Abstract
The European royal dynasties of the Early Modern Age provide a useful framework for human inbreeding research. In this article, consanguineous marriage, inbreeding depression and the purging of deleterious alleles within a consanguineous population are investigated in the Habsburgs, a royal dynasty with a long history of consanguinity over generations. Genealogical information from a number of historical sources was used to compute kinship and inbreeding coefficients for the Habsburgs. The marriages contracted by the Habsburgs from 1450 to 1750 presented an extremely high mean kinship (0.0628±0.009), which was the result of the matrimonial policy conducted by the dynasty to establish political alliances through marriage. A strong inbreeding depression for both infant and child survival was detected in the progeny of 71 Habsburg marriages in the period 1450–1800. The inbreeding load for child survival experienced a pronounced decrease from 3.98±0.87 in the period 1450–1600 to 0.93±0.62 in the period 1600–1800, but temporal changes in the inbreeding depression for infant survival were not detected. Such a reduction of inbreeding depression for child survival in a relatively small number of generations could be caused by elimination of deleterious alleles of a large effect according with predictions from purging models. The differential purging of the infant and child inbreeding loads suggest that the genetic basis of inbreeding depression was probably very different for infant and child survival in the Habsburg lineage. Our findings provide empirical support that human inbreeding depression for some fitness components might be purged by selection within consanguineous populations.
Keywords: royal inbreeding, Habsburg dynasty, consanguineous marriage, inbreeding depression, purging of inbreeding depression
Introduction
In humans, the most extreme cases of close inbreeding are frequently found in royal dynasties. Indeed, brother-sister and parent-child marriages were not unusual in ancient royal dynasties such as the Egyptian pharaohs or the Persian dynasty (Middleton, 1962; Bixler, 1982a, 1982b; Ager, 2005). Unfortunately, the study of inbreeding from ancient royal dynasties suffers from a number of limitations. First of all, it is difficult to construct full pedigrees from those dynasties because the genealogical information from the ancient record presents many gaps and uncertainties. In the royal families of Egypt, for example, the pharaoh had many wives and very many children in such a way that it was not easy to establish unequivocally in some cases who was the mother of his successor (Middleton, 1962; Bixler, 1982a, 1982b). Even in the most recent Egyptian royal family as the Ptolemaic dynasty, some full-sibling marriages are controversial because they are not well documented. Secondly, the adverse effects of consanguineous marriage on fitness traits such as survival and fertility cannot be easily investigated in those dynasties because information on such characters is hardly available from the ancient record. A way to circumvent some of the limitations in the study of inbreeding from ancient dynasties is exemplified in an analysis of microsatellite loci of a number of royal mummies of the 18th Egyptian dynasty (Hawass et al., 2010). The analysis of genetic relationships among individuals from those molecular markers allowed the identification of close relatives in such a way that a five-generation pedigree that included Akhenaton and Tutankhamun pharaohs was inferred. Further research on the ancient royal dynasties based on molecular genetic markers is needed before those royal dynasties can be useful for inbreeding studies.
The European royal dynasties of the Early Modern Age, unlike the ancient dynasties, provide a useful framework for human inbreeding research. Firstly, it is known that close kin marriages such as uncle-niece, first cousins and other consanguineous unions occurred frequently in such dynasties along prolonged periods of time. Secondly, the genealogical records for the European dynasties available in historical sources are very extensive and accessible in such a way that such dynasties may be considered as pedigreed populations, where inbreeding coefficients can be computed with extreme precision from extended pedigrees (Alvarez et al., 2009, 2011). Thirdly, the mortality and fertility data for European royal families are also complete in the historical sources so that such dynasties may be very useful for the study of inbreeding depression, that is, the reduced survival and fertility of offspring of related individuals caused by increased homozygosity for deleterious alleles (Charlesworth and Charlesworth, 1999; Charlesworth and Willis, 2009). In humans, most of the empirical evidence on inbreeding depression comes from the progeny of first cousins because this is the most common form of consanguineous union in current human populations (Khoury et al., 1987; Bittles and Neel, 1994; Bittles and Black, 2010; Hamamy et al., 2011). The magnitude of inbreeding depression for inbreeding levels higher than that corresponding to first cousins (inbreeding coefficient, F>0.0625) is less known so that, at present, the relationship between fitness and human inbreeding is still poorly understood (Robert et al., 2009; Postma et al., 2010; Bittles, 2012). In this regard, the European royal dynasties could supply substantial empirical evidence on human inbreeding depression from the inbreeding levels resulting of persistent consanguinity over generations.
Furthermore, as the European royal dynasties are pedigreed and are known to have experienced inbreeding for a long time, they are highly suited to investigate the effectiveness of natural selection in reducing inbreeding depression in consanguineous human populations. Empirical evidence in several animal and plant species, as well as theoretical studies shows that inbreeding depression can be reduced or purged within a consanguineous population because the selection efficiency against deleterious recessive alleles is increased by inbreeding (Hedrick, 1994; Ballou, 1997; Wang et al., 1999; Crnokrak and Barrett, 2002; Glémin, 2003; Boakes and Wang, 2005; Boakes et al., 2007; García-Dorado, 2012). Theoretical models predict that the degree of purging depends on the genetic basis of inbreeding depression. Thus, alleles of a large effect, those that are lethals or semilethals when in homozygous condition, will be easily purged from the population in a relatively small number of generations, while the genetic load resulting from mildly deleterious alleles will persist (Hedrick, 1994; Wang et al., 1999; Boakes and Wang, 2005). In humans, some studies have reported that populations with higher rates of inbreeding tend to display lower inbreeding effect on prereproductive mortality and such an effect was attributed to the successive elimination of deleterious alleles in the more inbred populations (Sanghvi, 1966; Khoury et al., 1987; Khlat and Khoury, 1991). However, this empirical evidence on purging of inbreeding depression in human populations has been questioned and it is considered as not conclusive (Chakraborty and Chakravarti, 1977; Radha Rama Devi et al., 1987; Bittles et al., 1991). Therefore, at present, little is known about the purging of inbreeding depression within consanguineous human populations even though this issue is of practical concern because the high prevalence of consanguineous marriages in many contemporary human populations (Bittles, 2010, 2012; Hamamy et al., 2011). In this context, the European royal dynasties as pedigreed populations could be very useful to detect temporal changes in inbreeding depression due to elimination of deleterious alleles. Furthermore, the rate of purging in such dynasties could shed light on the genetic basis of human inbreeding depression as the relative contribution of alleles with major and minor fitness effects to inbreeding depression could be revealed from the differential purging of such alleles (Charlesworth and Willis, 2009).
