Abstract
The BD GeneOhm MRSA assay could identify methicillin-resistant Staphylococcus aureus (MRSA) strains at a high ratio (97.8%). Analysis of 11 assay-negative MRSA strains suggested that insertion of non-mec staphylococcal cassette chromosome elements (SCCs) downstream of orfX, and carriage of SCCmecs with a left extremity that cannot be detected by the kit, might lead to their being given an incorrect negative status.
TEXT
Widely used commercially available real-time PCR-based methicillin-resistant Staphylococcus aureus (MRSA) detection kits rely on amplification of DNA with a combination of primers identifying the chromosomal region of S. aureus and the left extremity of staphylococcal cassette chromosome mec elements (SCCmecs). The great advantage of the system is that it can discriminate MRSA strains from other methicillin-resistant staphylococcal species. Although the kit was evaluated by many researchers as a useful tool (1–4), some concerns have been reported (5–7). Here, we report evaluation of the BD GeneOhm MRSA assay using 484 MRSA strains. The BD GeneOhm MRSA assay was carried out as recommended by the manufacturer.
The rate of detection of MRSA strains was 97.8%, leaving 11 (2.2%) MRSA strains that could not be identified (see Table S1 in the supplemental material). To clarify the causes of the false-negative results, we determined nucleotide sequences of the region downstream of orfX of 11 strains using DNA fragments amplified by long-range PCRs and nested PCRs as the templates (see Table S2 in the supplemental material). Representative results are illustrated in Fig. 1.
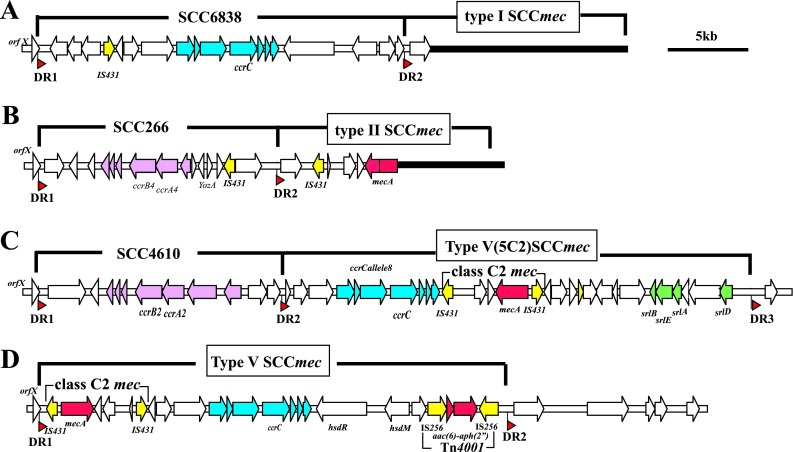
Fig 1.
Structures of SCCs and SCCmecs carried by BD GeneOhm MRSA assay-negative strains. Structures of elements are illustrated based on the nucleotide sequences deposited in the DDBJ/EMBL/GenBank databases as follows: panel A (JCSC6838), AB774373; panel B (J266), AB774374; panel C (JCSC4610), AB773816; panel D (JCSC7481), AB774378. Red arrowheads indicate the location of integration site sequences of SCC that comprise sequences of direct repeats (DRs). Open reading frames (ORFs) are colored as follows: yellow, transposase insertion sequences; red, resistance genes; green, ORFs related to sorbitol utilization; pink, ORFs in ccr gene complexes carrying ccrA and ccrB genes; blue, ORFs in ccr gene complexes carrying ccrC.
Type I SCCmec strain JCSC6838 possessed SCC6838 (22.8 kb) carrying ccrC (Fig. 1A). Type II SCCmec strain J266 possessed SCCJ266 (14.7 kb) carrying ccrA4 and ccrB4 (Fig. 1B). Type V SCCmec strain JCSC4610 possessed SCC4610 (15 kb) carrying ccrA2 and ccrB2. A type V SCCmec4610 (29.5 kb) was located just downstream of SCC4610. It is a novel subtype of type V SCCmec-encoding genes related to sorbitol utilization at the J3 region (Fig. 1C). Three other type V SCCmec strains were judged to carry SCC and SCCmec in a manner similar to that of JCSC4610 based on PCR experiments.
