Abstract
Genetic rearrangement by recombination is one of the major driving forces for genome evolution, and recombination is known to occur in non-random, discreet recombination sites within the genome. Mapping of recombination sites has proved to be difficult, particularly, in the human MHC region that is complicated by both population variation and highly polymorphic HLA genes. To overcome these problems, HLA-typed individuals from three representative populations: Asian, European and African were used to generate phased HLA haplotypes. Extended haplotype homozygosity (EHH) plots constructed from the phased haplotype data revealed discreet EHH drops corresponding to recombination events and these signatures were observed to be different for each population. Surprisingly, the majority of recombination sites detected are unique to each population, rather than being common. Unique recombination sites account for 56.8% (21/37 of total sites) in the Asian cohort, 50.0% (15/30 sites) in Europeans and 63.2% (24/38 sites) in Africans. Validation carried out at a known sperm typing recombination site of 45 kb (HLA-F-telomeric) showed that EHH was an efficient method to narrow the recombination region to 826 bp, and this was further refined to 660 bp by resequencing. This approach significantly enhanced mapping of the genomic architecture within the human MHC, and will be useful in studies to identify disease risk genes.
Keywords: MHC, recombination sites, HLA haplotypes, Asian
Introduction
The human major histocompatibility complex (MHC) genomic region is the most gene-dense and polymorphic in the human genome. However it is also known to exhibit lower recombination rates relative to the genome average, with human leukocyte antigen (HLA) alleles inherited as haplotypes and preserving a high degree of linkage disequilibrium (LD) (Traherne, 2008). Recombination is one of the key processes driving genome evolution and genetic polymorphism, with recombination events occurring in localized sites within a 1–2 kb region, characteristically bordered by segments of low recombination rate (The International HapMap Consortium, 2005). Occurrences of recombination events are found to be influenced by short DNA sequences in yeast (Petes, 2001), but such sequence motifs are only able to explain a fraction of all sites in humans (Myers et al., 2006, 2008).
Locating recombination sites by empirical means have been difficult and in the human MHC, with early detection of meiotic recombination sites being conducted using single sperm recombinant typing (Jeffreys et al., 2001; Cullen et al., 2002). To date, only six recombination sites have been definitively mapped within the 4 Mb human MHC region using this method (Cullen et al., 2002). An alternative approach is by in silico modeling of population-genotyped single nucleotide polymorphism (SNPs) data (Calabrese, 2007; Coop et al., 2008; Khil and Camerini-Otero, 2010), most notably from the International HapMap Project (The International HapMap Consortium, 2005), where the vast amount of data provide a means to estimate localized regions of elevated recombination rates from the patterns of LD. Yet, this method of using admixed population in data analysis, though useful in identifying recombination sites in many parts of the genome, is insufficient when applied to the MHC region. This is due to marked divergence in the haplotypic structure within the MHC across geographically distinct population, which is also influenced by the underlying HLA allelic combinations (Blomhoff et al., 2006; de Bakker et al., 2006; Smith et al., 2006). Population-specific reshuffling of the MHC genomic region driven by factors such as natural selection and neutral genetic drift may result in recombination breakpoints specific to each population. Without consideration of the chromosomal HLA allelic typing, the inference of recombination sites from admixed population-genotyped SNPs data may mask the identification of recombination sites (Maniatis et al., 2002), and possibly prevent the detection of population-specific recombination sites. Thus far, there has been no single approach that provides sufficient sensitivity and precision to detect recombination sites within the MHC region.
The human MHC is known to be associated with numerous autoimmune and inflammatory diseases (Trowsdale, 2011), but efforts to identify susceptible genes are often impeded due to its extreme LD. Hence, accurate inference of a recombination map in the MHC region will be highly advantageous for disease risk marker identification. In this study, we describe a method for identifying recombination sites within the MHC region that integrates both HLA-typed data and phased chromosomal SNPs data to generate extended haplotype homozygosity (EHH) plots (Sabeti et al., 2002) in a representative Asian population comprising Singapore Chinese (CHSG) (Yu et al., 2005). This will address the deficiency in current recombination inference methods, which does not incorporate information on HLA haplotype variants. More importantly, when LD is compared as phase-discrete haplotypes, it will significantly enhance the sensitivity to detect breakpoints, as it eliminates the ambiguity generated by heterozygous SNP calls. Furthermore, to validate and refine the presence of recombination breakpoints in these haplotypes, sequencing was performed across SNPs intervals that demonstrated a breakdown of haplotypes in the EHH analysis. We then applied this method to European- and African-descent populations to examine their recombination profiles, and thus provide a detailed comparative analysis of the recombination patterns within the human MHC.