The Habsburg dynasty (also known as the House of Austria) was one of the most important sovereign dynasties of Europe and had two main branches: the Austrian and the Spanish Habsburgs (Wandruszka, 1964; Ingrao, 2000; Sked, 2001; Encyclopedia Britannica, 2007; Bennassar et al., 2010). As a consequence of the matrimonial policy conducted by the dynasty to establish political alliances through marriage, an important number of consanguineous marriages such as uncle-niece and first-cousins were contracted by the Habsburgs. Some of those unions were marriages between members of the own dynasty in order to keep their heritage in their own hands and other consanguineous unions were marriages with members of related dynasties. The consequences of the prolonged consanguineous marriage have been investigated for the Spanish branch of the Habsburg dynasty (Alvarez et al., 2009, 2011). It was demonstrated that the inbreeding effects such as inbreeding depression for survival played a major role in the extinction of the Spanish Habsburg lineage at the end of the 17th century. In the present article, we extend the analysis of consanguineous marriage and inbreeding depression from the Spanish Habsburgs to the Habsburg dynasty as a whole. The analysis includes an important number of Habsburg families and covers a period of time of more than 300 years, from ∼1450 to 1800, so that potential changes in inbreeding depression due to purging of deleterious alleles could be detected. From this perspective, the Habsburg dynasty is considered as an inbreeding laboratory to investigate the effects of human consanguinity.
Materials and methods
Genealogical information for the Habsburg dynasty was introduced into a database that included more than 4 000 individuals along more than 20 parent-offspring generations. The genealogical information was obtained from several sources (The Geneall Project http://www.geneall.net; Wikipedia http://en.wikipedia.org/wiki; Encyclopedia Britannica, 2007). Kinship and inbreeding coefficients were computed from the database by means of the FSPEED computer program (Tenset Technologies, http://www.tenset.co.uk/fspeed/). The coefficient of kinship (θ, also called the coefficient of coancestry) between two individuals is the probability that two alleles at the same locus drawn at random, one from each individual, are identical by descent and the inbreeding coefficient (F) of an individual is the kinship coefficient of his or her parents (Cavalli-Sforza and Bodmer, 1971; Falconer and Mackay, 1996; Lynch and Walsh, 1998).
In order to evaluate the impact of inbreeding on survival, progeny mortality data of a number of Habsburg marriages were obtained from several sources (The Geneall Project http://www.geneall.net; Wikipedia http://en.wikipedia.org/wiki; Encyclopedia Britannica, 2007). The Habsburg families considered, correspond to the 11 Holy Roman Emperors, from Maximilian I (1459–1519) to Charles VI (1685–1740), and the Spanish Habsburg kings, their children and their grandchildren. Mortality data were classified into two categories: infant deaths (deaths in the first year of life, excluding miscarriages and stillbirths) and child deaths (deaths between years 1–10). Only deaths attributable to natural causes were considered for the analysis. Miscarriages and stillbirths were not included in the analysis because the information on such early deaths is sometimes contradictory in the historical sources. In addition, a statistically significant inbreeding effect on either miscarriages or stillbirths was not detected for the Spanish Habsburgs (Alvarez et al., 2009). On the other hand, only those families with three or more children were considered for the analysis of inbreeding depression because the estimates of survival from families with either one or two children will present large sampling errors due to small sample size. As a whole, there were 502 pregnancies in the 71 Habsburg families examined: 93 infant deaths, 76 child deaths and 333 survivals at age 10. The mean number of pregnancies per family was 7.07±0.46 and the average progeny mortality across families was 0.209±0.024 for infants, 0.188±0.026 for children and 0.359±0.029 for total mortality from birth to 10 years.
Different regression analyses were used to characterize the inbreeding depression for survival. Regression analyses of log-transformed survival as a function of inbreeding coefficient were performed because survival is expected to decrease linearly with inbreeding on a logarithmic scale according to the multilocus viability genetic model based on the assumption of multiplicative fitness interactions among loci (Morton et al., 1956; Cavalli-Sforza and Bodmer, 1971; Charlesworth and Charlesworth, 1999; Charlesworth and Willis, 2009). On this basis, the effect of inbreeding on survival is measured in terms of the inbreeding load (B) defined as the slope of the regression of the natural logarithm of survival on inbreeding coefficient, which is an estimate of the number of lethal equivalents in the population. The estimation of B allowed us to compare the results for the Habsburgs with the theoretical predictions from purging models, which usually express the decrease in inbreeding depression as a reduction in the number of lethal equivalents (Hedrick, 1994; Wang et al., 1999; Boakes and Wang, 2005). The significance of the difference between two B estimates corresponding to different periods was tested by the Fs-test for differences between two regression coefficients (Sokal and Rohlf, 1995). Analysis of departures from a linear relationship between log-transformed survival and F was performed by means of the nonlinearity t-test (Lynch and Walsh, 1998). This test compares the change in mean survival between two low levels of F and that between two high levels of F. Linearity deviations were also investigated by performing quadratic regressions of log-transformed survival on F. On the other hand, we have also performed linear regression analyses to compare our results with the estimates of human inbreeding depression from literature because the rate at which survival declines with increasing inbreeding is expressed in most studies on human inbreeding as absolute decrease in survival for inbred individuals, with a given value of F, relative to non-inbred individuals (see for example, Bittles and Neel, 1994; Jorde, 2001; Bittles and Black, 2010; Hamamy et al., 2011; Bittles, 2012).
