lab-made polyclonal anti-BlaZ antibodies. SbI indicates the band of IgG-binding protein SbI. Molecular weight standards are indicated on the left (kDa). (B) Effect of intact and proteinase K (PK)-treated S. aureus ATCC 14458 soluble BlaZ and EVs on the growth of ampicillin-susceptible E. coli DH5α in LB-ampicillin (100 μg/ml) medium. ***, P < 0.001. (C) Effect of intact and proteinase K-treated S. aureus ATCC 14458 soluble BlaZ and EVs on the growth of ampicillin-susceptible E. coli DH5α on LB-ampicillin (100 μg/ml) agar plate.
Concluding remarks.
Gram-negative bacterial EVs play many important roles, such as in the transfer of proteins and genetic materials, cell-to-cell signaling, and the formation of biofilm in polymicrobial communities (4, 5). However, no prior studies had reported on the role of Gram-positive bacterial EVs in interbacterial communication. In the present study, we showed that S. aureus EVs carried biologically active BlaZ, a β-lactamase protein, and these EVs enabled other ampicillin-susceptible Gram-negative and Gram-positive bacteria to survive in the presence of ampicillin. This is the first report demonstrating that Gram-positive bacterial EVs play functional roles in interbacterial communication like that of Gram-negative bacteria. Currently, however, limited data are available on Gram-positive bacterial EVs. We hope to stimulate further in-depth biochemical and microbiological studies in the field of Gram-positive bacterial EVs, like the rapid progress seen in studies of Gram-negative bacterial vesicles, to advance both basic and clinical sciences.
ACKNOWLEDGMENTS
This study was supported by a grant from the Korean Ministry of Education, Science, and Technology, grant FPR08B1-240 from the 21C Frontier Functional Proteomics Program, and by National Research Foundation of Korea grants funded by the Korea government (MEST; no. 20120005634 and no. 20110000215).
We thank Gwang-Pil Park for excellent assistance with writing in English.
Footnotes
Published ahead of print 25 March 2013
REFERENCES
- 1. Beveridge TJ. 1999. Structures of Gram-negative cell walls and their derived membrane vesicles. J. Bacteriol. 181:4725–4733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lee EY, Bang JY, Park GW, Choi DS, Kang JS, Kim HJ, Park KS, Lee JO, Kim YK, Kwon KH, Kim KP, Gho YS. 2007. Global proteomic profiling of native outer membrane vesicles derived from Escherichia coli. Proteomics 7:3143–3153 [DOI] [PubMed] [Google Scholar]
- 3. Lee EY, Choi DS, Kim KP, Gho YS. 2008. Proteomics in Gram-negative bacterial outer membrane vesicles. Mass Spectrom. Rev. 27:535–555 [DOI] [PubMed] [Google Scholar]
- 4. Mashburn-Warren LM, Whiteley M. 2006. Special delivery: vesicle trafficking in prokaryotes. Mol. Microbiol. 61:839–846 [DOI] [PubMed] [Google Scholar]
- 5. Kuehn MJ, Kesty NC. 2005. Bacterial outer membrane vesicles and the host-pathogen interaction. Genes Dev. 19:2645–2655 [DOI] [PubMed] [Google Scholar]
- 6. Mashburn LM, Whiteley M. 2005. Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature 437:422–425 [DOI] [PubMed] [Google Scholar]
- 7. Schooling SR, Beveridge TJ. 2006. Membrane vesicles: an overlooked component of the matrices of biofilms. J. Bacteriol. 188:5945–5957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bomberger JM, Maceachran DP, Coutermarsh BA, Ye S, O'Toole GA, Stanton BA. 2009. Long-distance delivery of bacterial virulence factors by Pseudomonas aeruginosa outer membrane vesicles. PLoS Pathog. 5:e1000382 doi:10.1371/journal.ppat.1000382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ciofu O, Beveridge TJ, Kadurugamuwa J, Walther-Rasmussen J, Hoiby N. 2000. Chromosomal beta-lactamase is packaged into membrane vesicles and secreted from Pseudomonas aeruginosa. J. Antimicrob. Chemother. 45:9–13 [DOI] [PubMed] [Google Scholar]
- 10. Wai SN, Lindmark B, Soderblom T, Takade A, Westermark M, Oscarsson J, Jass J, Richter-Dahlfors A, Mizunoe Y, Uhlin BE. 2003. Vesicle-mediated export and assembly of pore-forming oligomers of the enterobacterial ClyA cytotoxin. Cell 115:25–35 [DOI] [PubMed] [Google Scholar]
- 11. Lee EY, Choi DY, Kim DK, Kim JW, Park JO, Kim S, Kim SH, Desiderio DM, Kim YK, Kim KP, Gho YS. 2009. Gram-positive bacteria produce membrane vesicles: proteomics-based characterization of Staphylococcus aureus-derived membrane vesicles. Proteomics 9:5425–5436 [DOI] [PubMed] [Google Scholar]
