Abstract
The mammalian neocortical progenitor cell niche is composed of a diverse repertoire of neuroepithelial cells, radial glia (RG), and intermediate neurogenic progenitors (INPs). Previously, live-cell imaging experiments have proved crucial in identifying these distinct progenitor populations, especially INPs, which amplify neural output by undergoing additional rounds of proliferation before differentiating into new neurons. INPs also provide feedback to the RG pool by serving as a source of Delta-like 1 (Dll1), a key ligand for activating Notch signaling in neighboring cells, a well-known mechanism for maintaining RG identity. While much is known about Dll1-Notch signaling at the molecular level, little is known about how this cell–cell contact dependent feedback is transmitted at the cellular level. To investigate how RG and INPs might interact to convey Notch signals, we used high-resolution live-cell multiphoton microscopy (MPM) to directly observe cellular interactions and dynamics, in conjunction with Notch-pathway specific reporters in the neocortical neural stem cell niche in organotypic brain slices from embryonic mice. We found that INPs and RG interact via dynamic and transient elongate processes, some apparently long-range (extending from the subventricular zone to the ventricular zone), and some short-range (filopodia-like). Gene expression profiling of RG and INPs revealed further progenitor cell diversification, including different subpopulations of Hes1+ and/or Hes5+ RG, and Dll1+ and/or Dll3+ INPs. Thus, the embryonic progenitor niche includes a network of dynamic cell–cell interactions, using different combinations of Notch signaling molecules to maintain and likely diversify progenitor pools.
Introduction
During mammalian neocortical development, glutamatergic neurons are generated within a specialized neurogenic niche from diverse progenitor populations located in the ventricular zone (VZ) and subventricular zone (SVZ), including neuroepithelial cells, radial glia (RG) in the VZ (RGvz) and outer SVZ (RGosvz), and intermediate neurogenic progenitors (INPs) in the VZ (INPvz, also known as short neural precursors, SNPs) and SVZ (INPsvz) (Gal et al., 2006; Corbin et al., 2008; Pontious et al., 2008; Kriegstein and Alvarez-Buylla, 2009; Fietz et al., 2010; Hansen et al., 2010; Stancik et al., 2010; Taverna and Huttner, 2010; Shitamukai et al., 2011; Wang et al., 2011; Hevner and Haydar, 2012; Franco and Müller, 2013). Altogether, these diverse progenitors coordinate neurogenesis, including generating appropriate numbers of neurons, guiding migration, and establishing patterning information within and between radial units, disruption of which may underlie neurological disorders.
INPs amplify neural output from RG through additional rounds of mitoses before symmetrically differentiating into newborn glutamatergic neurons (Haubensak et al., 2004; Miyata et al., 2004; Noctor et al., 2004), contributing pyramidal neurons for all layers of the neocortex (Kowalczyk et al., 2009). INPs express Tbr2 (also known as Eomes), a T-box transcription factor important in INP specification (Englund et al., 2005; Arnold et al., 2008; Sessa et al., 2008), and for propagation of protomap patterning information from RG to newborn neurons within radial columns (Elsen et al., 2013). INPs are also a source for Delta-like 1 (Dll1) feedback signals to activate Notch signaling in RG (Mizutani et al., 2007; Kawaguchi et al., 2008b; Yoon et al., 2008). Thus, INPs simultaneously provide intrinsic neural differentiation information to new neurons, and extrinsic inhibitory signals that maintain the RG pool. How this latter signal is relayed at the cellular level is unknown.
Notch signaling is an evolutionarily conserved cell–cell contact-dependent mechanism for generating cellular diversity. While much about the molecular basis of Notch signaling has been elucidated in the mammalian neocortex (Fortini, 2009; Kopan and Ilagan, 2009; Ables et al., 2011; Pierfelice et al., 2011), virtually nothing is known about the actual types of cellular contacts that must convey this molecular information in the in vivo embryonic neurogenic niche. Interestingly, a handful of studies from Drosophila have revealed that Notch signaling occurs through the extension and retraction of dynamic long-range processes containing Delta protein in punctate distribution (De Joussineau et al., 2003; Rajan et al., 2009; Cohen et al., 2010). Previous live-cell imaging studies in the neocortex revealed that INPs undergo four sequential stages or modes of migration, each with unique morphological properties (Noctor et al., 2004). Since INPs are a source of Dll1, we hypothesized that INP morphological changes may be more than just migration stages, and might serve as a basis for INP-mediated Dll1 feedback to RG, similar to the long-range lateral inhibition events observed in Drosophila. We tested this hypothesis by using high-resolution two-color live-cell multiphoton imaging, FACS-based gene expression profiling, and immunocytochemistry to identify how INPs and RG interact to transmit differential Notch signaling in the neocortical neurogenic niche.
Materials and Methods
Terminology.
We adapted the progenitor nomenclature used by Kawaguchi et al. (2008a) based on their molecular profiling of neocortical progenitors, and modified it according to current cellular information. Neocortical embryonic neuroepithelial stem cell (eNSC) progenitors with apical and basal attachments are referred to as Radial Glia ventricular zone progenitors (RGvz); eNSC without apical, but retaining the basal process and attachment as RG outer subventricular progenitors (RGosvz); INPs residing in the subventricular zone as INPsvz; and INPs in the ventricular zone as INPvz. The latter progenitors have also been referred to as short neural precursors (SNPs) (Gal et al., 2006; Stancik et al., 2010).
Animals.
All animals were treated in accordance with IUCAC approved protocols at the Seattle Children's Research Institute. Wild-type CD1 mice were from Jackson Labs. Tbr2GFP BAC transgenic reporter mice were obtained from Gensat and maintained on the CD1 background (Kwon and Hadjantonakis, 2007; Kowalczyk et al., 2009). Hes5GFP transgenic reporter mice (Basak and Taylor, 2007) were maintained on a C57BL/6 background. Timed matings were considered as embryonic day (E) 0.5 from vaginal plug date, and embryos of either sex were used in this study.