Two logistic regression models were used to detect changes in inbreeding depression along time due to purging selection, the ancestral inbreeding regression model devised by Ballou (1997) and an alternative model developed by Boakes and Wang (2005):
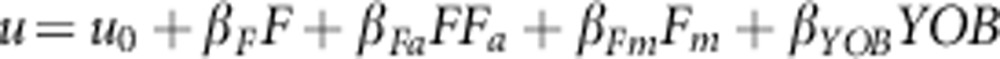 |
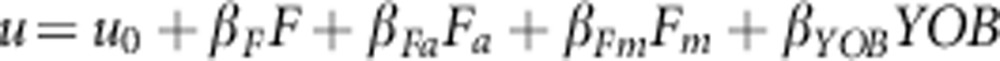 |
In these models, u is the logit transformation of a measure of fitness such as
survival, u0 is the mean fitness of non-inbred individuals, F
is the inbreeding coefficient of progeny, Fm is the maternal
inbreeding, Fa is the ancestral inbreeding coefficient, YOB is the
year of birth and the β parameters are the corresponding regression
coefficients. The ancestral inbreeding coefficient (Fa) is the
cumulative proportion of an individual's genome that has been previously exposed to
inbreeding in its ancestors as defined by Ballou (1997), and
was computed as 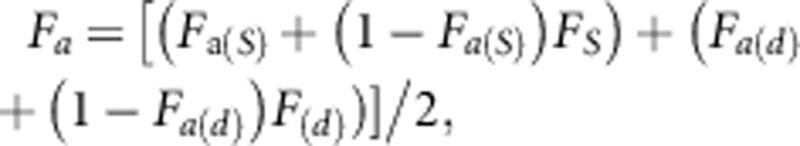 where the
subscripts s and d are used to denote coefficients for the sire and dam of the individual.
In the Ballou's model, βFa is the regression coefficient
corresponding to the interaction between inbreeding and ancestral inbreeding (F
× Fa). In both models, a negative value of
βF indicated inbreeding depression and a positive value of
βFa indicated purging. Regression analyses and other
statistical methods were performed by means of either the statistical software R (R
Foundation for Statistical Computing 2011, http://www.R-project.org) or the SPSS 15.0 statistical software
system (SPSS inc., http://www.spss.com).
where the
subscripts s and d are used to denote coefficients for the sire and dam of the individual.
In the Ballou's model, βFa is the regression coefficient
corresponding to the interaction between inbreeding and ancestral inbreeding (F
× Fa). In both models, a negative value of
βF indicated inbreeding depression and a positive value of
βFa indicated purging. Regression analyses and other
statistical methods were performed by means of either the statistical software R (R
Foundation for Statistical Computing 2011, http://www.R-project.org) or the SPSS 15.0 statistical software
system (SPSS inc., http://www.spss.com).
Results
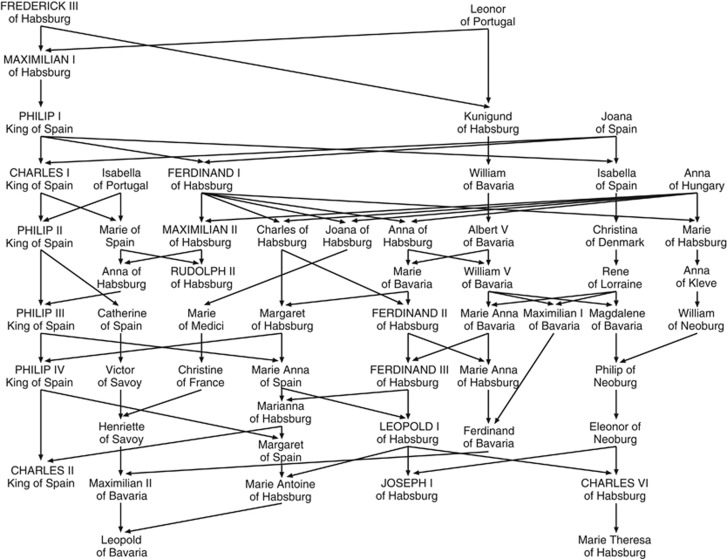
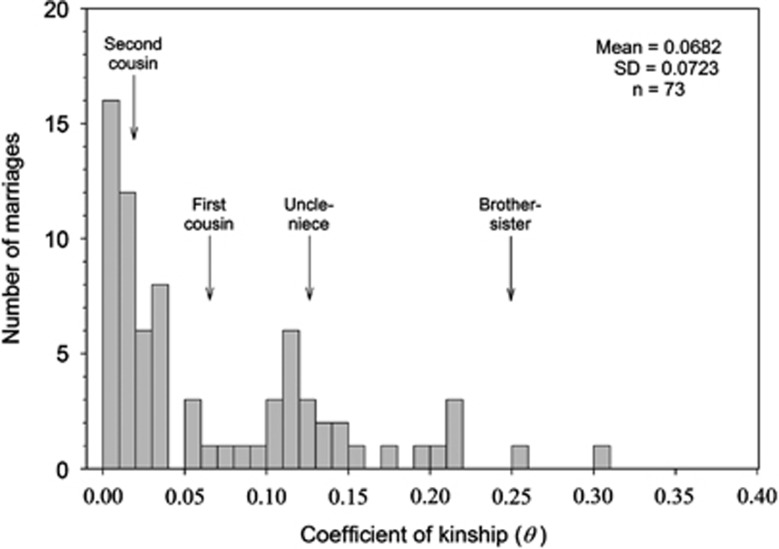
Kinship and inbreeding coefficients for the Habsburgs were calculated from a database that included more than 4000 individuals belonging to more than 20 parent-offspring generations. A partial pedigree of the Habsburg dynasty is represented by chains of descent in Figure 1. A total number of 73 marriages were contracted by the Holy Roman Emperors and the Spanish Habsburg kings, their children and their grandchildren, in the period of 300 years from 1450 to 1750. The children of Habsburg women married to individuals of other different dynasties did not belong to the Habsburg dynasty and therefore these individuals were not considered for the kinship analysis. Many of the Habsburg marriages were consanguineous unions. Thus, there were four uncle-niece marriages (Philip II of Spain with his niece Anna of Habsburg, Archduke Charles of Habsburg with Marie of Bavaria, Philip IV of Spain with Marianna of Habsburg and Leopold I of Habsburg with Margaret of Spain, as shown in Figure 1), two double first cousins, nine first cousins, four first cousins once removed (marriage of an individual with the offspring of his/her first cousin), one quadruple second cousins, seven second cousins and many other marriages with more remote kinship. The coefficient of kinship of the Habsburg marriages showed a skewed distribution, as shown in Figure 2. The mean kinship coefficient was 0.0682, which means that the Habsburg marriages had, on average, a kinship higher than that of a first cousin couple (θ=0.0625). In a total number of 73 marriages, 49 (67.1%) had a kinship higher than that of a second cousin marriage (θ=0.0156), 29 (39.7%) a kinship higher than first cousins (θ=0.0625), 13 (17.8%) a kinship higher than uncle-niece (θ=0.125) and 2 (2.7%) a kinship higher than an incestuous union (parent-child or brother-sister, θ=0.250). Only 24 (32.9%) of the Habsburg marriages had a coefficient of kinship lower than that corresponding to a second cousin marriage.