Materials and methods
DNA samples and sequence-based HLA typing
DNA was extracted from peripheral blood lymphocytes obtained with prior consent from 247 blood donors who are Singapore Chinese (CHSG). They were typed for four HLA loci, HLA-A, -B, -C and -DRB1 by sequence-based typing. For the HLA-A, -B and -C genes, this involved PCR amplification of hypervariable exons 2 and 3 using specific primers, followed by direct DNA sequencing of the PCR products in opposite directions. HLA-DRB1 was sequenced and typed as previously described (Sayer et al., 2004). The purified PCR templates were sequenced with the corresponding primers using the ABI BigDye Terminator v3.1 Cycle Sequencing Kit, as per manufacturer's instructions. The extension products were then purified using an ethanol/EDTA/sodium acetate precipitation protocol to remove excess dye terminators. Sequencing was performed by electrophoresis on 96-well plates in an ABI Prism 3100 Genetic Analyser (Applied Biosystems, Foster City, CA, USA).
SNPs genotyping and selection
The analysis of SNPs of the MHC region was performed on the Illumina GoldenGate MHC Panel platform (Illumina, San Diego, CA, USA). The SNPs coordinates were mapped to the Human Reference Sequence Assembly 36.1 (NCBI 36.1). Genotyping results were filtered using the following criteria: SNP loci deviating from Hardy–Weinberg equilibrium using a Fisher's exact test at a significance level of 0.001; SNPs loci with a call rate of <95% and SNP loci with a minor allele frequency of <5% were discarded. In addition, for familial data, SNP genotypes that were discordant with the parental structure in more than one family were discarded. After the quality control checks, 1877 SNP loci were left for further analysis.
Haplotypes reconstruction and analysis
Haplotype reconstruction was performed using a Bayesian-based algorithm, implemented in the software PHASE 2.1 (Stephens et al., 2001). To maximize the phase-unambiguous information from 12 families, haplotype phasing for the high-resolution SNP map was done in two stages. First the phase-unambiguous family haplotypes were obtained by running PHASE with pedigree information. These produced a set of 24 distinct phase-unambiguous haplotypes. Next, these haplotypes were treated as ‘known' haplotypes and seeded together with the unrelated individuals for haplotype reconstruction with PHASE. HLA alleles were translated into unique digits and phased together with the SNPs.
EHH and LD analysis
EHH analysis was performed independently on chromosomal SNPs haplotypes tagged to their HLA alleles. Briefly, EHH calculated at a position x is defined as the probability that two chromosomes, carrying an allele (or haplotype) of interest at an anchor locus, have identical SNPs sequence from the anchor locus to the position x (Sabeti et al., 2002).
LD maps were assembled from four-loci HLA chromosomal SNPs haplotypes using LDMAP (Maniatis et al., 2002). LD maps constructed reflect the LD pattern characterized in form of accumulative LD unit (LDU), where the map length is relative to the cumulative recombination over many generations. The historical recombination rates for HLA chromosomal SNPs haplotypes were estimated using the program LDHat (McVean et al., 2004). LDHat implements a coalescent-based approach to infer recombination rates from the SNPs genetic variation patterns with a block penalty of 5 and 10 000 000 iterations. To translate the population-scaled recombination rates (4Ner) presented by LDHat to per-generation recombination rates (r), the effective population size (Ne) was set at 10 000 (Takahata, 1993; Erlich et al., 1996; Conrad et al., 2006).
Probable recombination sites were scored when either EHH decay is observed in two or more HLA haplotypes across the SNPs interval or when at least 10% of a single HLA haplotype diverged from the core haplotype.
PCR amplification for sequencing of recombination sites
For resequencing of recombination sites, PCR primers were designed such that overlapping PCR fragments tile across the recombination site interval. Primers were designed from repeat-masked human genome sequence with the help of the Primer3 application. Supplementary Table 1 lists the pairs of PCR primers used for each recombination site and includes the annealing temperatures that were specific to the thermal cycling profiles for each PCR reaction. Sequencing was carried out as previously described.
Recombination sites variation between populations
Genotype data consisting of 30 European (CEU) trios and 30 Yoruban (YRI) trios were obtained from the HapMap phase II collection. SNPs genotype data within the MHC region interrogated by the Illumina GoldenGate assay were selected and subjected to the screening procedure as described above. The HapMap populations HLA-A, -B and -C typing were attained from (Erlich et al., 2011) and phased together with the selected SNPs. A total of 1360 common SNPs loci found across the CEU, YRI and CHSG populations were chosen for further downstream EHH analysis and population recombination sites comparison.
Results
LD pattern and identification of recombination sites across MHC region
HLA SNP haplotypes were derived using PHASE 2.1 (Stephens et al., 2001), from 247 CHSG individuals (211 unrelated and 36 comprising members of family trios) and the resulting 470 chromosomes were grouped according to their HLA alleles. Among the most frequently occurring HLA haplotypes, those detected were A*02:07-C*01:02-B*46:01 (12.8%), C*01:02-B*46:01-DRB1*09:01 (9.6%), A*33:03-C*03:02-B*58:01 (9.0%), C*03:02-B*58:01-DRB1*03:01 (7.7%), A*02:03-C*07:02-B*38:02 (4.3%), A*11:01-C*07:02-B*40:01 (4.0%), and C*08:01-B*15:02-DRB1*12:02 (3.6%) (Supplementary Table 2 and 3). These HLA haplotypes are subjected to EHH analysis where the computed EHH scores were then plotted against the physical distance at varying positions from the defined core region (Figure 1). The results showed that at incremental distances away from a defined HLA locus, the EHH scores of a haplotype registers a drop whenever a recombination event disrupted the integrity of the LD. Genomic segments with high EHH scores are stretches of conserved regions that have relatively low numbers of recombination events. In addition, the recombination rates based on the pooled 470 chromosomes were also estimated using LDhat (McVean et al., 2004).
