Abstract
Two homosexual men were colonized in the urethra with Haemophilus parainfluenzae nonsusceptible to ampicillin (MIC, 8 μg/ml), amoxicillin-clavulanate (MIC, 4 μg/ml), cefotaxime (MIC, 1.5 μg/ml), cefepime (MIC, 3 μg/ml), meropenem (MIC, 0.5 μg/ml), cefuroxime, azithromycin, ciprofloxacin, tetracycline, and chloramphenicol (all MICs, ≥32 μg/ml). Repetitive extragenic palindromic PCR (rep-PCR) showed that the strains were indistinguishable. The isolates had amino acid substitutions in PBP3, L4, GyrA, and ParC and possessed Mef(A), Tet(M), and CatS resistance mechanisms. This is the first report of extensively drug-resistant (XDR) H. parainfluenzae.
TEXT
Haemophilus influenzae isolates producing TEM-1/ROB-1 β-lactamases and/or with altered PBP3 (β-lactamase negative ampicillin resistant [BLNAR]) are nowadays increasingly reported. This species can also be multidrug resistant (MDR) due to expression of coassociated mechanisms involving quinolones, macrolides, trimethoprim-sulfamethoxazole, tetracyclines, and chloramphenicol (1–3). In contrast, little attention has been paid to Haemophilus parainfluenzae. Only a few BLNAR and/or TEM-producing isolates have been described (4, 5), and those with resistance to quinolones are rarely reported (6, 7). To our knowledge, MDR H. parainfluenzae isolates (e.g., simultaneously resistant to β-lactams, quinolones, and macrolides) have never been found.
In April 2012, a homosexual man was treated with penicillin intramuscularly for primary syphilis. Four months later, he presented with urethritis. A pansusceptible Neisseria gonorrhoeae strain (numerous colonies) of sequence type (ST) 7616 (www.ng-mast.net) and an H. parainfluenzae strain (few colonies) were isolated from the urethral swab, implementing blood and chocolate agar plates. The infection was treated empirically with ciprofloxacin, leading to complete resolution of symptoms. Retrospectively, the H. parainfluenzae strain (AE-2096513) was considered a colonizer, but its very unusual MDR phenotype observed using the standardized disk diffusion method drew our attention (http://www.eucast.org/mic_distributions/.)
Species identification was routinely achieved by indole test (negative) and direct analysis of colonies by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (Bruker Daltonics). It was also confirmed by sequencing of the 16S rRNA (MicroSeq 500; Applied Biosystem). Production of β-lactamases was initially tested using the Cefinase paper disc (BBL) and further evaluated by assessing the hydrolytic activity against nitrocefin (100 μM) for 1 h. Although it is not a standardized methodology, the Etest (bioMérieux) was used to obtain the MICs for antibiotics. Mueller-Hinton agar plus 5% defibrinated horse blood and 20-mg/liter β-NAD plates (Oxoid) were used, and results were interpreted according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) criteria (8).
Genomic extraction was performed using the QIAamp DNA minikit (Qiagen). Two microarray platforms (AMR-ve 0.5m and AMR+ve-3; Alere Technologies) were implemented to detect numerous antibiotic resistance genes (9). Some of them may potentially be present in Haemophilus spp., including the following (targeted antibiotics): blaTEM (β-lactams); qnrA/B/S, qepA, and aac6-Ib-cr (quinolones); erm(A)/(B)/(C)/(D)/(F) and mef(A) (macrolides); tet(A)/(B)/(C)/(D)/(E)/(G)/(K)/(M) (tetracyclines); dfrA/D/G/K and sul1/2/3 (trimethoprim-sulfamethoxazole); and catA/B/D/P/Q/S/III, fexA, floR, and cmlA1 (chloramphenicol) (2, 3). To amplify and sequence further resistance traits (targeted antibiotics), previously reported or designed primers were also used: the PBP3 transpeptidase domain of ftsI (5), blaTEM (primers TEM-F1 [5′-CGTGTCGCCCTTATTCCC-3′] and TEM-B1 [5′-AGGCACCTATCTCAGCGATC-3′]), the blaTEM promoter region (primers proTEM-F1 [5′-AATTCTTGAAGACGAAAGGG-3′] and proTEM-R2 [5′-CGCTGTTGAGATCCAGTTCG-3′]), and blaROB (β-lactams) (10); the quinolone resistance-determining region of gyrA and parC (quinolones) (6); rplD and rplV (L4 and L22 proteins, respectively) of the 50S ribosomal subunit (11) and mef(A) (macrolides) (12); tet(M) (tetracyclines) (13); catD, catP, and catS (primers catDPS-F and catDPS-R [9], catHP-F [5′-GAGATGATGCAGCCTTTG-3′], and catHP-R [5′-AGTCCGACAAACTGGAAG-3′]) (chloramphenicol). Presence of mutations in the copies of the 23S rRNA (resistance to macrolides) was assessed by amplifying the genes from base 1902 to base 2956 (Escherichia coli numbering) (primers 23S-3 [11] and 23S-R [5′-CCGCCAGGATTATTCCTTTA-3′]), cloning them with the TOPO XL PCR kit (Invitrogen), and sequencing five randomly selected colonies. Results for all of the above-mentioned genes were compared to the deposited genome of H. parainfluenzae T3T1 (GenBank accession number NC_015964) or homologies were searched for in BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Repetitive extragenic palindromic PCR (rep-PCR) was performed as previously described and interpreted using the Agilent Bioanalyzer (14).