Figure 1.
Partial pedigree of the Habsburg dynasty represented by chains of descent. Holy Roman Emperors and Spanish Habsburg kings are shown in capital letters.
Figure 2.
Distribution of the kinship coefficient (θ) in 73 Habsburg marriages. Expected kinships for second cousin (θ=0.0156), first cousin (θ=0.0625), uncle-nice (θ=0.125) and brother-sister (θ=0.25) are indicated by arrows.
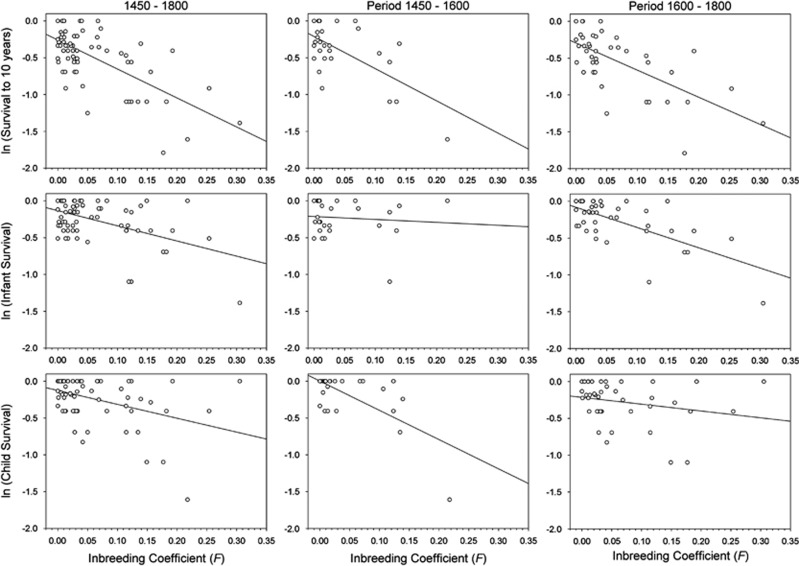
The inbreeding effect on survival was investigated in the progeny of 71 Habsburg marriages corresponding to the Holy Roman Emperors, from Maximilian I (1459–1519) to Charles VI (1685–1740), and the Spanish Habsburg kings, their children and their grandchildren. The children of all Habsburg women (including those married to individuals from different dynasties) were considered for the analysis of inbreeding effect on survival. The analysis covered a period of more than 300 years approximately from 1450 to 1800. Multiple regression analyses of survival as a function of inbreeding coefficient of progeny (F) and mother (Fm), and mean year of birth of progeny (YOB) for the 71 Habsburg families are shown in Table 1 (for log-transformed data, the total number of families was 69 because the progeny survival was zero in two families). A statistically significant negative regression of survival on F was detected for both infant and child survival as well as for total survival from birth to 10 years. The regression was statistically significant for both log-transformed and non-transformed data. On the contrary, an effect of Fm on progeny survival was not detected. YOB had a statistically significant effect on child survival for non-transformed data. A statistically significant effect of YOB on infant and total survival was not detected. The strong decline experienced by log-transformed survival with increasing F for both infant and child survival, as well as for total survival is graphically depicted in Figure 3 (first column). The inbreeding load (B) measured as the regression slope of log-transformed survival on F was 2.050±0.440 for infant and 1.886±0.516 for child survival, whereas the estimate of B for total survival was 3.937±0.568 (Table 2). Thus, the inbreeding effects on infant and child survival supposed the 52.1% and 47.9%, respectively, of the total inbreeding load from birth to 10 years. The impact of inbreeding as absolute decrease in survival for first cousin offspring (F=0.0625) inferred from simple regression analyses for non-transformed survival was 8.63%±7.90 for infant survival, 8.50%±8.39 for child survival and 13.54%±5.40 for survival to 10 years. The estimate of the cost of inbreeding for total survival to 10 years in the Habsburg dynasty was not very different from a previous estimate of 17.8%±12.3 obtained from eight families of the Spanish branch of the dynasty (Alvarez et al., 2009). It appears from our result that the extent of inbreeding depression in the Habsburgs was very similar for infant and child survival, irrespective of the measurement scale. It is also interesting to note that the inbreeding effects on infant and child mortality seem to be basically independent as the correlation between infant and child survival among families was quite low for log-transformed survival (r=−0.075, P=0.539), as well as for non-transformed survival (r=−0.018, P=0.885). This result suggest that effects of common environment (environmental covariance) for infant and child survival resulting of the fact that both fitness components are estimated from the same families should be of little importance in the Habsburg families.
Table 1. Multiple regression analyses of survival (S) as a function of inbreeding coefficient of progeny (F) and mother (F m), and year of birth (YOB) in 71 Habsburg families.
| S | P | ln S | P | |
|---|---|---|---|---|
| Survival to 10 years | ||||
| Constant | 1.399 | 0.005 | 0.483 | 0.527 |
| F | −2.130 | 0.000 | −3.890 | 0.000 |
| Fm | 0.085 | 0.853 | 0.084 | 0.908 |
| YOB | 0.000 | 0.195 | 0.000 | 0.331 |
| Infant survival | ||||
| Constant | 0.634 | 0.170 | −0.388 | 0.520 |
| F | −1.231 | 0.001 | −1.902 | 0.000 |
| Fm | −0.419 | 0.337 | −0.588 | 0.306 |
| YOB | 0.000 | 0.588 | 0.000 | 0.651 |
| Child survival | ||||
| Constant | 1.950 | 0.000 | 0.886 | 0.212 |
| F | −1.323 | 0.001 | −2.002 | 0.001 |
| Fm | 0.658 | 0.163 | 0.686 | 0.309 |
| YOB | −0.001 | 0.031 | −0.001 | 0.148 |
Figure 3.
Log-transformed survival as a function of inbreeding coefficient (F) in the Habsburg dynasty.