Gene expression profiling.
To analyze gene expression in proliferating INPs and RG, progenitor cells were isolated from E14.5 neocortex using fluorescence activated cell sorting (FACS). Dorsal neocortices were microdissected from Tbr2GFP+ embryos and triturated with a polished glass pipette in DMEM and 1X trypsin (Invitrogen) until a single cell suspension was obtained. Trypsin was then quenched with FBS (Invitrogen), cells were pelleted and gently resuspended in fresh DMEM/FBS, and then passed through 100 μm cell strainer. Cell suspensions were vitally labeled with Hoechst 33342 (Invitrogen) before FACS sorting (Dako MoFlo). Cells were sorted according to DNA content (2N or >2N) as indicated by Hoechst labeling, GFP fluorescence, and scatter to limit cell doublets. Tbr2GFP+ cells with >2N DNA were considered INPs; Tbr2GFP− cells with >2N DNA were considered RG; cells with 2N DNA content were discarded. Approximately 20,000 cells were collected per Tbr2GFP+ embryo, and ∼105 cells were pooled from multiple embryos to make each sample. Cells were collected into RNA later (Ambion) and RNA was isolated as previously described (Bedogni et al., 2010). Samples were hybridized to Affymetrix Mouse Gene 430 2.0 chips (N = 4 chips per group). Enrichments were considered significant based on the false-discovery rate adjusted p values (padj < 0.05). Searches of literature and online public high-throughput embryonic mouse brain gene expression databases were used to validate gene expression patterns in the VZ, SVZ, or cortical plate (GenePaint, www.genepaint.org; Developing Mouse Brain Atlas, http://developingmouse.brain-map.org; Brain Gene Expression Map, http://www.stjudebgem.org).
Reporter constructs.
Mouse 4.3KB Dll1 promoter driving LacZ [Dll1LacZ (Beckers et al., 2000)] was subcloned into pd2YFP (promoterless vector with destabilized T1/2 = 2 h YFP reporter, Clontech) generating Dll1d2YFP. Mouse Dll3 promoter driving d2CFP has been described previously [Dll3d2CFP (Nelson et al., 2009)]. Mouse Hes1 and Hes5 promoter constructs driving d2GFP have been described previously [Hes1d2GFP, Hes5d2GFP (Ohtsuka et al., 2006)], which were subcloned into pd2YFP and pd2CFP, respectively (Hes1d2YFP, Hes5d2CFP). The entire Hes1d2YFP expression cassette was subcloned upstream of the Hes5d2CFP expression cassette, generating a single plasmid backbone containing dual Hes1 and Hes5 contextual Notch reporters (H1YH5C). Constitutively expressed (cytomegalovirus promoter) stable tandem-dimer Tomato red fluorescent protein expression construct (CMV-tdTomato) was gift from R. Wong (University of Washington, Seattle, WA). CAGGS-mTom membrane targeted tdTomato construct was generated from subcloning the mTom ORF from the Rosa26-mT/mG targeting vector [Plasmid 17787, Addgene (Muzumdar et al., 2007)] into the CAGG-retroviral backbone (Zhao et al., 2006).
Neocortical transfections and slice culture.
E12.5–14.5 transgenic or wild-type embryos were collected, DNA (1–2 μg/μl) was microinjected into a telencephalic ventricle, and ex utero electroporation (3–5 pulses, 35 mV, square wave BTX generator, 3 mm paddle electrodes) was used to target transfection into progenitors at the ventricular surface of the neocortex. Brains were dissected, embedded in 4% low-melting point agarose, cut into 250 μm organotypic slices with a vibratome (Leica), transferred to 35 mm-well culture insert (Millicell-CM filter insert, Millipore), and cultured in Neurobasal media supplemented with N2/B27 (Invitrogen), FBS (5%, Invitrogen), l-glutamine (Invitrogen), penicillin/streptomycin (Invitrogen), and incubated at 37°C, 5%CO2/95%O2. DAPT treatment [N-[N-(3,5-Difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester, 10 μm, Sigma] was used to synchronize progenitor differentiation (Nelson et al., 2007).
Live-cell imaging and analysis.
Slices were examined for reporter activity with a fluorescent stereomicroscope (Olympus), and transferred to heated imaging flow chambers (34°C). A specialized split-beam femtosecond pulsed MaiTai DeepSee laser (Spectra Physics) driving dual Olympus FV1000 upright microscopes each equipped with IR-corrected 25× 1.05 NA Ultra-objectives (Olympus FV1000 MPM) was used to collect live-imaging data. Stabilized slices were imaged under low flow conditions with bubbled media from above (supplemented with 3 mm HEPES, 0.5% FBS, 5%CO2/95%O2). Laser wavelength was tuned to 890 nm excitation. Live-cell multiphoton microscopy (MPM) imaging was typically done 50–75 μm below the slice surface through a depth of 50–100 μm along the z-axis, imaged in 2–3 μm optical slices. Z-stacks were collected every 10–12 min with n = 3–4 successfully imaged slices per reporter/experiment from 2 to 3 animals at a given age (Table 1), except for mTom experiments where n = 2. One Olympus FV1000 microscope was also equipped as a single-photon laser-scanning confocal microscope (LSCM) with tunable emission filters, and was used to visualize/separate YFP/CFP fluorescence from H1YH5C dual reporter (515/458 nm, respectively). Four-dimensional datasets were corrected for thermal/tissue drift, analyzed as individual slices/frames and as volume rendered datasets using a combination of Olympus FV1000, Imaris (Bitplane), and Fiji software packages, including cell counts, process measurements, rotations, and manual reconstruction/segmentation of individual neocortical progenitors cells in Imaris. Snapshots of maximum intensity Z-projections (MIP) from selected regions of interests within 4D-datasets were converted into QuickTime files, and individual time point panels were assembled in Photoshop and/or Illustrator (Adobe) for display purposes in multipanel figures. All imaging data are summarized in Table 1.
Table 1.