- 12. Gurung M, Moon DC, Choi CW, Lee JH, Bae YC, Kim J, Lee YC, Seol SY, Cho DT, Kim SI, Lee JC. 2011. Staphylococcus aureus produces membrane-derived vesicles that induce host cell death. PLoS One 6:e27958 doi:10.1371/journal.pone.0027958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hong SW, Kim MR, Lee EY, Kim JH, Kim YS, Jeon SG, Yang JM, Lee BJ, Pyun BY, Gho YS, Kim YK. 2011. Extracellular vesicles derived from Staphylococcus aureus induce atopic dermatitis-like skin inflammation. Allergy 66:351–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim MR, Hong SW, Choi EB, Lee WH, Kim YS, Jeon SG, Jang MH, Gho YS, Kim YK. 2012. Staphylococcus aureus-derived extracellular vesicles induce neutrophilic pulmonary inflammation via both Th1 and Th17 cell responses. Allergy 67:1271–1281 [DOI] [PubMed] [Google Scholar]
- 15. Rivera J, Cordero RJ, Nakouzi AS, Frases S, Nicola A, Casadevall A. 2010. Bacillus anthracis produces membrane-derived vesicles containing biologically active toxins. Proc. Natl. Acad. Sci. U. S. A. 107:19002–19007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Volardri RK, Kernodle DS. 1998. Characterization of a chromosomal gene encoding type B β-lactamase in phage group II isolates of Staphylococcus aureus. Antimicrob. Agents Chemother. 42:3163–3168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kernodle DS, Zygmunt DJ, McGraw PA, Chipley JR. 1990. Purification of Staphylococcus aureus beta-lactamases by using sequential cation-exchange and affinity chromatography. Antimicrob. Agents Chemother. 34:2177–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Scherl A, Francois P, Converset V, Bento M, Burgess JA, Sanchez JC, Hochstrasser DF, Schrenzel J, Corthals GL. 2004. Nonredundant mass spectrometry: a strategy to integrate mass spectrometry acquisition and analysis. Proteomics 4:917–927 [DOI] [PubMed] [Google Scholar]
- 19. Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 20. Coquet L, Junter GA, Jouenne T. 1998. Resistance of artificial biofilms of Pseudomonas aeruginosa to imipenem and tobramycin. J. Antimicrob. Chemother. 42:755–760 [DOI] [PubMed] [Google Scholar]
- 21. Nesin M, Svec P, Lupski JR, Godson GN, Kreiswirth B, Kornblum J, Projan SJ. 1990. Cloning and nucleotide sequence of a chromosomally encoded tetracycline resistance determinant, tetA(M), from a pathogenic, methicillin-resistant strain of Staphylococcus aureus. Antimicrob. Agents Chemother. 34:2273–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Horinouchi S, Weisblum B. 1982. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J. Bacteriol. 150:815–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gray GS, Fitch WM. 1983. Evolution of antibiotic resistance genes: the DNA sequence of a kanamycin resistance gene from Staphylococcus aureus. Mol. Biol. Evol. 1:57–66 [DOI] [PubMed] [Google Scholar]
- 24. Granieri L, Baret JC, Griffiths AD, Merten CA. 2010. High-throughput screening of enzymes by retroviral display using droplet-based microfluidics. Chem. Biol. 17:229–235 [DOI] [PubMed] [Google Scholar]
- 25. Azucena E, Mobashery S. 2001. Aminoglycoside-modifying enzymes: mechanisms of catalytic processes and inhibition. Drug Resist. Update 4:106–117 [DOI] [PubMed] [Google Scholar]
- 26. Charette SJ, Lambert H, Nadeau PJ, Landry J. 2010. Protein quantification by chemiluminescent Western blotting: elimination of the antibody factor by dilution series and calibration curve. J. Immunol. Methods 353:148–150 [DOI] [PubMed] [Google Scholar]
- 27. Ricci V, Chiozzi V, Necchi V, Oldani A, Romano M, Solcia E, Ventura U. 2005. Free-soluble and outer membrane vesicle-associated VacA from Helicobacter pylori: two forms of release, a different activity. Biochem. Biophys. Res. Commun. 337:173–178 [DOI] [PubMed] [Google Scholar]
- 28. Lee HM, Choi EJ, Kim JH, Kim TD, Kim YK, Kang C, Gho YS. 2010. A membranous form of ICAM-1 on exosomes efficiently blocks leukocyte adhesion to activated endothelial cells. Biochem. Biophys. Res. Commun. 397:251–256 [DOI] [PubMed] [Google Scholar]
- 29. Zeelenberg IS, Ostrowski M, Krumeich S, Bobrie A, Jancic C, Boissonnas A, Delcayre A, Le Pecq JB, Combadiere B, Amigorena S, Thery C. 2008. Targeting tumor antigens to secreted membrane vesicles in vivo induces efficient antitumor immune responses. Cancer Res. 68:1228–1235 [DOI] [PubMed] [Google Scholar]
- 30. Ellis TN, Kuehn MJ. 2010. Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol. Mol. Biol. Rev. 74:81–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kadurugamuwa JL, Beveridge TJ. 1996. Bacteriolytic effect of membrane vesicles from Pseudomonas aeruginosa on other bacteria including pathogens: conceptually new antibiotics. J. Bacteriol. 178:2767–2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rohde H, Burdelski C, Bartscht K, Hussain M, Buck F, Horstkotte MA, Knobloch JK, Heilmann C, Herrmann M, Mack D. 2005. Induction of Staphylococcus epidermidis biofilm formation via proteolytic processing of the accumulation-associated protein by staphylococcal and host proteases. Mol. Microbiol. 55:1883–1895 [DOI] [PubMed] [Google Scholar]