Table 2. Estimates of inbreeding load (B) in the Habsburg dynasty.
| All data set 1450–1800 | Period 1450–1600 | Period 1600–1800 |
Test for differences between periods
|
||
|---|---|---|---|---|---|
| F S | P | ||||
| Survival to 10 years | |||||
| B | 3.937 | 4.373 | 3.674 | 0.331 | 0.360 |
| SE | ±0.568 | ±1.032 | ±0.672 | ||
| P | 0.000 | 0.000 | 0.000 | ||
| Infant survival | |||||
| B | 2.050 | 0.396 | 2.745 | 3.737 | 0.060 |
| SE | ±0.440 | ±0.858 | ±0.481 | ||
| P | 0.000 | 0.649 | 0.000 | ||
| Child survival | |||||
| B | 1.886 | 3.975 | 0.929 | 7.945 | 0.005 |
| SE | ±0.516 | ±0.868 | ±0.618 | ||
| P | 0.001 | 0.000 | 0.140 | ||
The extensive data set for progeny mortality in the Habsburg dynasty allows us some opportunity to deal with the relationship between survival and inbreeding coefficient. According to the population genetic theory, survival is expected to decrease linearly with inbreeding coefficient on a logarithmic scale under the assumption of multiplicative fitness interactions among loci (Morton et al., 1956; Cavalli-Sforza and Bodmer, 1971; Charlesworth and Charlesworth, 1999; Wang et al., 1999; Charlesworth and Willis, 2009). Statistically significant departures from a linear relationship between log-transformed survival and inbreeding coefficient were not detected by the nonlinearity t-tests in the Habsburgs (Supplementary Table S1). Such departures were not found for both infant and child survival as well as for total survival. However, these results must be taken with caution because the statistical power of the nonlinearity test was probably not very high. First of all, it is necessary to take into account that the number of data for inbreeding coefficients higher than 0.125 was not very large in our sample and the deviations of linearity caused by epistatic interactions among loci are expected to be important precisely for high inbreeding levels (Crow and Kimura, 1970). Secondly, the non-linearity in log-transformed fitness caused by purging selection is not expected to be strong when only a small proportion of the genetic load is effectively purged (Wang et al., 1999). Therefore, although our results suggest a linear relationship between log-transformed survival and F, at least as a first approximation, quadratic regressions of log-transformed survival on F were also performed from the Habsburg mortality data (Supplementary Table S1). The quadratic coefficients were not statistically significant for both infant and child regressions, as well as for total survival. However, the quadratic coefficients for both infant and child regressions were close to statistical significance. In these cases, the quadratic functions of F gave a better fit to data than linear regressions, as the adjusted R2 were higher and the AIC (Akaike information criterion) were lower than those values corresponding to the linear regressions, but the differences in R2 and AIC between quadratic and linear regressions were small. Therefore, although a concave function for infant survival and a convex function for child survival produced better-fitting regression lines, there is no strong evidence of lack of linearity in the regressions of log-transformed survival on F. In any case, the occurrence of either epistatic interactions among loci or purging selection as agents promoting departures from linearity in the inbreeding depression detected in the Habsburg families cannot be completely ruled out.
Temporal changes in the inbreeding depression for progeny survival of the Habsburg marriages were investigated by multiple regression models specifically devised to detect purging effects (Supplementary Table S2). The multiple logistic regression model developed by Boakes and Wang (2005) detected a strong and highly significant effect of inbreeding depression (negative value for the βF coefficient) in the progeny of Habsburg marriages for both infant and child survival, as well as for total survival to 10 years. A slight and non significant purging effect (positive value for βFa) was found for each one of the three fitness components. On the other hand, the ancestral inbreeding regression model (Ballou, 1997) did not detect a statistically significant inbreeding depression for any fitness component and, according to this model, the coefficient βFa presented non-significant negative values, which indicated a trend of ancestral inbreeding to increase inbreeding depression for all fitness components. The lack of consistency in the results obtained from the two regression models was not unexpected as these models have rather different properties to detect purging (Boakes and Wang, 2005). Thus, although both models are conservative, the Ballou's model is more appropriate in the detection of purging when inbreeding depression is caused by mutations of a large effect, whereas the alternative model has a higher statistical power in the detection of purging of mildly deleterious alleles.
In order to circumvent these inconsistencies associated with the two regression models, a simple alternative approach for detecting purging was performed. The complete data set corresponding to the progeny of the Habsburg marriages was split into two groups. The first group included those families belonging to the period of 1450–1600 (28 families) and the second group to those families of the period 1600–1800 (41 families). If a purging effect reducing inbreeding depression occurred in the Habsburg lineage, then the inbreeding load (B) estimated from the second group of data would be lower than the inbreeding load estimated from the first group as a consequence of removing recessive deleterious alleles. The analysis of inbreeding depression for log-transformed survival carried out separately for the two different periods showed some remarkable temporal changes in the inbreeding load (Table 2, Figure 3, second and third columns). Thus, the inbreeding load for child survival experienced a strong reduction from 3.975±0.868 in the first period to 0.929±0.618 in the second period, and this reduction in inbreeding load between periods was statistically significant (Fs-test=7.945, P=0.005). This finding reveals that the linearity of log-transformed survival as a function of the inbreeding coefficient observed for child survival in the whole data set was more apparent than real as the whole data set is really composed by two groups with different regression slope. For infant survival, the inbreeding load exhibited an increase from 0.396±0.858 in the first period to 2.745±0.481 in the second period, and this increase between periods was very close to the statistical significance (Fs-test=3.737, P=0.06). The total inbreeding load for survival from birth to 10 years was 4.373±1.032 for the first period and 3.674±0.672 for the second period, and the difference between periods was not statistically significant.