As shown in Table 1, the H. parainfluenzae isolate was resistant to β-lactams, macrolides, quinolones, tetracyclines, and chloramphenicol, leaving only trimethoprim-sulfamethoxazole and rifampin in the susceptible range. The MIC of meropenem (0.5 μg/ml) was interpreted as susceptible, though this value is defined as intermediate when the isolate causes meningitis (8). Thus, the H. parainfluenzae can be defined as an extensively drug-resistant (XDR) isolate (1).
Table 1.
Antimicrobial phenotypes and molecular mechanisms of resistance found in the two XDR H. parainfluenzae isolatesa
| Antimicrobial | MIC (μg/ml) and interpretationb | Molecular mechanism(s) of resistance |
|---|---|---|
| β-lactams | PBP3: Lys276Asn, Ala307Asn, Val329Ile, Ser385Thr, Ile442F, Val511Ala, Asn526Lys (TEM-1 not expressed) | |
| Ampicillin | 8, R | |
| Amoxicillin | 6, R | |
| Amoxicillin-clavulanate | 4, R | |
| Cefuroxime | 32, R | |
| Ceftriaxone | 0.25, R | |
| Cefotaxime | 1.5, R | |
| Cefepime | 3, R | |
| Meropenem | 0.5, S (I) | |
| Macrolides | Mef(A); L4: Ala69Serc | |
| Erythromycin | >256, R | |
| Clarithromycin | >256, R | |
| Azithromycin | >256, R | |
| Quinolones | GyrA: Ser84Phe, Asp88Tyr; ParC: Ser84Phe | |
| Ciprofloxacin | >32, R | |
| Levofloxacin | >32, R | |
| Tetracycline | 32, R | Tet(M) |
| Trimethoprim-sulfamethoxazole | 0.25, S | |
| Rifampin | 0.75, S | |
| Chloramphenicol | 96, R | CatS |
Since the isolates are indistinguishable, the phenotypic and molecular backgrounds of resistance are the same.
Interpretation of MICs according to the EUCAST criteria (S, susceptible; I, intermediate; R, resistant) (8): for ampicillin, cefuroxime, rifampin, clarithromycin, levofloxacin, and tetracycline, S is a MIC of ≤1 μg/ml; for amoxicillin, amoxicillin-clavulanate, and chloramphenicol, S is a MIC of ≤2 μg/ml; for ceftriaxone, cefotaxime, and azithromycin, S is a MIC of ≤0.12 μg/ml; for cefepime, S is a MIC of ≤0.25 μg/ml; for meropenem, S is a MIC of ≤2 μg/ml, but in cases of meningitis (interpretation indicated in parentheses), S is a MIC of ≤0.25 μg/ml and R is a MIC of ≥2 μg/ml; for erythromycin, ciprofloxacin, and trimethoprim-sulfamethoxazole, S is a MIC of ≤0.5 μg/ml.
The Ala69Ser substitution was observed after comparison with H. influenzae Rd but not with H. parainfluenzae T3T1.