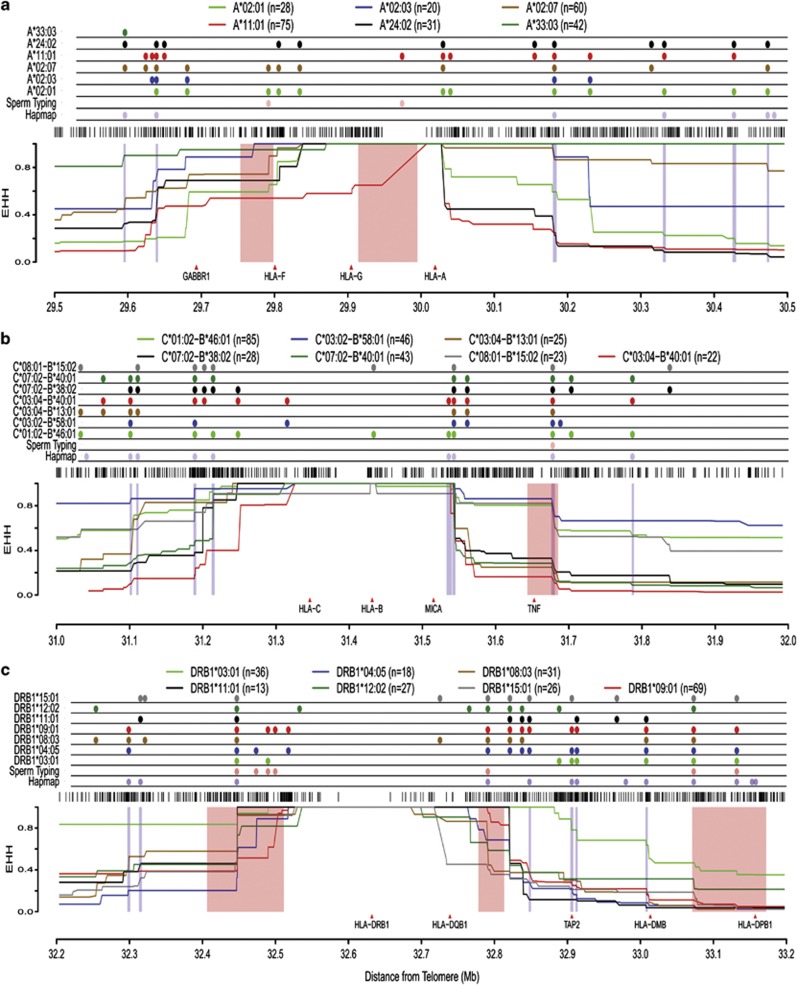
Figure 1.
EHH plots of SNP haplotypes for common HLA alleles. Plots covering 1 Mb region with the classical HLA loci/haplotypes used as anchor positions: (a) HLA-A, (b) HLA-C-B and (c) HLA-DRB1 respectively. Positions of recombination sites and their relative sizes are mapped onto the plots as follows: six recombination segments identified by sperm recombinants (Cullen et al., 2002) (highlighted in pink columns) and HapMap-inferred recombination sites (The International HapMap Consortium, 2005) that coincide with EHH drops (highlighted in blue columns). The black bars above each EHH plot represent the density of SNP coverage in the region.
Generally, strong homozygosity with EHH ⩾0.9, indicating the preservation of an extended haplotype, can be observed in extended regions proximate to the HLA-A, -B and -DRB1 genes. Strong homozgosity or regions of high LD were observed at least 190 kb (position 29 838 709–30 027 753) at the HLA-A region, 213 kb (position 31 325 794–31 538 700) for all the common HLA-C-B haplotypes and 181 kb (position 32 536 263–32 717 405) at the HLA-DRB1 region (Figure 2). In most instances, these segments were flanked by recombination events characterized by EHH drops in several HLA haplotypes. The recombination rates estimated from the pooled chromosomes generally were in agreement with the EHH analysis, whereby the strong homozygosity segments coincided with low recombination rates. The few recombination rates peaks observed in regions near the HLA-G and HLA-B genes are likely to be SNPs sequence variation between different allelic haplotypes rather than the differences within same allelic haplotype.