Discussion
The study of inbreeding in the European royal dynasties of the Early Modern Age has received very little attention in spite of the advantages of using such dynasties as human inbreeding models. In this respect, the Habsburgs were one of the most interesting dynasties because their matrimonial policy led to an extreme case of persistent consanguinity over generations. Our research on consanguineous marriage in the Habsburg dynasty covered a period of 300 years from 1450 to1750 and was performed from the extensive genealogical information available in historical sources. The mean kinship coefficient of 73 marriages contracted by the Habsburgs along those three centuries was 0.0628, higher than the kinship coefficient of a first cousin marriage (Figure 2). Approximately 40% of the Habsburg marriages had a kinship coefficient higher than that corresponding to a first-cousin union (θ=0.0625) and almost 20% of the marriages had a kinship higher than that corresponding to an uncle-niece union (θ=0.125). Consequently, the inbreeding coefficient of both the Austrian Emperors and Spanish kings of the dynasty were very high particularly from the middle of the 16th century. In general, Emperors had inbreeding coefficients lower than Spanish kings. Average of inbreeding coefficients was 0.0752 for the Holy Roman Emperors and 0.1287 for the Spanish kings. The Emperor with the highest inbreeding coefficient was Leopold I (F=0.1568) and the second highest was Ferdinand II (F=0.1390), while the Spanish king with the highest inbreeding was Charles II (F=0.2538) and the second highest was Philip III (F=0.2177). The highest inbreeding coefficient in the Habsburg dynasty occurred in the Austrian branch where Marie Antoine of Habsburg, daughter of Emperor Leopold I and his niece Margaret of Spain (sister of Charles II of Spain), had an inbreeding coefficient of 0.3053, which is higher than the inbreeding coefficient of the progeny of an incestuous union (parent-offspring or brother-sister).
A strong inbreeding depression for both infant and child survival was detected in the progeny of 71 marriages contracted by the Habsburgs in a period of more than 300 years from 1450 to 1800 (Table 1, Figure 3, first column). In the Habsburgs, the inbreeding depression measured as absolute decrease in survival for an F value of 0.0625, corresponding to first cousin offspring, was 8.63%±7.90 for infant survival, 8.50%±8.39 for child survival and 13.54%±5.40 for total survival from birth to 10 years. As compared with the average value of inbreeding depression observed in contemporary human populations, these figures reveal the strong impact of inbreeding on progeny survival in the Habsburgs. Thus, the most recent estimates of the degree of inbreeding depression for prereproductive survival in humans have been obtained from first cousin offspring in two meta-analyses performed from a large number of current human populations (Bittles and Neel, 1994; Bittles and Black, 2010). The absolute decrease in survival to a median age of 10 years in the progeny of first cousin marriages relative to unrelated parents was 4.4%±4.6 from a compilation of 38 populations (Bittles and Neel, 1994). Later, this average estimate was revised to a value of 3.5% from 69 human populations resident in 15 countries located across four continents (Bittles and Black, 2010). It is necessary to take into account, however, that the magnitude of inbreeding depression detected in the Habsburg families lies within the range of variation observed in current human populations as the difference in mortality between the offspring of first cousins and those of unrelated parents ranged from nearly zero to ∼19% in the meta-analyses mentioned above (Bittles and Neel, 1994). In addition, the extent of inbreeding depression in the Habsburgs was not very different from that estimated in other particular inbreeding studies such as those performed in the Utah Mormon population or the Darwin/Wedgwood dynasty. The study of the Utah Mormon population since 1847 until 1945 showed a 9% of absolute increase in prereproductive mortality before age 16 among the offspring of first-cousin marriages (Jorde, 2001), while the analysis of consanguineous marriage of 25 Darwin/Wedgwood families, which included the Charles Darwin's one revealed an absolute increase in mortality until 10 years for first cousin offspring in the range of 5.4–9.5% (Berra et al., 2010).
The detection of such inbreeding effects in the Habsburg dynasty was probably favoured by the fact that our research is based on royal families. In such families, a significant reduction of environmental variation for progeny survival is expected as socioeconomic variation, as well as differences in health conditions had to be of little importance in such families compared with the general population. In fact, demographic studies have shown that both infant and child mortality remained rather stable among Europe's ruling families in the period 1500–1800 even though births and deaths rates in the European population experienced remarkable changes in that period (Bourgeois-Pichat, 1965; Peller, 1965; Kümmel, 1990; Lancaster, 1990; Bennassar et al., 2010). The results found for the Habsburgs are consistent with these observations. Thus, a significant regression effect of YOB on either infant or total survival was not detected for the Habsburg families. Regarding child survival, a statistically significant negative slope was observed only for non-transformed data (Table 1).
The prevalence of consanguineous marriages in the Habsburg dynasty during a period of more than 300 years (∼10 generations) offers the opportunity to investigate whether a part of the inbreeding depression for survival was purged by natural selection as the result of elimination of deleterious alleles. In fact, temporal changes in the inbreeding depression for child survival were detected in the Habsburg dynasty. Thus, the inbreeding load (B) for child survival experienced a strong and statistically significant decline from 3.975±0.868 in the period 1450–1600 to 0.929±0.618 in the period 1600–1800 (Table 2, Figure 3, second and third columns). This temporal change in inbreeding depression supposed a reduction of almost 80% of the inbreeding load for child survival in a period of ∼10 generations of close kin marriage. Evidently, environmental factors could be involved in such a reduction of inbreeding depression but it is necessary to consider that temporal changes expressed in terms of a statistically significant regression effect of YOB on log-transformed child survival were not observed in the Habsburg data set, as mentioned above. Although the contribution of environmental effects to the reduction of inbreeding depression cannot be completely discarded, the reduction in inbreeding load for child survival detected in the Habsburg dynasty is in accordance with theoretical evidence from models of purging. Thus, simulation studies of deleterious mutations in finite populations for multilocus viability models have shown that an important part of the inbreeding depression can be purged in the initial period of inbreeding, involving a relatively small number of generations, due to elimination of lethals and deleterious alleles of a large effect (Hedrick, 1994; Wang et al., 1999; Boakes and Wang, 2005). In fact, computer simulations from continuous full-sib mating models showed a sharp reduction of ∼70–80% of the number of lethal equivalents in a few generations when the genetic load is mainly caused by deleterious of a large effect (Figure 1, Figure 2, Figure 3 and 5 in Hedrick, 1994; Figure 5 in Wang et al., 1999). Therefore, our findings for the Habsburg dynasty suggest that the inbreeding depression for some fitness components such as child survival could be effectively purged in consanguineous human populations. Indirectly, the results here presented for the Habsburgs also indicate that an important part of the inbreeding depression for child survival could be the result of alleles with a major effect on fitness, which were effectively purged along the period of ∼10 generations of close kin marriage.