Live imaging summary
| Reporter | Slices | Age | Duration | Interval | Divisions# |
Dynamics |
RGosvz | ∼Number of cells t0 | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Apical | Basal | Radial | Tangential | Hooks | |||||||
| Tbr2GFP | n = 4 | E12/14 | 37 h:00 m | 10 m | + | ++ | ++++ | ++ | ++ | na | nd |
| tdTom* | n = 4 | E14.5 + 1 | 10 h:00 m | 10 m | 8 | 3 | +++ | + | ++ | 6 | 92 |
| mTom** | n = 2 | E15.5 + 1 | 19 h:48 m | 12 m | 4 | 4 | ++++++ | ++++ | ++++ | 3 | nd |
| Dll1d2YFP | n = 4 | E12/13 + 1 | 23 h:36 m | 12 m | 30 | 10 | +++ | ++ | ++ | 1 | 219 |
| Hes5d2GFP | n = 2 | E14.5 + 1 | 14 h:50 m | 10 m | 12 | 1 | + | nd | ++ | 10 | 127 |
| n = 5 | E14.5 + 1 | na | na | ++ | na | na | ++ | 17 | 402 | ||
Slice hours and cells are combined for total; radial dynamics include INPsvz/INPvz/RGvz/RGosvz interactions; tangential dynamics refer to INPvz/RGvz apical processes; na, not applicable; nd, not determined; f, fixed slices;
*CMVtdTomato transfected into Tbr2GFP,
**CAGGS-membrane-tdTomato, all other slices from wildtype mice;
#divisions were noted, but dynamics were the main focus.
Immunolabeling.
All immunolabeling was performed on freshly harvested tissue fixed briefly (∼30 min) in 4% PFA, cryoprotected in 30% sucrose-PBS, cryosectioned at 12 μm, rehydrated in PBS, blocked with 10% horse serum in PBS-0.01% Triton X-100 for 1 h at room temp, with primary antibodies diluted in block and incubated overnight at room temp, washed 3–5× with PBS, incubated in species-specific ALEXA 488/568/647 conjugated secondary antibodies (Invitrogen), washed, counterstained with Dapi (Invitrogen), and mounted in Flouromount-G (Southern Biotech). Primary antibodies include anti-Dll1 (sheep, R&D Systems); anti-Dll3 (rb), and anti-Neurog2, anti-Neurog1, anti-NeuorD1, anti-Jag1, anti-Sox2, anti-Bhlhb5 (all goat, Santa Cruz Biotechnology); anti-Tuj1 (ms, Covance); anti-b-Gal (rb, Cappel), anti-Hes1 (rb, gift from N. Brown), house anti-Tbr1 and Tbr2 (rb), anti-Pax6 (rb, Covance), anti-Sox9 (rb, Millipore Bioscience Research Reagents); transgenic Tbr2GFP and Hes5GFP signals represent endogenous reporter levels. All sections were analyzed using a Zeiss 710 Quasar 34-channel LSCM (Carl Zeiss), and a MIP of three 1 μm optical slices was used to facilitate visualization of fine Tbr2GFP processes in stained sections.
Results
INPs have dynamic and transient long-range filopodia-like processes
Previous live-cell imaging studies in the embryonic rodent neocortex revealed that INPs exhibited varied morphologies (Haubensak et al., 2004; Miyata et al., 2004; Noctor et al., 2004, 2008; Attardo et al., 2008), considered to possibly reflect stages of INP migration. Furthermore, most studies used predominately single-cell or single-marker labeling strategies to facilitate the identification of clones for lineage or cleavage-plane analyses. However, this approach precludes observation of interactions between diverse types of progenitors, which include different types of RG and INPs in the VZ, SVZ, and OSVZ (Franco and Müller, 2013).
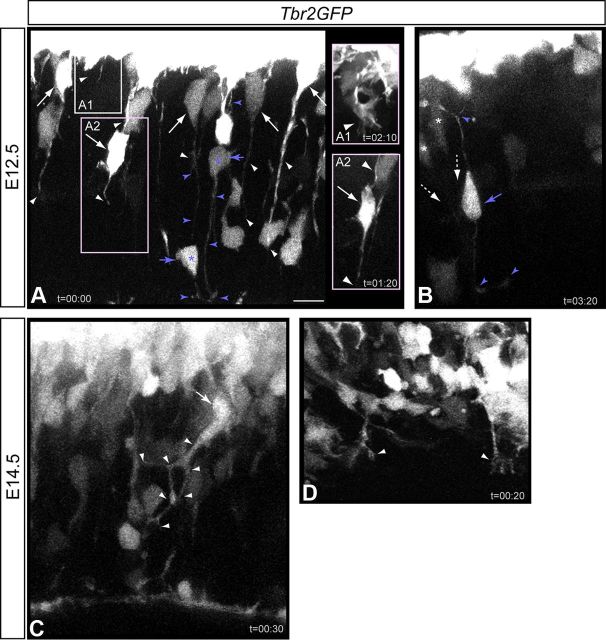
As a first step toward understanding how these networks of diverse progenitors may interact at the cellular level, we took advantage of a transgenic Tbr2GFP reporter mouse to label INPs (Kwon and Hadjantonakis, 2007), and used high-resolution live-cell MPM to visualize INPs and their processes in organotypic brain slice cultures prepared from Tbr2GFP embryos. Live-cell imaging at the SVZ/VZ interface readily detected Tbr2GFP+ INPvz and INPsvz processes at E12.5 (Fig. 1A,B) that exhibited extensive process dynamics including extensions, retractions, and branching. For example, Tbr2GFP+ INPsvz frequently extended long-range radial processes toward the ventricular surface (∼50–100 μm) (Fig. 1A,A2,B), as well as shorter and more complex filopodia-like processes from INPsvz clustered at the SVZ/VZ interface (Fig. 1A1). INPvz exhibited dynamic interactions via their short basal process with overlying INPsvz (Fig. 1A,B, blue asterisks, arrows, arrowheads) forming transient radially oriented cell contacts, and also extended short tangential filopodial processes from their apical attachment (Fig. 1B). Live-cell MPM imaging in acute E14.5 Tbr2GFP slices revealed similar behaviors, including direct radially-oriented INPsvz-INPvz contacts (Fig. 1C). Additionally, some INPsvz migrated apically with extensive process extension, retraction, and branching near the ventricular surface (Fig. 1C), undergoing complex morphological changes over short time periods (Fig. 1D). These results, together with earlier observations (Noctor et al., 2004; Kowalczyk et al., 2009), suggest that dynamic and transient long-range filopodia are a fundamental cellular property of INPs throughout neurogenesis in the neocortex.