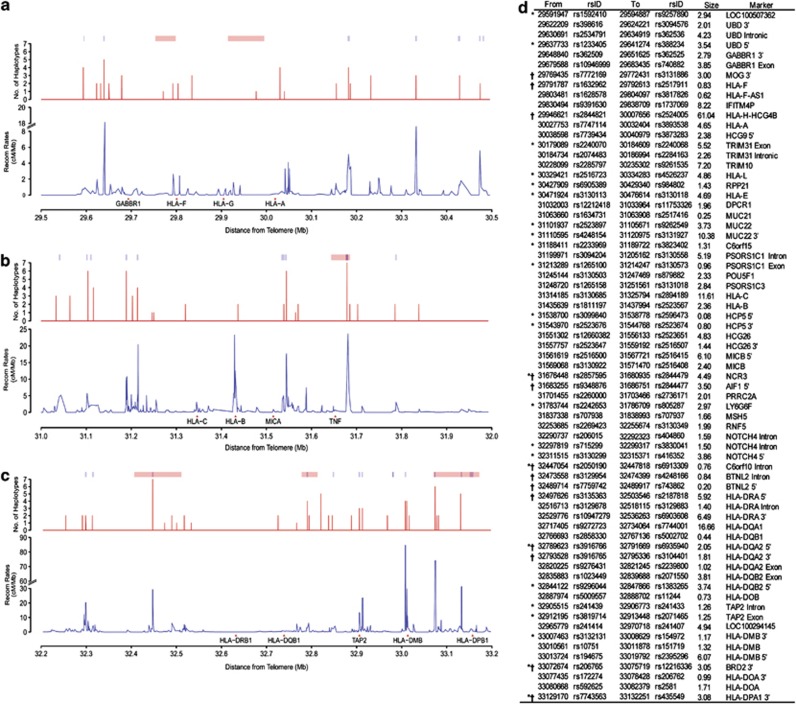
Figure 2.
Recombination regions identified by SNPs interval displaying EHH decay. Plots illustrate the number of unique HLA allelic haplotypes independently registered drop in EHH (red) and the recombination rates (blue) across (a) HLA-A, (b) HLA-C-B and (c) HLA-DRB1. Recombination segments identified by sperm recombinants (Cullen et al., 2002) are indicated by the pink colored bars and the HapMap-inferred recombination sites (The International HapMap Consortium, 2005) are indicated by the blue colored bars. (d) List of the recombination sites. ‘†' indicates intervals that reside within sperm typing segments and ‘*' indicates intervals that overlapped with the HapMap recombination sites. The recombination site was mapped to the Human Reference Sequence Assembly 36.1 (NCBI 36.1) and assigned to a marker that is in the closest proximity to the site.
Decay in EHH values across SNPs intervals is indicative of probable recombination sites and is characterized by distinct step-wise drops that can be seen to accumulate with greater distance from the defined locus position. These drops occur at discreet intervals and in a non-random manner, suggesting the presence of specific genomic sequences or recombination sites. Furthermore, multiple haplotypes independently exhibited EHH drops at the same genomic location, providing additional support for the presence of a recombination site. As an example, it can be seen that four different haplotypes: HLA-A*02:01, A*02:07, A*11:01 and A*24:02 haplotypes all registered decay in EHH at the position 30 027 753–30 032 404, indicative of a recombination site (Figure 1). In total, 69 EHH drops were captured across 4.91 Mb of the extended MHC (Figure 2d). In comparison, other approaches such as sperm typing and Hapmap prediction identified only 6 and 29 recombination sites respectively across the same 4.1 Mb region. Thirty seven EHH drop sites were found to be entirely novel, that is, not previously identified by sperm typing or by HapMap inference. There were also EHH-identified sites that occurred in regions with low recombination rates. This observation is likely because these recombination sites occur only in specific HLA haplotype variants and the number is not large enough to translate into a spike in the recombination rates estimated from the population-pooled chromosomes. This highlights the importance of accounting for the HLA haplotypes variants to improve the sensitivity and specificity in the recombination sites and LD pattern analyses within the MHC region. The EHH-defined recombination sites are relatively precise, with 28/69 (41%) mapped to <2 kb in size and another 29/69 (42%) falling between 2 and 5 kb. This illustrates the enhanced precision of the EHH mapping protocol. Moreover, more than a third of the recombination sites were found in the MHC class II region, consistent with reports that this region was subjected to more intense segments shuffling as compared with the class I region (Miretti et al., 2005).
Refining haplotypes breakdown intervals
To validate that EHH decays reflect the occurrence of recombination events leading to breakdown in the haplotypes, several EHH drop sites were selected for sequencing. The allelic pattern of the polymorphic sites in the recombinant haplotypes was used to mark out the boundary where recombination occurred. These haplotypes are conserved either telomeric or centromeric of the breakpoint; combined, they reveal the chromosomal crossover region at the putative recombination site.