Molecular analysis indicated that AE-2096513 was resistant to β-lactams and amoxicillin-clavulanate because of seven amino acid substitutions in the PBP3 domain (Table 1). A similar pattern of substitutions has been recently described in one BLNAR H. parainfluenzae isolate found in Spain (4). Remarkably, AE-2096513 carried blaTEM-1, but the β-lactamase was not expressed based on both nitrocefin tests implemented. This observation was partially supported by the finding that blaTEM-1 possessed a P3 promoter, which is recognized as the weakest in driving gene expression among those previously described (15).
High-level resistance to macrolides was attributed to the presence of the efflux-mediated resistance mechanism Mef(A) and to the Ala69Ser substitution in the L4 protein, a substitution previously associated with macrolide resistance in H. influenzae (11). The L22 protein did not contain substitutions. Two different copies of the 23S rRNA were cloned and sequenced (GenBank accession numbers KC559885 and KC559886), but previously reported mutations conferring macrolide resistance were not recorded (11, 16).
AE-2096513 was also highly resistant to quinolones due to the classic GyrA (Ser84Phe and Asp88Tyr) and ParC (Ser84Phe) amino acid substitutions (6). Finally, tetracycline and chloramphenicol resulted in resistance due to production of the ribosomal protective protein Tet(M) and the CatS acetyltransferase enzyme, respectively (Table 1) (13).
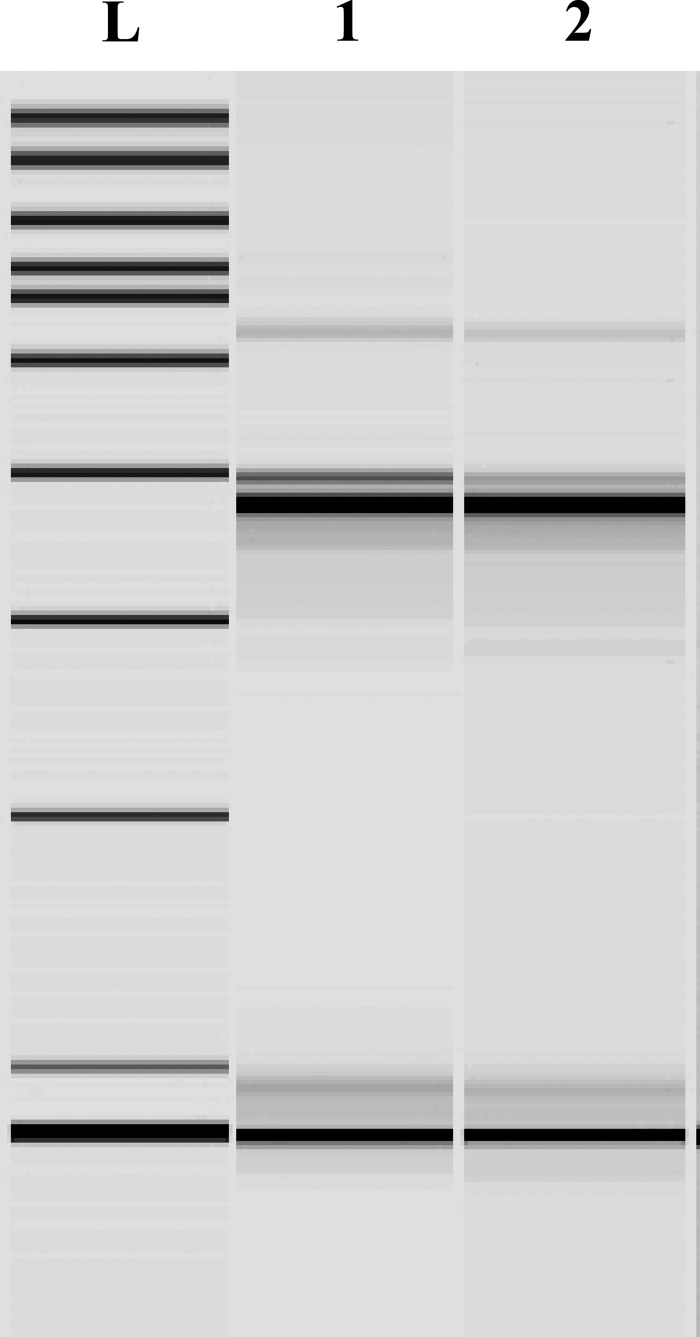
Surprisingly, 5 months after the identification of the above-mentioned strain, a further XDR H. parainfluenzae strain (few colonies) was isolated from a urethral swab of another homosexual man. Again, it was together with an N. gonorrhoeae strain (numerous colonies) of ST8371, resulting in resistance to penicillin, tetracycline, and ciprofloxacin but susceptibility to azithromycin and ceftriaxone. This second case was empirically treated with ciprofloxacin and doxycycline without clinical improvement and then with ceftriaxone plus azithromycin with resolution of the infection. Interestingly, the strain (AE-2137638) possessed exactly the same phenotypic and molecular characteristics as AE-2096513 (Table 1). More importantly, the rep-PCR analysis showed that the two H. parainfluenzae isolates were indistinguishable (Fig. 1). This finding was also supported by identical DNA sequences of the gyrA and parC housekeeping genes.