Figure 1.
INPs extend and retract dynamic processes. High-resolution live-cell MPM was used to visualize INP cell dynamics in E12.5 (A, B) and E14.5 (C, D) Tbr2GFP neocortical slices. Still images of selected time points from live-cell MPM imaging experiments depict varied morphological transitions, process dynamics, and INPsvz-INPvz contacts. Images are typically snapshots of maximum intensity Z-projections (MIP) of ∼15–30 μm analyzed from within ∼70–100 μm imaging volumes, imaged ∼50–75 μm from the surface in 2–3 μm optical steps every 10–12 min (t = hours:min): due to strong expression of stable GFP from this BAC transgene, GFP reporter persists beyond downregulation of endogenous Tbr2, and due to the imaging setting necessary to visualize fine processes in the VZ (∼1 μm or less), GFP signal in the upper SVZ is often saturated, obscuring analyses. A, B, E12.5 Tbr2GFP+ INPsvz extend and retract filopodial-like processes (50–100 μm; white long arrows/arrowheads; B, asterisks, white dashed arrows). INPvz (A, B; blue short arrows/arrowheads) exhibit different morphological states and exhibit both apical (tangential process extension) and basal process dynamics (B). INPsvz and INPvz processes interact, forming a transient type of radially oriented cell-contact (A; INPvz, blue asterisks, arrows/arrowheads; INPsvz, white arrows/arrowheads). C, D, Similar Tbr2GFP+ INP dynamics are observed in E14.5 acute neocortical slices. Note the highly branched processes and dynamics of the INPsvz in C, and in a higher power view, the extensive morphological changes in some INPsvz apical processes near the ventricular surface in D. Scale bars: A–C, 15 μm; D, ∼7 μm. INPs, Intermediate neurogenic progenitors; MPM, multiphoton microscopy; vz, ventricular zone; svz, subventricular zone.
INPs and RG create a dynamic network of transient cellular interactions in the embryonic neocortical stem cell niche
Our next step toward understanding what types of cellular interactions might underlie Dll1-mediated signaling from INPs to RG, we needed to define the different types of cellular interactions that occur between INPs and RG. To better understand how the ensemble of INPs interact with neighboring RG progenitors, we used ex utero electroporation to target transfection of a constitutively expressed stable fluorescent reporter CMV-tdTomato (tdTom) into the neocortex of E14.5 Tbr2GFP embryos, and cultured brain slices overnight for reporter expression. This approach allowed high-resolution two-color live-cell MPM imaging of mosaic, densely labeled VZ progenitors and progeny (tdTom+) within the transgenically labeled INPvz/svz (Tbr2GFP+) background.
These experiments revealed several novel types of cell–cell interactions and morphological distinctions between INPs and RG within their apical and basal neurogenic niches. For example, the apical region of the VZ was rich in dynamic INPsvz long apical processes that extended to the ventricular surface (Fig. 2A–C). INPsvz apical processes were observed in close juxtaposition to RGvz apical processes and attachments, and frequently contacted M-phase RGvz (tdTom+/Tbr2GFP−) (Fig. 2A,B). Similarly, INPsvz apical processes contacted INPvz progenitors along their basally located somas and their apical processes (Fig. 2A,B), in a “radial relay” type of arrangement. Furthermore, RGvz or INPvz mitotic events displaced apical processes of neighboring RGvz, INPvz, and INPsvz, creating characteristic contact-dependent hook-like morphologies (Fig. 2A–E), similar to those described (Miyata, 2007, 2008). Numerous examples of these hook-like morphologies were evident throughout our live-imaging experiments (see below). Digital isolation and reconstruction of apical processes surrounding an early M-phase RGvz cell illustrated the extensive nature and apparent multiplicity of filopodia and interactions surrounding mitotic events in the apical VZ progenitor niche (Fig. 2B).
Figure 2.