On the other hand, the inbreeding depression for infant survival did not display statistically significant temporal variation in the Habsburg dynasty (Table 2, Figure 3, second and third columns). The inbreeding load for infant survival varied from 0.396±0.858 in the period 1450–1600 to 2.745±0.481 in the last period. This difference in the inbreeding load between periods was not statistically significant, even though the increase was very close to statistical significance. Therefore, it appears that inbreeding depression for infant survival was not purged in the dynasty and it could be explained if inbreeding depression for this fitness component were mainly due to mildly deleterious alleles. Simulation studies have shown that detrimental alleles of relatively small effect may become fixed by random genetic drift in a small population and, in this case, the genetic load is not purged (Hedrick, 1994; Wang et al., 1999; Boakes and Wang, 2005). On this basis, the differential purging of the infant and child genetic loads in the Habsburg lineage could be explained in terms of different relative contributions of alleles with minor and major effects to the inbreeding depression for the two survival components. However, it is also necessary to take into account the degree of dominance of the deleterious alleles involved in inbreeding depression. Thus, empirical evidence from Drosophila shows that large effect mutations as lethals are nearly fully recessive, whereas deleterious mutations of relatively small effect are partially recessive (Simmons and Crow, 1977; Charlesworth and Charlesworth, 1987). The dominance level is particularly critical for the purging process when inbreeding is due to small population size as in this case only highly recessive mutations can be efficiently purged (Kirkpatrick and Jarne, 2000; Glémin, 2003). In this way, both the magnitude of deleterious effect and its dominance level could be involved in the differential purging of inbreeding depression for infant and child survival detected in the Habsburgs. In any case, the results taken as a whole suggest that the genetic basis of inbreeding depression was probably very different for infant and child survival in the Habsburg lineage.
As a consequence of the opposite temporal change in the inbreeding load for infants and children, the total inbreeding load from birth to 10 years was very similar in quantitative terms in the two periods (4.373±1.032 and 3.674±0.672 in the first and second periods, respectively), but presented remarkable differences in its composition (Table 2). Thus, most (90.9%) of the inbreeding load from birth to 10 years in the period 1450–1600 was due to inbreeding effects on child mortality, whereas in the period 1600–1800 most (74.7%) of the total inbreeding load was caused by inbreeding effects on infant survival. The different responses to inbreeding showed by the different survival components reveal that purging of genetic load might be a very complex process in consanguineous human populations as purging effects on particular fitness components can be counterbalanced by changes in other fitness components. Further research on other European royal dynasties is needed to confirm whether our findings are valid exclusively for the Habsburg dynasty or they reflect basic genetic aspects of human inbreeding depression.
Data archiving
Data deposited in the dryad repository: doi:10.5061/dryad.gt64c.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Ager SL. Familiarity breeds: Incest and the Ptolemaic dynasty. JHS. 2005;125:1–34. doi: 10.1017/s0075426900007084. [DOI] [PubMed] [Google Scholar]
- Alvarez G, Ceballos FC, Quinteiro C. The role of inbreeding in the extinction of a European royal dynasty. PLoS ONE. 2009;4 (4:e5174. doi: 10.1371/journal.pone.0005174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez G, Quinteiro C, Ceballos FC.2011Inbreeding and genetic disorderIn: Ikehara K, (ed.).Advances in the Study of Genetic Disorders InTech: Croatia.21–44. [Google Scholar]
- Ballou JD. Ancestral inbreeding only minimally affects inbreeding depression in mammalian populations. J Hered. 1997;88:169–178. doi: 10.1093/oxfordjournals.jhered.a023085. [DOI] [PubMed] [Google Scholar]
- Bennassar B, Blayau N, Denis M, Jacquart J, Lebrun F. Historia Moderna. Akal: Madrid; 2010. [Google Scholar]
- Berra TM, Alvarez G, Ceballos FC. Was the Darwin/Wedgwood dynasty adversely affected by consanguinity. BioScience. 2010;60:376–383. [Google Scholar]
- Bittles AH.2010Consanguinity, genetic drift, and genetic diseases in populations with reduced numbers of foundersIn: Speicher MR, Antonarakis SE, Motulsky AG, (eds).Vogel and Motulskýs Human Genetics: Problems and Approaches Springer-Verlag: Berlin; 507–528. [Google Scholar]
- Bittles AH. Consanguinity in Context. Cambridge University Press: Cambridge; 2012. [Google Scholar]
- Bittles AH, Black ML. Consanguinity, human evolution, and complex diseases. Proc Natl Acad Sci USA. 2010;107 ((suppl 1)::1779–1786. doi: 10.1073/pnas.0906079106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittles AH, Mason WM, Greene J, Appaji Rao N. Reproductive behaviour and health in consanguineous marriages. Science. 1991;252:789–794. doi: 10.1126/science.2028254. [DOI] [PubMed] [Google Scholar]
- Bittles AH, Neel JV. The costs of human inbreeding and their implications for variations at the DNA level. Nat Genet. 1994;8:117–121. doi: 10.1038/ng1094-117. [DOI] [PubMed] [Google Scholar]
- Bixler RH. Sibling incest in the royal families of Egypt, Peru and Hawaii. JSR. 1982a;18:264–281. [Google Scholar]
- Bixler RH. Comment on the incidence and purpose of royal sibling incest. Am Ethnol. 1982b;9:580–582. [Google Scholar]
- Boakes E, Wang J. A simulation study on detecting purging of inbreeding depression in captive populations. Genet Res. 2005;86:139–148. doi: 10.1017/S001667230500772X. [DOI] [PubMed] [Google Scholar]
- Boakes EH, Wang J, Amos W. An investigation of inbreeding depression and purging in captive pedigreed populations. Heredity. 2007;98:172–182. doi: 10.1038/sj.hdy.6800923. [DOI] [PubMed] [Google Scholar]
