Abstract
Summary: We present a new, accurate and efficient tool for mapping short reads obtained from the Illumina Genome Analyzer following sodium bisulfite conversion. Our tool, BRAT, supports single and paired-end reads and handles input files containing reads and mates of different lengths. BRAT is faster, maps more unique paired-end reads and has higher accuracy than existing programs. The software package includes tools to end-trim low-quality bases of the reads and to report nucleotide counts for mapped reads on the reference genome.
Availability: The source code is freely available for download at http://compbio.cs.ucr.edu/brat/ and is distributed as Open Source software under the GPLv3.0.
Contact: elenah@cs.ucr.edu
1 INTRODUCTION
Methylation of DNA is involved in a variety of biological processes, including embryogenesis and development, silencing of transposable elements and regulation of gene transcription. The gold standard method to detect cytosine methylation is sodium bisulfite treatment of DNA (Frommer et al., 1992), which converts unmethylated cytosines to uracils, but leaves the vast majority of methylated cytosines unchanged. The combination of bisulfite conversion and next generation sequencing has already enabled some genome-wide studies of DNA methylation (Cokus et al., 2008; Lister et al., 2008). The success of these methods critically depends on the availability of accurate and time-efficient tools capable of mapping millions of bisulfite-treated short reads to a reference genome.
This latter task, called BS-mapping, can be computationally intensive. Due to the effect of the bisulfite conversion, BS-mapping must allow Ts in the sequenced reads to align to Cs in the reference genome and similarly As in the reads to align to Gs in the genome. Hereafter, these types of T–C and A–G allowed mismatches are called BS-mismatches. In order to allow for BS-mismatches during the mapping, one can (i) allow a large number of mismatches, about  of the read length assuming that methylation is rare; (ii) use an exhaustive search where for each read all possible combinations of Ts are converted to Cs or (iii) apply different kinds of reference/reads conversions, usually involving the reduction of the alphabet cardinality. Allowing a large number of mismatches introduces many false positives due to non-BS-mismatches and can be very computationally expensive, which makes this strategy impractical. Similarly, the second option generates a very large number of candidates and presents similar problems.
of the read length assuming that methylation is rare; (ii) use an exhaustive search where for each read all possible combinations of Ts are converted to Cs or (iii) apply different kinds of reference/reads conversions, usually involving the reduction of the alphabet cardinality. Allowing a large number of mismatches introduces many false positives due to non-BS-mismatches and can be very computationally expensive, which makes this strategy impractical. Similarly, the second option generates a very large number of candidates and presents similar problems.
The conversion of a genome and/or reads has been shown to be a successful strategy. For instance, in (Lister et al., 2008) the authors mapped sequenced reads to three versions of the genome: the original genome, the genome in which Cs are replaced with Ts and finally the genome in which Gs are changed to As. Reads were allowed up to two mismatches to capture methylated Cs. The shortcoming of this method is that it does not handle instances where a read contains both unmethylated and methylated Cs with the number of Cs higher than the number of allowed mismatches. Another strategy was proposed in Cokus et al. (2008), where the reads are transformed in position–weight matrices and alignment is carried out in probability space. Due to its computational complexity, the authors suggest that their approach is not practical unless the reference genome is small.
To meet these challenges, several BS-mapping tools have been designed such as mrsFAST (Hormozdiari et al., 2009), BSMAP (Xi and Li, 2009), VerJInxer (Zeschnigk et al., 2009) and RMAP-bs (Smith et al., 2009). The description of the algorithm used in mrsFAST is not publicly available. VerJinxer uses q-grams that simulate all possible methylation patterns. RMAP-bs uses hashing on the reads and employs wildcard matching to allow BS-mismatches. BSMAP uses hashing on the reference genome, where seeds are words of a fixed length expanded to account for all possible combinations of substitutions Cs to Ts. This latter approach can be very slow due to the large search space induced by the additional seeds.