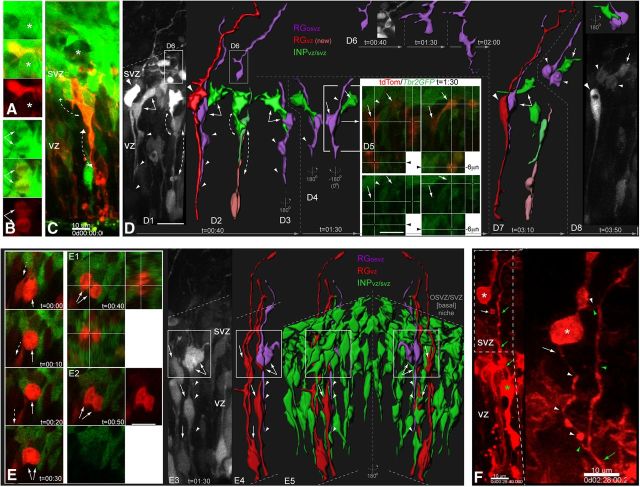
INP and RG interactions in the apical -neocortical neural stem cell niche. A–C, Constitutively active soluble RFP (CMV-tdTomato, tdTom) was electroporated ex utero into E14.5 Tbr2GFP neocortex, slices were cultured overnight, and analyzed by high-resolution two-color live-cell MPM imaging, which reveals the types of interactions between diverse neocortical progenitors underlying the basis for cell contact-dependent signaling in the apical embryonic stem cell niche (VZ and ventricular surface). Neocortical progenitors include RGvz (tdTom+/Tbr2GFP−, apical+/basal+ attachment), INPvz (tdTom+/−/Tbr2GFP+, apical attachment/short basal/VZ), and INPsvz (tdTom+/−/Tbr2GFP+, apical process, no apical attachment/multipolar/basal VZ, SVZ). A, In addition to INMs of RGvz (asterisks and dashed arrows) and INPvz, live-imaging reveals contact between INPsvz apical processes (arrowheads) and dividing RGvz daughters (cells 1, 2; cell 3). Additionally, contacts between INPsvz/INPvz/RGvz apical processes with neighboring apical divisions resulted in a transient hook-like morphology (especially apparent around non-labeled apical divisions, asterisks and small arrows). B, Reconstruction of an apical RGvz division (yellow cell, arrows indicate cell body and thinned basal process) with its neighboring labeled cells and their apical processes including RGvz (red), newly divided progenitor (new RGvz or new INPvz; light red, tdTom+/Tbr2GFP−), INPvz, and INPsvz (green or green/red) demonstrates the extensive contacts of neighboring apical processes surrounding dividing progenitors. C–H, Electroporation of constitutively active of membrane-targeted tdTomato (CAGGS-memTomato, mTom) reveals a much more extensive network of cellular processes and interactions, represented in C as a rotated ortho-slice view showing xyz-planes (x plane is 22 μm projection): note the basal INPsvz and the apical INPvz mitoses (left and right arrows, respectively), and the transient hook-like apical process displacements in RGvz and INPvz surrounding unlabeled RGvz divisions (asterisks), small D–H indicate regions shown at higher power below, although at different z levels and/or time points. D, High-power view of digitally isolated RGvz (mTom+/Tbr2GFP−, red channel only,) reveals more extensive membrane contact during hook-like formation (small arrow) surrounding an adjacent unlabeled RGvz division (mTom−/Tbr2GFP−, asterisk): note the dynamic tangential filopodia-like processes emanating from an upper region of the apical process (arrowheads). E, Hook-like formations (arrows) surround an INPvz division (asterisks). F, Higher-power view of boxed region in C reveals long-range dynamic filopodia extending tangentially from an apical RGvz process (F; arrows, arrowheads, mTom+/Tbr2GFP−), as well as from INPvz apical processes and attachment (G). H, Higher-power view of a digitally isolated segment from an apical RGvz attachment revels multiple filopodia-like protrusions from the endfoot itself. D–E, All panels from different z-planes or time points in C. RG, Radial glia; RFP, red fluorescent protein; INMs, interkinetic nuclear migrations.
Although our two-color approach using soluble fluorescent reporters revealed much about the nature of cell–cell interactions and larger-scale cell behaviors, to better visualize the finer morphologic properties of processes and filopodia, we developed a membrane targeted tdTomato construct (CAGGS:membrane-tdTom, mTom). The mTom construct was transfected into E14.5 Tbr2GFP neocortices, and slices were prepared for live-cell MPM imaging as described above. Live-cell MPM revealed intense mTom+ fluorescent signal localized to the plasma membrane (Fig. 2C–H), confirming our targeting strategy.
In addition to more clearly confirming the cell–cell interactions described above, such as INPsvz (mTom+/Tbr2GFP+) progenitors extending/retracting/reextending apical processes into the VZ (Fig. 2C), mTom labeling revealed more extensive process dynamics, especially within the VZ and from the apical processes and/or the apical attachments of RGvz and INPvz. For example, digital isolation of a mTom+/Tbr2GFP− RGvz revealed extensive morphological changes induced in the apical process upon displacement by a neighboring apical division of (presumably) another RGvz progenitor (asterisk, mTom−/Tbr2GFP−) (Fig. 2D). These mitotic events seem to do more than simply displace neighboring apical processes, as membrane labeling clearly revealed that the process itself responds by extending transient filopodia and by extensive membrane ruffling to apparently partially encapsulate the neighboring mitotic progenitor (Fig. 2D). INPvz (mTom−/Tbr2GFP+) also induced hook-like morphologies in both RGvz (mTom+/Tbr2GFP−) and INPsvz neighboring apical processes (Fig. 2E), Transient hook-like morphologies were apparent in every live-cell imaging experiment (Table 1), especially those with dense mosaic membrane labeling, indicative of normal proliferation in the imaged slice culture. For example, within the isolated volume shown in Figure 2C (241×152×22 μm, xyz, respectively) at least 8 hook-like formations were apparent at the indicated time point (Fig. 2C, asterisks).
Interestingly, membrane labeling also revealed extensive RGvz (mTom+/Tbr2GFP−) and INPvz (mTom+/Tbr2GFP+) long-range tangential dynamic filopodia-like extensions from their apical process (Fig. 2D,F,G). For example, numerous and dynamic filopodia extended tangentially from the RGvz apical process above the segment exhibiting hook-like morphology (Fig. 2D). These RGvz tangential processes extended over ≥30–40 μm, and contacted neighboring labeled radial progenitors (Fig. 2F). INPvz also extended numerous thin tangential filopodia that emanated from both their apical process as well as their soma, in addition to their thicker dynamic short basal process that could be observed interacting with cells in the overlying SVZ (Fig. 2G). Interestingly, we also observed extensive, highly dynamic processes emanating from the INPvz apical attachment at the VZ surface (mTom+/Tbr2GFP+) (Fig. 2G) confirming our previous observations (Fig. 1). Apical endfoot dynamics were especially apparent in RGvz. For example, high-power views of a digitally isolated RGvz apical endfoot (mTom+/Tbr2GFP−) revealed that multiple (∼3–10) different dynamic processes transiently projected (∼5–20 μm) into the periventricular VZ and toward neighboring RGvz endfeet within an ∼4 h period (Fig. 2H). Membrane labeling was essential to fully visualize the dynamics of transient filopodia from both INPvz and RGvz, but they could still be distinguished in all of our live imaging experiments using a soluble GFP transgenic label (Tbr2GFP, Fig. 1), as well as later experiments using a destabilized soluble YFP label (Dll1d2YFP reporter; see Fig. 8). Altogether, it appears that RGvz and INPvz can cycle through different morphological stages of apical process morphology, ranging from simple (straight process, no tangential filopodia, simple endfoot) to complex (straight process, multiple dynamic tangential filopodia, complex endfoot) to responsive (hook process, transient encapsulation, and membrane ruffling).