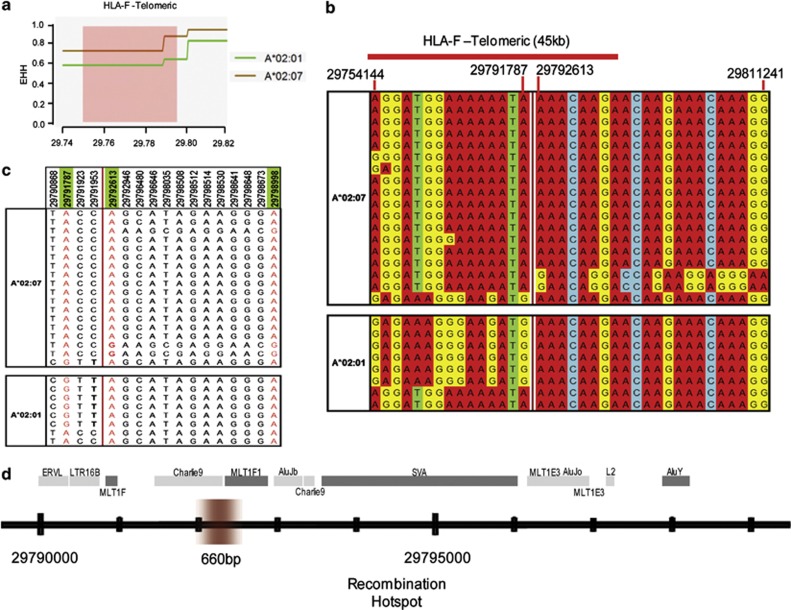
A sperm-mapped 45-kb recombination segment between STR markers MOG-C to RF (HLA-F-telomeric) was narrowed down to a 0.8-kb interval defined by an EHH drop at position 29 791 787–29 792 613 (Figure 3a). Two HLA haplotypes A*02:01 and A*02:07 independently registered this identical EHH drop. Individuals carrying at least 1 copy of the HLA-A haplotypes that break across this interval were selected for sequencing, giving a total of 8 HLA-A*02:01 haplotypes and 18 HLA-A*02:07 haplotypes.
Figure 3.
Recombination site within the sperm-typed recombination region between STR markers MOG-C to RF. (a) EHH plot of HLA-A haplotypes across the HLA-F telomeric recombination region (45 kb) previously defined by sperm typing. (b) The allelic calls of the Illumina GoldenGate MHC genotyped SNPs on the HLA-A*02:01 and HLA-A*02:07 haplotypes telomeric and centromeric of the recombination interval. (c) Resequencing across an 8 kb segment (position 29 790 868 to 29 798 998) that overlaps with the SNPs recombination interval. HLA-A*02:01 and HLA-A*02:07 haplotypes are derived from SNPs ascertained through resequencing. Positions highlighted in green correspond to SNPs in the Illumina GoldenGate MHC panel and their allelic calls are highlighted in red while the others are polymorphic sites revealed by resequencing. (d) Location of 660 bp recombination site refined by resequencing and the repetitive elements in close proximity to the site.
Of 18 HLA-A*02:07 haplotypes, 14 are telomeric-conserved before 29 791 787; of these 14, 2 diverged from the others across this interval. Meanwhile, 16/18 HLA-A*02:07 haplotypes are centromeric-conserved after 29 792 613; of these, 1 diverged from the others across the interval. For HLA-A*02:01, all 8 haplotypes are centromere-conserved, and 2 diverged across the interval (Figure 3b). For sequencing, a long 8-kb stretch was selected in order to reveal DNA elements flanking the recombination site. The sequencing results in Figure 3c uncovered 17 polymorphic SNP sites, including 3 sites assayed by the Illumina GoldenGate MHC SNPs (29 791 787, 29 792 613 and 29 798 998). Sequencing calls for all three sites corroborated with the Illumina GoldenGate MHC SNPs array output, independently validating the existence of haplotypes breakdown. The telomeric end of the EHH drop was thus further refined to position 29 791 953, defining a 660-bp window for the recombination site. The targeted sequencing further revealed a number of transposon elements, such as AluY, L2 elements (Witherspoon et al., 2009), LTR16B and MLT1F (Gaudieri et al., 1999), flanking the identified site. Two other SNPs intervals (position 31 676 448–31 680 935 and position 33 132 110–33 132 251) within the two sperm-mapped recombination site segments (LTA–BAT2 and BRD2–HLA-DPB1) were selected and sequenced. Using the above-described method, we are able to refine the haplotype crossover sites of LTA–BAT2 from 42.1 to 1.7 kb (Supplementary Figure 1), and BRD2–HLA-DPB1 from 101.6 to 1.7 kb (Supplementary Figure 2).