While the mapping method plays an important role, increasing the read length and employing paired-end sequencing further improves the number of uniquely mapped reads (Lister and Ecker, 2009). To accommodate users who prefer paired-end sequencing, we have developed a new time-efficient BS-mapping tool called BRAT. Our tool supports single and paired-end short reads. BRAT uses a specially designed binary representation of the reference genome and reads that allows for BS-mismatches without affecting the search space. Our tool seamlessly handles input files containing reads/mates of various lengths aligning all the bases of the reads/mates. Experimental results show that (i) on paired-end reads, our tool is much faster, maps more unique pairs and has higher mapping accuracy than BSMAP and mrsFAST and (ii) on single reads, BRAT's performance is comparable to the performance of RMAP-bs, which to our knowledge is currently the best BS-mapping tool for single reads.
2 METHODS AND EXPERIMENTAL RESULTS
BRAT uses hashing of the reference genome, which effectively reduces the search space and allows simultaneous mapping of mates in paired-end alignment. First, BRAT constructs two binary representations, namely the TA- and CG-references (each reference uses one bit per base). Then fixed-length words (seeds) from the two references are hashed into a hash table, storing references names and positions within the references where the seeds occur. Pairs or single reads as well as their reverse complements are also converted and mapped in binary representations directly to a forward strand of the genome (See Supplementary Material for additional details).
Due to the reduced complexity of the converted genome and/or the reads, the chances of false positives increase dramatically with the number of allowed non-BS-mismatches. To ensure the highest possible accuracy, BRAT maps reads/pairs with up to one non-BS-mismatch in the first 36 bases of reads to compensate for sequencing errors. The number of non-BS-mismatches beyond the first 36 bases is unlimited. In addition, BRAT handles sequencing errors at the preprocessing stage. Users can select to employ another tool in the software suite that trims the low base quality ends of reads, thus reducing the chance of sequencing errors in the reads (the majority of sequencing errors tend to occur at the ends). After trimming, reads might have different lengths, but BRAT supports the mapping of all the bases in the reads even if given a mix of reads of different lengths.
We have compared our tool with RMAP-bs, mrsFAST and BSMAP using real bisulfite-treated reads on Plasmodium falciparum obtained with Illumina GAII and in silico reads on Homo sapiens and P.falciparum. Homo sapiens has long CpG islands whereas P.falciparum is AT rich. Table 1 reports the results of these experiments. Our real dataset contains 21.5 M reads, whereas for the simulation we generated 1 and 10 M randomly chosen pairs/reads with 90% of Cs converted to Ts (no sequencing errors were introduced for this experiment). Only perfect matches and BS-mismatches were allowed in this experiment. Parameter options used with the programs were for RMAP-bs (m 0, S 1, h 26/32), BSMAP (s 9, v 0, r 0, m 106, x 306, OLIGOLEN 36) and mrsFAST (e 0, n 2, min 106, max 306).
Table 1.
Comparing the performance and sensitivity of BS-mapping tools when non-BS-mismatches are not allowed
| Genome, read length and number of reads/pairs | Time | RAM (MB) | Total mapped unique reads/pairs | Correctly mapped unique reads/pairs | ||
|---|---|---|---|---|---|---|
| Single read | RMAP | P.falciparum, 26 bp, 21.5 M | 8 m 3 s | 1500 | 7 413 261 | n/a |
| BRAT | 1 m 59 s | 982 | 7 379 870 | n/a | ||
| RMAP | H.sapiens, chr X, 32 bp, 10 M | 4 m 52 s | 2100 | 7 906 395 | 7 906 395 | |
| BRAT | 6 m 28 s | 2000 | 7 915 050 | 7 915 050 | ||
| Paired end | BSMAP | P.falciparum, 32 bp, 1 M | 1160 m 0 s | 171 | 402 602 | 393 810 |
| BRAT | 0 m 40 s | 982 | 913 225 | 913 225 | ||
| mrsFAST | 48 m 10 s | 687 | 635 784 | 620 622 |
With single reads, both RMAP-bs and BRAT had 100% mapping accuracy. The mapping accuracy is calculated as the ratio between unique reads/pairs mapped correctly and total number of unique reads/pairs, where unique reads/pairs are reads/pairs that are mapped perfectly or with BS-mismatches to a single location.