Figure 8.
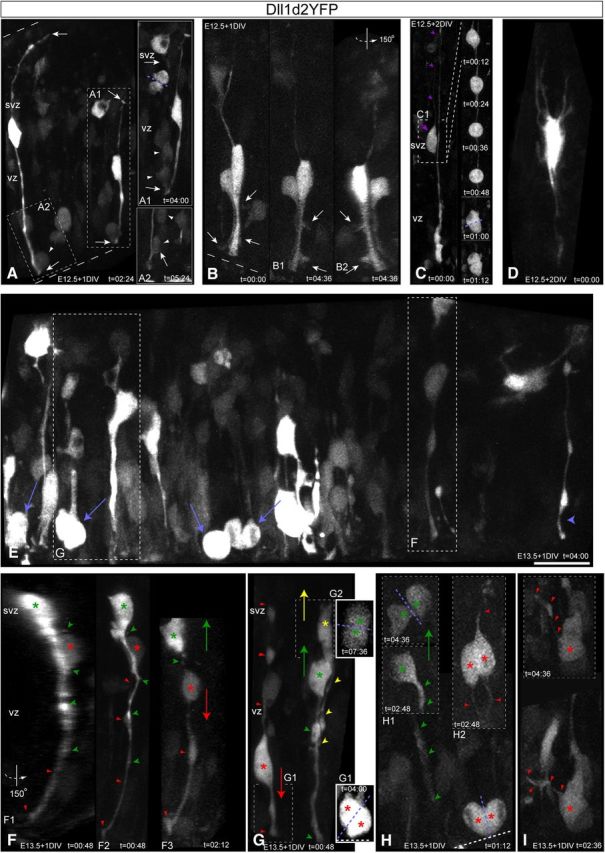
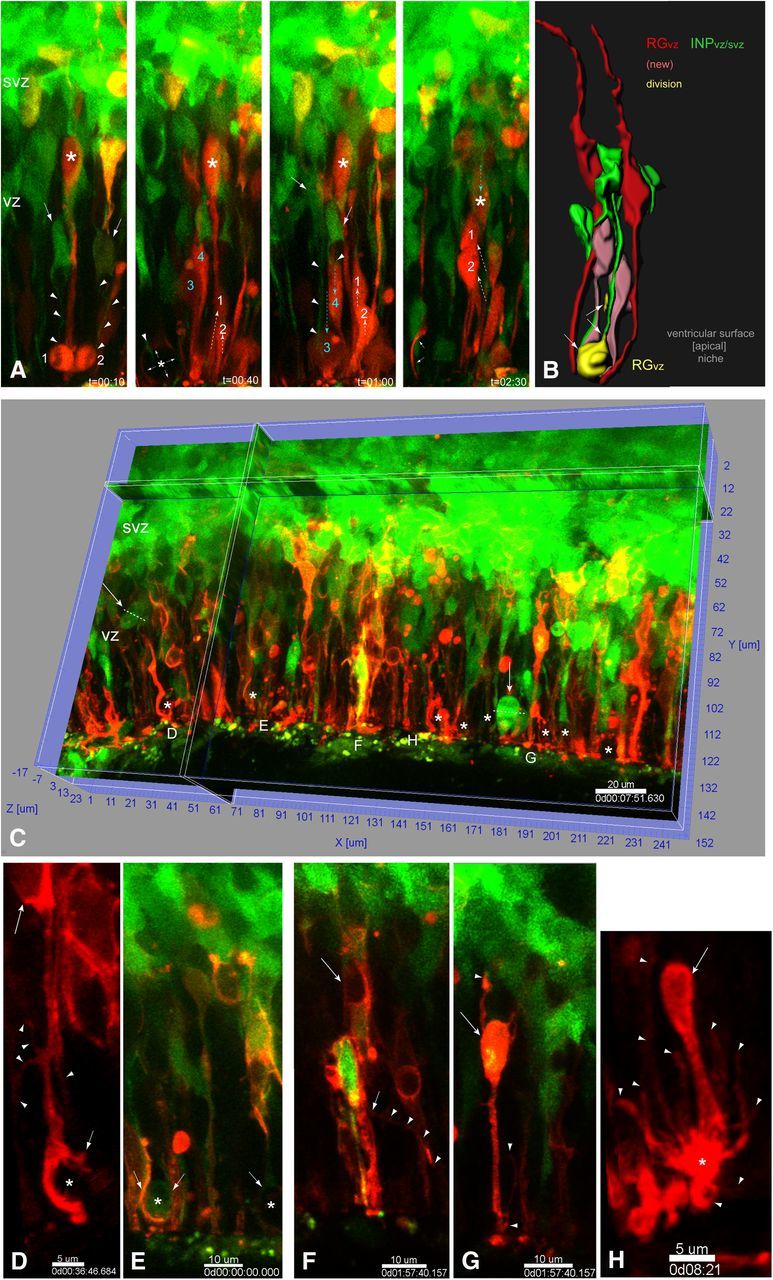
Dll1-signaling progenitors in the NSC niche interact with neighboring cells through dynamic cell–cell contacts. Ex utero electroporation of the mouse Dll1 promoter [4.3KB, Beckers et al. (2000)] driving destabilized YFP (t1/2 = 2 h, d2YFP) reporter construct into wild-type E12.5 (A–D) or E13.5 (E–H) neocortices cultured 1DIV (A, B, E–H) or 2DIV (C, D) and then analyzed by high-resolution live-cell MPM revealed Dll1 activity within all neocortical progenitor subtypes, all of which extended/retracted dynamic radial, multipolar, and/or tangential processes. A, Both RGvz (Dll1d2YFP+, apical+/basal+ attachments) and INPvz (Dll1d2YFP+, apical+/basal− attachments) apical processes maintained contact with neighboring VZ apical divisions through the hook-phase via thin processes contacting the daughter cell remaining closest and longest near the ventricular surface (A1, A2). INPsvz (top) project apically directed filopodia that contact underlying mitotic INPsvz, which then also begin to project apically directed filopodia into the VZ post-division (A1). B, Isolated Dll1d2YFP+/+ RGvz-INPvz pair tracked over time reveals the extension and retraction of tangentially oriented filopodia from their apical processes (B1, B2). C, After 2DIV, Dll1 activity was still detected in RGosvz (Dll1d2YFP+ apical-/basal+ attachment) undergoing basal somal mitotic translocation next to a Dll1 active RGvz (C1), as well as dynamic INPsvz (D). E–I, Extensive Dll1 activity is detected within the eNSC niche (E), including apical dividing progenitors (blue arrows): boxed regions F and G in detail below, Dll1d2YFP+ INPsvz hook-like radial process (blue arrowhead). F, Withdrawal of apical radial contact (F1, F2) from overlying Dll1-active INPsvz (green asterisk, arrowheads) precedes INM/apical division in the underlying Dll1-active INPvz (F3, red asterisks, arrowheads). G, By contrast, in a different region in the same slice, a similar INPsvz-INPvz interaction (G2, yellow-green asterisks, arrowheads, respectively) results in the retraction of the INPvz apical process and INM/basal division in the SVZ, compared with the neighboring Dll1 active INPvz (G1, red asterisk, arrowheads). H, Dll1 is active in both basal (H1, green) and symmetric apical dividing progenitors (H2, RGvz, red), and rapid tangential process extension post-division in apical progenitor at the ventricular surface (I). Scale bars: A–C, 20μn, D–I, 15μn. eNSC, Embryonic neural stem cell.
Due to high expression of stable GFP from the Tbr2GFP reporter and dense INP cellularity, progenitor interactions in the basal niche (basal region of VZ, the SVZ, and OSVZ) were more difficult to distinguish. Nevertheless, labeled INPsvz (tdTom+/Tbr2GFP+) were observed to divide and contact neighboring mitotic INPsvz (Fig. 3A,B). Also, membrane labeled INPsvz displayed extensive contacts with neighboring INPsvz (Fig. 3C). Multipolar INPsvz (tdTom+/Tbr2GFP+) exhibited dynamic interactions with more apically located INPsvz, which in turn interacted with additional VZ progenitors, including INPvz somas and short basal processes (tdTom+/Tbr2GFP+), as well as likely newly generated RG daughters (tdTom+/Tbr2GFP−) in a radial-relay manner (Fig. 3C, see also Fig. 2).
Figure 3.