Population-specific recombination sites
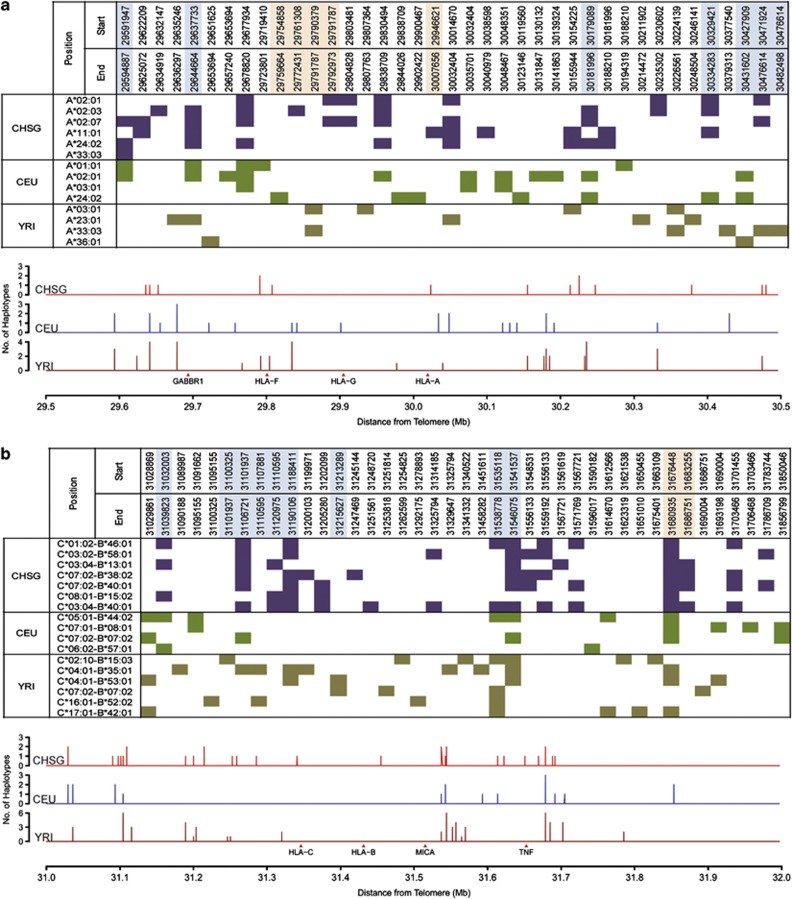
Our knowledge of recombination variation between populations is limited and not well studied. The EHH approach integrated with the HLA information present an excellent opportunity to investigate the MHC region recombination profiles of different populations. Here, we applied our method on two HapMap populations, the Europeans (CEU) and the Yorubans (YRI), of which the HLA-A and HLA-B typings were available. To maintain consistency, 1360 SNPs that were common and found in all three populations were used for the EHH analysis (Supplementary Figure 3 and 4). We were able to map back to >90% of the recombination sites in the CHSG population, albeit at larger recombination sites interval, as identified in the previous section. Together, there were 37 intervals identified as probable recombination sites in the CHSG, 30 in the CEU and 38 in YRI (Figure 4). Interestingly, >50% of the identified sites in each population (CHSG—56.8%, CEU—50.0% and YRI—63.2%) were uniquely population-specific. In fact, <16% of the sites were shared among the three populations and all of these sites fell within either the HapMap determined recombination sites or the segments determined by sperm typing (Supplementary Table 4). Furthermore, recombination sites specific to a population can also be found to be relatively close to other recombination sites unique to another population group resulting in a genomic segment where recombination events are likely to be found across populations. This was especially evident in regions near HLA-B. For example, in the genomic segment near MUC 22, the population-specific recombination sites identified in the three populations were within a 31-kb segment (position 31 089 987–31 120 975). This observation is in agreement with the previous reports where recombination sites tend to occur within a cluster surrounded by regions with reduced recombination (Jeffreys et al., 2001).
Figure 4.
Recombination sites across the CHSG, CEU and YRI population. (a) HLA-A (b) HLA-C-B. The upper section of each panel displays the HLA haplotypes recombination sites interval across the three populations. The lower section of each panel displays plots that indicate the number of unique HLA allelic haplotypes independently registered EHH drop in the CHSG, CEU and YRI.
DiscusSion
From a genomic perspective, the human MHC region is unique as it is characterized by diverse haplotypic variation and structure. The contribution of individual HLA haplotypes towards recombination is overlooked by in silico approaches that used pooled global population data (Stumpf and McVean, 2003). The admixture of a vast number of diverse HLA haplotypes complicates the inference process and masks potential recombination sites. To address the problem, we set out to construct EHH plots using phased HLA haplotypes to identify recombination sites. We utilized phased HLA haplotypes from a single population comprising CHSG and were able to identify novel recombination sites, thus enhancing the recombination map resolution. Furthermore, the strong concordance of the identified recombination sites in CHSG population with the HapMap and sperm typing recombination segments (Cullen et al., 2002; The International HapMap Consortium, 2005), reinforced by the sequencing of selected regions to illustrate haplotypes breakpoints, demonstrates the reliability of this approach.