- Bourgeois-Pichat J.1965The general development of the population of France since the eighteenth centuryIn: Glass DV, Eversley DEC, (eds).Population in History Edward Arnold: London; 87–100. [Google Scholar]
- Cavalli-Sforza LL, Bodmer WF. The Genetics of Human Populations. W H Freeman and Company: San Francisco; 1971. pp. 358–362. [Google Scholar]
- Chakraborty R, Chakravarti A. On consanguineous marriages and the genetic load. Hum Genet. 1977;36:47–54. doi: 10.1007/BF00390435. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D. The genetic basis of inbreeding depression. Genet Res. 1999;74:329–340. doi: 10.1017/s0016672399004152. [DOI] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B. Inbreeding depression and its evolutionary consequences. Ann Rev Ecol Syst. 1987;18:237–268. [Google Scholar]
- Charlesworth D, Willis JH. The genetic basis of inbreeding depression. Nature Rev Genet. 2009;10:783–796. doi: 10.1038/nrg2664. [DOI] [PubMed] [Google Scholar]
- Crnokrak P, Barrett SCH. Purging the genetic load: a review of the experimental evidence. Evolution. 2002;56:2347–2358. doi: 10.1111/j.0014-3820.2002.tb00160.x. [DOI] [PubMed] [Google Scholar]
- Crow JF, Kimura M. An Introduction to Population Genetics Theory. Harper & Row: New York; 1970. pp. 77–81. [Google Scholar]
- Encyclopedia Britannica, Inc 2007The New Encyclopedia Britannica15th edn.The House of Habsburg. Macropaedia. Encyclopedia Britannica: Chicago [Google Scholar]
- Falconer DS, Mackay TFC. Introduction to Quantitative Genetics. Pearson/ Prentice Hall: Harlow; 1996. [Google Scholar]
- García-Dorado A. Understanding and predicting the fitness decline of shrunk populations: inbreeding, purging, mutation, and standard selection. Genetics. 2012;190:1461–1476. doi: 10.1534/genetics.111.135541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glémin S. How are deleterious mutations purged? Drift versus non-random mating. Evolution. 2003;57:2678–2687. doi: 10.1111/j.0014-3820.2003.tb01512.x. [DOI] [PubMed] [Google Scholar]
- Hamamy H, Antonarakis SE, Cavalli-Sforza LL, Temtamy S, Romeo G, Kate LP, et al. Consanguineous marriages, pearls and perils: Geneva international consanguinity workshop report. Genet Med. 2011;13:841–847. doi: 10.1097/GIM.0b013e318217477f. [DOI] [PubMed] [Google Scholar]
- Hawass Z, Gad YZ, Ismail S, Khairat R, Fathalla D, et al. Ancestry and pathology in king Tutankhamun's family. J Am Med Assoc. 2010;303:638–647. doi: 10.1001/jama.2010.121. [DOI] [PubMed] [Google Scholar]
- Hedrick PW. Purging inbreeding depression and the probability of extinction: full-sib mating. Heredity. 1994;73:363–372. doi: 10.1038/hdy.1994.183. [DOI] [PubMed] [Google Scholar]
- Ingrao CH. The Habsburg Monarchy, 1618-1815. Cambridge University Press: Cambridge; 2000. [Google Scholar]
- Jorde LB. Consanguinity and prereproductive mortality in the Utah Mormon population. Hum Hered. 2001;52:61–65. doi: 10.1159/000053356. [DOI] [PubMed] [Google Scholar]
- Khlat M, Khoury M. Inbreeding and diseases: demographic, genetic, and epidemiologic perspectives. Epidemiol Rev. 1991;13:28–41. doi: 10.1093/oxfordjournals.epirev.a036072. [DOI] [PubMed] [Google Scholar]
- Khoury MJ, Cohen BH, Chase GA, Diamond EL. An epidemiologic approach to the evaluation of the effect of inbreeding on prereproductive mortality. Am J Epidemiol. 1987;125:251–262. doi: 10.1093/oxfordjournals.aje.a114524. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick M, Jarne P. The effects of a bottleneck on inbreeding depression and the genetic load. Am Nat. 2000;155:154–167. doi: 10.1086/303312. [DOI] [PubMed] [Google Scholar]
- Kümmel WF.1990De morbis aulicis: on diseases found at courtIn: Nutton N, (ed.).Medicine at the Courts of Europe Routledge: London; pp 1500-1837. [Google Scholar]
- Lancaster HO. Expectations of Life. Springer-Verlag: New York; 1990. [Google Scholar]
- Lynch M, Walsh B. Genetics and Analysis of Quantitative Traits. Sinauer Associates: Sunderland; 1998. pp. 267–268. [Google Scholar]
- Middleton R. Brother-sister and father-daughter marriage in ancient Egypt. Am Sociol Rev. 1962;27:603–611. [Google Scholar]
- Morton NE, Crow FJ, Muller HJ. An estimate of the mutational damage in man from data on consanguineous marriages. Proc Natl Acad Sci USA. 1956;42:855–863. doi: 10.1073/pnas.42.11.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peller S.1965Births and deaths among Europe's ruling families since 1500In: Glass DV, Eversley DEC, (eds).Population in History Edward Arnold: London [Google Scholar]
- Postma E, Martini L, Martini P. Inbred women in a small and isolated Swiss village have fewer children. J Evol Biol. 2010;23:1468–1474. doi: 10.1111/j.1420-9101.2010.02013.x. [DOI] [PubMed] [Google Scholar]
- Radha Rama Devi A, Appaji Rao N, Bittles AH. Inbreeding and the incidence of childhood genetic disorders in Karnakata, South India. J Med Genet. 1987;24:362–365. doi: 10.1136/jmg.24.6.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert A, Toupance B, Tremblay M, Heyer E. Impact of inbreeding on fertility in a pre-industrial population. Eur J Hum Genet. 2009;17:673–681. doi: 10.1038/ejhg.2008.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanghvi LD. Inbreeding in India. Eugen Quart. 1966;13:291–301. doi: 10.1080/19485565.1966.9987682. [DOI] [PubMed] [Google Scholar]
- Simmons MJ, Crow JF. Mutations affecting fitness in Drosophila populations. Ann Rev Genet. 1977;11:49–78. doi: 10.1146/annurev.ge.11.120177.000405. [DOI] [PubMed] [Google Scholar]
- Sked A. The Decline and Fall of the Hapsburg Empire. Longman: London; 2001. [Google Scholar]
- Sokal RR, Rohlf FJ. Biometry. Freeman: New York; 1995. pp. 498–499. [Google Scholar]
- Wandruszka A. The House of Habsburg: Six Hundred Years of a European Dynasty. Greenwood Press: Westport; 1964. [Google Scholar]
- Wang J, Hill WG, Charlesworth D, Charlesworth B. Dynamics of inbreeding depression due to deleterious mutations in small populations: Mutation parameters and inbreeding rate. Genet Res. 1999;74:165–178. doi: 10.1017/s0016672399003900. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.