INP and RG interactions in the basal/outer neocortical neural stem cell niche. tdTom (A, B, D, E) or mTom (C, F) was electroporated ex utero into E14.5 Tbr2GFP neocortex, slices were cultured overnight, and analyzed by high-resolution two-color live-cell MPM imaging, which revealed the types of interactions between diverse neocortical progenitors underlying the basis for cell contact-dependent signaling in the basal embryonic stem cell niche (SVZ and basal regions of VZ, as well as OSVZ). Neocortical progenitors include RGvz (Tom+/Tbr2GFP−, apical+/basal+ attachment), RGosvz (Tom+/Tbr2GFP−, apical−/basal+ attachment, located in basal VZ-SVZ-OSVZ), INPvz (Tom+/−/Tbr2GFP+, apical attachment/short basal process/VZ), and INPsvz (Tom+/−/Tbr2GFP+, apical process, no apical attachment/multipolar/basal VZ, SVZ). A, B, Basally dividing INPsvz cells (A, asterisk; B, arrows) are contacted over short-ranges by neighboring INPsvz cells (A, tdTom+/Tbr2GFP+; B, tdTom−/Tbr2GFP+). C, INPsvz cells (mTom+/Tbr2GFP+) form clusters in the SVZ, making radially oriented cell contacts with more apically located INPsvz (top dashed arrows) that in turn make additional radial contacts via its dynamic apical process with underlying INPvz (mTom−/Tbr2GFP+) and presumptive RGosvz (mTom+/Tbr2GFP−), which subsequently withdraws its apical attachment and undergoes basal somal translocation and mitosis in the basal region of the VZ: also note that vasculature (C, asterisk) is present in the basal niche. D, Radial relays of cell–cell interactions from INPsvz and RGosvz to underlying INPvz and RGvz are frequent in the basal niche. D1–7, Time series of tdTom+ labeled progenitors in the Tbr2GFP background (D1) with selected radial relay interactions (dashed arrows) between progenitors with dynamic apical processes were digitally isolated, reconstructed, and color-coded (D2): RGvz, red; RGosvz, purple; INPsvz and INPvz, green and lighter green, respectively; recently divided RGvz, light red (tdTom+/Tbr2GFP−, apical attachment but thin or incomplete/indefinable basal process). D2, Radial relay of cell–cell contacts are detailed for an INPsvz-RGosvz-RGvz interaction, and INPsvz-INPsvz-RGvz (newly divided cell; light red, tdTom+/Tbr2GFP−, apical attachment, thin/incomplete basal process). Note that in D1, two morphologically and behaviorally distinct RGosvz (purple) were apparent. Reconstruction, rotation, and tracking of cell interactions and morphologies over time reveal specific and maintained contact from INPsvz to RGosvz, as well as changes in RGosvz apical process (D3–4). D5, Single optical sections with orthogonal views at different z levels confirm cell identities: note cross-hairs and arrows are positioned over the cell bodies in top panels (red/green channels) and bottom panels (green channel only) of RGosvz (left panels, tdTom+/Tbr2GFP−) and INPsvz (right panels, tdTom+/Tbr2GFP+); arrowhead points to RGosvz basal process in this single plane. D7–8, Continued tracking of this cell pair reveals that the INPsvz maintains contact during RGosvz basal somal translocation and mitosis. D6, A second RGosvz migrates apically and extends a dynamic apical process into SVZ. E, Tracking of single RGosvz within the transgenic background in general reveals the basal niche itself responds to RGosvz mitotic basal somal translocations by clustering around their divisions. E, Two-color single optical slices with orthogonal (E1) and single channel (E2) views of boxed region in E3 (grayscale, tdTom+) neocortical progenitors, with reconstruction of the post-mitotic RGosvz (purple) and neighboring RGvz (red) (E4) embedded within the INPs at the VZ/SVZ interface (E5). F, Live-imaging of interactions between RGosvz (white asterisks and arrows) and the basal process of neighboring RGvz (green asterisk and arrows) reveals extensive RGosvz apical process dynamics, as well as shorter transient tangential filopodial-like protrusions from its basal process, both of which contact the neighboring RGvz basal process (green arrow) and induce reciprocal transient protrusions. Scale bars: D1–4, D6–8, E3–5, 15 μm; D5, E, E1, E2, 20 μm; A, B, 10 μm. osvz, Outer subventricular zone.