Earlier studies from Walsh et al. (2003) and Ahmad et al. (2003) have characterized the LD patterns of the Caucasian population. Though these studies highlighted the extent of LD within the MHC region, the resultant LD maps are limited in scope due to the relatively small amount of polymorphic markers used to derive them. A more recent study created a comprehensive haplotype map by investigating the LD between the SNPs and the HLA alleles across the extended MHC region (7.5 Mb) in four populations (de Bakker et al., 2006). This study effectively demonstrated that the extent of LD along the chromosomes is dependent on the underlying HLA allelic haplotypes and provided a panoramic view of the MHC genomic architecture. In comparison, our study provides a more detailed and precise description on the change in the LD structure, as well as LD breakages along the HLA haplotypes localized to the HLA class I and class II gene regions where the LD breakpoints are inferred as putative recombination sites. In the HapMap study, recombination sites were inferred from CEU, YRI and an Asian population comprising both Han Chinese (CHB) and Japanese (JPT). A recombination site was classified when at least two out of the three populations displayed evidence of recombination events, and therefore, these sites are often generic across populations, not population-specific (Khil and Camerini-Otero, 2010). Currently, there has been no extensive examination to characterize the recombination profile across different populations within the MHC region. The study by de Bakker et al., 2006 estimated the recombination rates independently from four populations and combined to provide a single estimate for the MHC region, but did not go further to compare the recombination rates variation between populations. A further study by (Kong et al., 2010), which compared the LD-based maps derived from three different populations revealed variation in the recombination rate between populations at the genome level without detailed information on the MHC region. To provide a clearer insight into the population-specific recombination profile within the highly polymorphic MHC region, we applied our approach to the CHSG, CEU and YRI population and examined their recombination profiles across the HLA-A and HLA-C-B region. Our study shows low number of recombination sites shared among the three populations: >50% of the recombination sites identified are specific to a single population. Recombination activities result in the breakage of haplotypes and have a direct influence on the genome haplotype diversity (Kauppi et al., 2004). Our findings of limited recombination sites shared among distinct populations suggest that the population-specific recombination sites may be an important mechanism that contributes to this high diversity of haplotypes within the MHC region. In contrast, in other parts of human genome, sites of haplotypes breakage are generally shared across distinct populations resulting in limited diversification of haplotypes (Daly et al., 2001; Conrad et al., 2006). Studies on other populations will be able to reveal additional population-specific recombination patterns that characterize the evolution of the MHC haplotypes. In conclusion, our data demonstrates the important role of HLA haplotypes in the identification of recombination sites within the human MHC region, and that discovery of unique recombination sites is possible only through single population analysis.
Data archiving
Genotype data have been submitted to Dryad: doi:10.5061/dryad.65v85.
Acknowledgments
This work was supported by Singapore Immunology Network, A*STAR.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Ahmad T, Neville M, Marshall SE, Armuzzi A, Mulcahy-Hawes K, Crawshaw J, et al. Haplotype-specific linkage disequilibrium patterns define the genetic topography of the human MHC. Hum Mol Genet. 2003;12:647–656. [PubMed] [Google Scholar]
- Blomhoff A, Olsson M, Johansson S, Akselsen HE, Pociot F, Nerup J, et al. Linkage disequilibrium and haplotype blocks in the MHC vary in an HLA haplotype specific manner assessed mainly by DRB1*03 and DRB1*04 haplotypes. Genes Immun. 2006;7:130–140. doi: 10.1038/sj.gene.6364272. [DOI] [PubMed] [Google Scholar]
- Calabrese P. A population genetics model with recombination hotspots that are heterogeneous across the population. Proc Natl Acad Sci USA. 2007;104:4748–4752. doi: 10.1073/pnas.0610195104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, Jakobsson M, Coop G, Wen X, Wall JD, Rosenberg NA, et al. A worldwide survey of haplotype variation and linkage disequilibrium in the human genome. Nat Genet. 2006;38:1251–1260. doi: 10.1038/ng1911. [DOI] [PubMed] [Google Scholar]
- The International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coop G, Wen X, Ober C, Pritchard JK, Przeworski M. High-resolution mapping of crossovers reveals extensive variation in fine-scale recombination patterns among humans. Science. 2008;319:1395–1398. doi: 10.1126/science.1151851. [DOI] [PubMed] [Google Scholar]
- Cullen M, Perfetto SP, Klitz W, Nelson G, Carrington M. High-resolution patterns of meiotic recombination across the human major histocompatibility complex. Am J Hum Genet. 2002;71:759–776. doi: 10.1086/342973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly MJ, Rioux JD, Schaffner SF, Hudson TJ, Lander ES. High-resolution haplotype structure in the human genome. Nat Genet. 2001;29:229–232. doi: 10.1038/ng1001-229. [DOI] [PubMed] [Google Scholar]