Fig 1.
Results of the rep-PCR analysis. L, ladder; 1, strain AE-2096513 (first patient, August 2012); 2, strain AE-2137638 (second patient, January 2013). The two XDR H. parainfluenzae isolates are indistinguishable.
This is the first report of XDR H. parainfluenzae isolates. The strains possess a pattern of multiple resistance traits that has never been described in a single isolate. To our knowledge, this is also the first time that mef(A), tet(M), catS, and the Ala69Ser substitution in L4 have been found in H. parainfluenzae.
H. parainfluenzae can be part of the nasopharyngeal flora and a possible cause of urethritis among homosexual men (17, 18). In our cases, contact between the two carriers was not explored to preserve the patients' privacy. Therefore, though this H. parainfluenzae phenotype was previously neither observed nor reported, we can only speculate that the two men had direct or indirect (via a third or fourth person) sexual contact. However, we highlight that H. parainfluenzae can be responsible for serious infections, such as meningitis, sepsis, septic arthritis, and endocarditis (19–22). Thus, larger studies should be planned to establish the epidemiologic relevance of these difficult-to-treat XDR organisms.
ACKNOWLEDGMENTS
We deeply thank Parham Sendi for the critical revision of the paper. We also thank John W. Looney and Alexandra Collaud for the technical help and Gilles Wandeler and Alexander Schweiger for providing the clinical data.
This work was supported by internal funds of the Institute of Infectious Diseases, Bern, Switzerland.
Footnotes
Published ahead of print 1 April 2013
REFERENCES
- 1. Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. 2012. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18:268–281 [DOI] [PubMed] [Google Scholar]
- 2. Tristram S, Jacobs MR, Appelbaum PC. 2007. Antimicrobial resistance in Haemophilus influenzae. Clin. Microbiol. Rev. 20:368–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pfeifer Y, Meisinger I, Brechtel K, Grobner S. 2013. Emergence of a multidrug-resistant Haemophilus influenzae strain causing chronic pneumonia in a patient with common variable immunodeficiency. Microb. Drug Resist. 19:1–5 [DOI] [PubMed] [Google Scholar]
- 4. Garcia-Cobos S, Arroyo M, Campos J, Perez-Vazquez M, Aracil B, Cercenado E, Orden B, Lara N, Oteo J. 2013. Novel mechanisms of resistance to β-lactam antibiotics in Haemophilus parainfluenzae: β-lactamase-negative ampicillin resistance and inhibitor-resistant TEM β-lactamases. J. Antimicrob. Chemother. doi:10.1093/jac/dks525 [DOI] [PubMed] [Google Scholar]
- 5. Tristram SG, Pitout MJ, Forward K, Campbell S, Nichols S, Davidson RJ. 2008. Characterization of extended-spectrum β-lactamase-producing isolates of Haemophilus parainfluenzae. J. Antimicrob. Chemother. 61:509–514 [DOI] [PubMed] [Google Scholar]
- 6. Law DK, Shuel M, Bekal S, Bryce E, Tsang RS. 2010. Genetic detection of quinolone resistance in Haemophilus parainfluenzae: mutations in the quinolone resistance-determining regions of gyrA and parC. Can. J. Infect. Dis. Med. Microbiol. 21:e20–e22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rodriguez-Martinez JM, Lopez-Hernandez I, Pascual A. 2011. Molecular characterization of high-level fluoroquinolone resistance in a clinical isolate of Haemophilus parainfluenzae. J. Antimicrob. Chemother. 66:673–675 [DOI] [PubMed] [Google Scholar]