Five other strains carried SCCmecs; the left extremities of those five strains could not be detected by the kit. Four of them carried novel or variant versions of SCCmecs. Those strains consisted of a type I strain and two type II SCCmec strains carrying the same J3 region as that of the type X SCCmec identified in the ST398 strain and two type V SCCmec strains, WIS (WBD3813) and a ST772 Panton-Valentine leukocidin (PVL)-positive MRSA strain, JCSC7481. The type V SCCmec of JCSC7481 (29.2 kb) is distinct from that of WIS in the direction of the mec gene complex as well as in the structures of the J1 and J3 regions (Fig. 1D).
Our data showed that a failure of the assay to identify SCCmecs might be due to two causes: insertion of SCC (6 strains) and carriage of SCCmecs with a novel left extremity (5 strains). We have reported that three distinct SCCmecs can be identified by PCRs with primer pairs designed for the chromosomal region (orfX) and extremities of SCCmecs flanking orfX (formally called mec right extremity polymorphism [MREP] typing) (8). The kit can identify 32 MRSA strains that were negative in the MREP typing, suggesting that the BD GeneOhm MRSA assay has an improved ability to identify SCCmecs having various extremities. Ten assay-negative strains other than WIS were isolated rather recently in Japan. It seems that MRSA strains carrying novel structures of SCCmecs as well as SCCs downstream of orfX might be emerging currently, resulting in increases in false-negative results in the assays and suggesting that further improvement is required.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a Grant-in-Aid from the MEXT (Ministry of Education, Culture, Sports, Science and Technology)-Supported Program for the Strategic Research Foundation at Private Universities and by BD Diagnostics (Quebec, Canada).
Footnotes
Published ahead of print 9 April 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00174-13.
REFERENCES
- 1. Boyce JM, Havill NL. 2008. Comparison of BD GeneOhm methicillin-resistant Staphylococcus aureus (MRSA) PCR versus the CHROMagar MRSA assay for screening patients for the presence of MRSA strains. J. Clin. Microbiol. 46:350–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Boyle-Vavra S, Daum RS. 2010. Reliability of the BD GeneOhm methicillin-resistant Staphylococcus aureus (MRSA) assay in detecting MRSA isolates with a variety of genotypes from the United States and Taiwan. J. Clin. Microbiol. 48:4546–4551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hombach M, Pfyffer GE, Roos M, Lucke K. 2010. Detection of methicillin-resistant Staphylococcus aureus (MRSA) in specimens from various body sites: performance characteristics of the BD GeneOhm MRSA assay, the Xpert MRSA assay, and broth-enriched culture in an area with a low prevalence of MRSA infections. J. Clin. Microbiol. 48:3882–3887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lucke K, Hombach M, Hug M, Pfyffer GE. 2010. Rapid detection of methicillin-resistant Staphylococcus aureus (MRSA) in diverse clinical specimens by the BD GeneOhm MRSA assay and comparison with culture. J. Clin. Microbiol. 48:981–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen WT, Wang JT, Lee WS, Huang CH, Liao CH, Chen YC, Chang SC. 2010. Performance of the BD GeneOhm methicillin-resistant Staphylococcus aureus (MRSA) PCR assay for detecting MRSA nasal colonization in Taiwanese adults. J. Microbiol. Immunol. Infect. 43:372–377 [DOI] [PubMed] [Google Scholar]
- 6. Farley JE, Stamper PD, Ross T, Cai M, Speser S, Carroll KC. 2008. Comparison of the BD GeneOhm methicillin-resistant Staphylococcus aureus (MRSA) PCR assay to culture by use of BBL CHROMagar MRSA for detection of MRSA in nasal surveillance cultures from an at-risk community population. J. Clin. Microbiol. 46:743–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Blanc DS, Basset P, Nahimana-Tessemo I, Jaton K, Greub G, Zanetti G. 2011. High proportion of wrongly identified methicillin-resistant Staphylococcus aureus carriers by use of a rapid commercial PCR assay due to presence of staphylococcal cassette chromosome element lacking the mecA gene. J. Clin. Microbiol. 49:722–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ito T, Katayama Y, Asada K, Mori N, Tsutsumimoto K, Tiensasitorn C, Hiramatsu K. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1323–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.