- de Bakker PIW, McVean G, Sabeti PC, Miretti MM, Green T, Marchini J, et al. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat Genet. 2006;38:1166–1172. doi: 10.1038/ng1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlich HA, Bergstrom TF, Stoneking M, Gyllensten U. HLA sequence polymorphism and the origin of humans. Science. 1996;274:1552b–1554b. doi: 10.1126/science.274.5292.1552b. [DOI] [PubMed] [Google Scholar]
- Erlich RL, Jia X, Anderson S, Banks E, Gao X, Carrington M, et al. Next-generation sequencing for HLA typing of class I loci. BMC Genomics. 2011;12:42. doi: 10.1186/1471-2164-12-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudieri S, Kulski JK, Dawkins RL, Gojobori T. Different evolutionary histories in two subgenomic regions of the major histocompatibility complex. Genome Res. 1999;9:541–549. [PubMed] [Google Scholar]
- Jeffreys AJ, Kauppi L, Neumann R. Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nat Genet. 2001;29:217–222. doi: 10.1038/ng1001-217. [DOI] [PubMed] [Google Scholar]
- Kauppi L, Jeffreys AJ, Keeney S. Where the crossovers are: recombination distributions in mammals. Nat Rev Genet. 2004;5:413–424. doi: 10.1038/nrg1346. [DOI] [PubMed] [Google Scholar]
- Khil PP, Camerini-Otero RD. Genetic crossovers are predicted accurately by the computed human recombination map. PLoS Genet. 2010;6:e1000831. doi: 10.1371/journal.pgen.1000831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Thorleifsson G, Gudbjartsson DF, Masson G, Sigurdsson A, Jonasdottir A, et al. Fine-scale recombination rate differences between sexes, populations and individuals. Nature. 2010;467:1099–1103. doi: 10.1038/nature09525. [DOI] [PubMed] [Google Scholar]
- Maniatis N, Collins A, Xu CF, McCarthy LC, Hewett DR, Tapper W, et al. The first linkage disequilibrium (LD) maps: delineation of hot and cold blocks by diplotype analysis. Proc Natl Acad Sci USA. 2002;99:2228–2233. doi: 10.1073/pnas.042680999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean GAT, Myers SR, Hunt S, Deloukas P, Bentley DR, Donnelly P. The fine-scale structure of recombination rate variation in the human genome. Science. 2004;304:581–584. doi: 10.1126/science.1092500. [DOI] [PubMed] [Google Scholar]
- Miretti MM, Walsh EC, Ke X, Delgado M, Griffiths M, Hunt S, et al. A high-resolution linkage-disequilibrium map of the human major histocompatibility complex and first generation of tag single-nucleotide polymorphisms. Am J Hum Genet. 2005;76:634–646. doi: 10.1086/429393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers S, Freeman C, Auton A, Donnelly P, McVean G. A common sequence motif associated with recombination hot spots and genome instability in humans. Nat Genet. 2008;40:1124–1129. doi: 10.1038/ng.213. [DOI] [PubMed] [Google Scholar]
- Myers S, Spencer CC, Auton A, Bottolo L, Freeman C, Donnelly P, et al. The distribution and causes of meiotic recombination in the human genome. Biochem Soc Trans. 2006;34:526–530. doi: 10.1042/BST0340526. [DOI] [PubMed] [Google Scholar]
- Petes TD. Meiotic recombination hot spots and cold spots. Nat Rev Genet. 2001;2:360–369. doi: 10.1038/35072078. [DOI] [PubMed] [Google Scholar]
- Sabeti PC, Reich DE, Higgins JM, Levine HZP, Richter DJ, Schaffner SF, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- Sayer D, Whidborne R, De Santis D, Rozemuller E, Christiansen F, Tilanus M. A multicenter international evaluation of single-tube amplification protocols for sequencing-based typing of HLA-DRB1 and HLA-DRB3,4,5. Tissue Antigens. 2004;63:412–423. doi: 10.1111/j.0001-2815.2004.00214.x. [DOI] [PubMed] [Google Scholar]
- Smith WP, Vu Q, Li SS, Hansen JA, Zhao LP, Geraghty DE. Toward understanding MHC disease associations: partial resequencing of 46 distinct HLA haplotypes. Genomics. 2006;87:561–571. doi: 10.1016/j.ygeno.2005.11.020. [DOI] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpf MPH, McVean GAT. Estimating recombination rates from population-genetic data. Nat Rev Genet. 2003;4:959–968. doi: 10.1038/nrg1227. [DOI] [PubMed] [Google Scholar]
- Takahata N. Allelic genealogy and human evolution. Mol Biol Evol. 1993;10:2–22. doi: 10.1093/oxfordjournals.molbev.a039995. [DOI] [PubMed] [Google Scholar]
- Traherne JA. Human MHC architecture and evolution: implications for disease association studies. Int J Immunogenet. 2008;35:179–192. doi: 10.1111/j.1744-313X.2008.00765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trowsdale J. The MHC, disease and selection. Immunol Lett. 2011;137:1–8. doi: 10.1016/j.imlet.2011.01.002. [DOI] [PubMed] [Google Scholar]
- Walsh EC, Mather KA, Schaffner SF, Farwell L, Daly MJ, Patterson N, et al. An integrated haplotype map of the human major histocompatibility complex. Am J Hum Genet. 2003;73:580–590. doi: 10.1086/378101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witherspoon DJ, Watkins WS, Zhang Y, Xing J, Tolpinrud WL, Hedges DJ, et al. Alu repeats increase local recombination rates. BMC Genomics. 2009;10:530. doi: 10.1186/1471-2164-10-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HX, Chia JM, Bourque G, Wong MV, Chan SH, Ren EC. A population-based LD map of the human chromosome 6p. Immunogenetics. 2005;57:559–565. doi: 10.1007/s00251-005-0002-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.