There is a slight difference in the number of mapped reads because RMAP-bs, in addition to BS-mismatches, allows a C in the reads to align to a T in a genome only when C is followed by a G. On paired-end reads, BRAT mapped 1.47 and 2.3 times more unique pairs (correctly) than mrsFAST and BSMAP, respectively, while retaining higher accuracy: BRAT had a mapping accuracy of 100%, whereas mrsFAST was 97.6% and BSMAP was 97.81%.
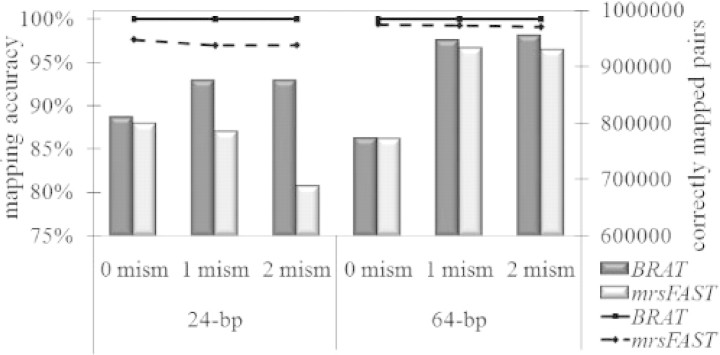
To compare our tool with the better performing tool for paired-end reads (mrsFAST) in the presence of sequencing errors, we used in silico 1 M paired-end 24 bases reads and 64 bases reads from P.falciparum with 90% of bisulfite conversion and 1% of sequencing errors. Figure 1 shows the number of correctly mapped unique pairs (bars) as well as mapping accuracy of both tools (lines). When mapping with non-BS-mismatches, we define a pair to be unique if it maps to a single location with the smallest number of non-BS-mismatches in both mates. BRAT mapped up to 21% more unique pairs than mrsFAST on 24 bases reads. In both experiments, BRAT had higher mapping accuracy. BRAT was also significantly faster than mrsFAST: on 24 bases reads, BRAT was 67, 12 and 18 times faster with 0, 1 and 2 mismatches, respectively, and on 64 bases reads it was 55, 20 and 37 times faster with 0, 1 and 2 mismatches, respectively.
Fig. 1.
BRAT versusmrsFAST: the number of correctly mapped unique pairs depends on reads length and the number of allowed non-BS-mismatches.
Supplementary Material
ACKNOWLEDGEMENTS
We thank V. Vacic and A. Smith for helpful comments and discussions.
Funding: NSF CAREER (IIS-0447773) to S.L., and UCR Regents' Faculty Fellowship to K.L.R.
Conflict of Interest: none declared.
REFERENCES
- Cokus S, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frommer M, et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl Acad. Sci. USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hormozdiari F, et al. Combinatorial algorithms for structural variation detection in high-throughput sequenced genomes. Genome Res. 2009;19:1270–1278. doi: 10.1101/gr.088633.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Ecker J. Finding the fifth base: genome-wide sequencing of cytosine methylation. Genome Res. 2009;19:959–966. doi: 10.1101/gr.083451.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A, et al. Updates to the RMAP short-read mapping software. Bioinformatics. 2009;25:2841–2842. doi: 10.1093/bioinformatics/btp533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Y, Li W. BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinformatics. 2009;10:232–240. doi: 10.1186/1471-2105-10-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeschnigk M, et al. Massive parallel bisulfite sequencing of CG-rich DNA fragments reveals that methylation of many X-chromosomal CpG islands in female blood DNA is incomplete. Hum. Mol. Genet. 2009;18:1439–1448. doi: 10.1093/hmg/ddp054. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.