- 8. EUCAST 2013. Clinical breakpoints, version 3.0. www.eucast.org/clinical_breakpoints/
- 9. Perreten V, Vorlet-Fawer L, Slickers P, Ehricht R, Kuhnert P, Frey J. 2005. Microarray-based detection of 90 antibiotic resistance genes of gram-positive bacteria. J. Clin. Microbiol. 43:2291–2302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Scriver SR, Walmsley SL, Kau CL, Hoban DJ, Brunton J, McGeer A, Moore TC, Witwicki E. 1994. Determination of antimicrobial susceptibilities of Canadian isolates of Haemophilus influenzae and characterization of their β-lactamases. Antimicrob. Agents Chemother. 38:1678–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Peric M, Bozdogan B, Jacobs MR, Appelbaum PC. 2003. Effects of an efflux mechanism and ribosomal mutations on macrolide susceptibility of Haemophilus influenzae clinical isolates. Antimicrob. Agents Chemother. 47:1017–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Soge OO, Tivoli LD, Meschke JS, Roberts MC. 2009. A conjugative macrolide resistance gene, mef(A), in environmental Clostridium perfringens carrying multiple macrolide and/or tetracycline resistance genes. J. Appl. Microbiol. 106:34–40 [DOI] [PubMed] [Google Scholar]
- 13. Strommenger B, Kettlitz C, Werner G, Witte W. 2003. Multiplex PCR assay for simultaneous detection of nine clinically relevant antibiotic resistance genes in Staphylococcus aureus. J. Clin. Microbiol. 41:4089–4094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jordens JZ. 1998. Characterisation of non-capsulate Haemophilus influenzae by repetitive extragenic palindromic (REP)-PCR. J. Med. Microbiol. 47:1031–1034 [DOI] [PubMed] [Google Scholar]
- 15. Tristram SG, Hawes R, Souprounov J. 2005. Variation in selected regions of blaTEM genes and promoters in Haemophilus influenzae. J. Antimicrob. Chemother. 56:481–484 [DOI] [PubMed] [Google Scholar]
- 16. Weisblum B. 1995. Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother. 39:577–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tian GZ, Zhang LJ, Wang XL, Zhang L, Li SF, Gu CM, Sun J, Cui BY. 2012. Rapid detection of Haemophilus influenzae and Haemophilus parainfluenzae in nasopharyngeal swabs by multiplex PCR. Biomed. Environ. Sci. 25:367–371 [DOI] [PubMed] [Google Scholar]
- 18. Hsu MS, Wu MY, Lin TH, Liao CH. 2013. Haemophilus parainfluenzae urethritis among homosexual men. J. Microbiol. Immunol. Infect. doi:10.1016/j.jmii.2012.11.007 [DOI] [PubMed] [Google Scholar]
- 19. Nwaohiri N, Urban C, Gluck J, Ahluwalia M, Wehbeh W. 2009. Tricuspid valve endocarditis caused by Haemophilus parainfluenzae: a case report and review of the literature. Diagn. Microbiol. Infect. Dis. 64:216–219 [DOI] [PubMed] [Google Scholar]
- 20. Cardines R, Giufre M, Ciofi degli Atti ML, Accogli M, Mastrantonio P, Cerquetti M. 2009. Haemophilus parainfluenzae meningitis in an adult associated with acute otitis media. New Microbiol. 32:213–215 [PubMed] [Google Scholar]
- 21. Mora A, Marimon I, Mesquida J, Perez A. 2011. Haemophilus parainfluenzae septic arthritis: report of a case and review of the literature. Enferm. Infecc. Microbiol. Clin. 29:472–473 [DOI] [PubMed] [Google Scholar]
- 22. Govind B, Veeraraghavan B, Anandan S, Thomas N. 2012. Haemophilus parainfluenzae: report of an unusual cause of neonatal sepsis and a literature review. J. Infect. Dev. Ctries. 6:748–750 [DOI] [PubMed] [Google Scholar]