Our two-color live-cell MPM approach further enabled us to observe cell–cell interactions between INPsvz and the newly described subpopulation of translocating RG in the SVZ and OSVZ (RGosvz). RGosvz were identified by the absence of an apical attachment but presence of a basal process attached to the pial surface; by somal translocation before mitosis; and by the lack of transgenic Tbr2GFP reporter expression (Miyata et al., 2004; Noctor et al., 2004, 2008; Shitamukai et al., 2011; Wang et al., 2011; Martínez-Cerdeño et al., 2012). For example, we observed divisions in the basal VZ and SVZ from progeny of tdTom+ or mTom+ transfected VZ progenitors that were not Tbr2GFP positive (Fig. 3C–E). Consistent with previous reports, basal processes to the pial surface were not always visible within the imaging volumes. In the sequence depicted in Figure 3D, multipolar INPsvz (tdTom+/Tbr2GFP+) contacted what first appeared to be another type of INPsvz (tdTom+/Tbr2GFP− with a long radial apical process) (Fig. 3D1–D5). However, reconstruction of these two cells, along with a neighboring RGvz (tdTom+/Tbr2GFP−) for comparison, revealed the presence of a long thin basal process in the tdTom+/Tbr2GFP− SVZ cell (Fig. 3D2–D5). As observed by MPM, this tdTom+/Tbr2GFP− SVZ cell retracted its apical process, migrated basally (∼15–20 μm), and divided (Fig. 3D7,D8), similar to that observed in Figure 3C. This type of basal mitotic somal translocation cellular behavior has been described in recent studies (Hansen et al., 2010; Shitamukai et al., 2011; Wang et al., 2011), identifying this Tbr2GFP− progenitor as RGosvz. Notably, the new daughter cell (tdTom+/Tbr2GFP−) that did not inherit the basal process maintained contact with the INPsvz (Fig. 3D7–D8). A second RGosvz was apparent in this timelapse sequence that underwent apical somal translocation toward the SVZ, extending a dynamic apical filopodia and tangential protrusions from its basal process (Fig. 3D6). Basally translocating, dividing RGosvz seemed to promote clustering of neighboring INPsvz (Fig. 3E,E1–E2). Digital isolation and reconstruction of neighboring labeled RGvz, RGosvz, and INPs reveals that the basal niche is composed of abundant INPsvz, RGvz basal processes, and RGosvz that interact extensively (see Fig. 5E3–E5). Interestingly, studies of fixed tissue have also shown that RG radial fibers extend short tangential filopodia-like processes, previously termed lamellate expansions (Rakic, 1972). Membrane labeling and live-cell MPM imaging of RGosvz revealed that both their long dynamic radially-directed apical processes and finer tangential filopodia from basal processes interacted with neighboring RGvz radial fibers and induced reciprocal outgrowth of processes (Fig. 3F).
Figure 5.
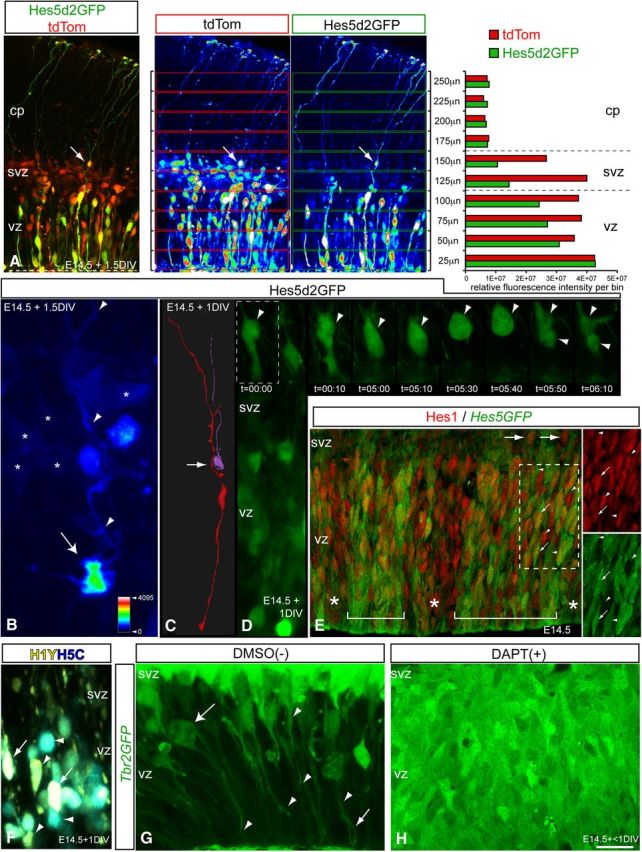
Differential and diverse output in Notch activity levels between INPs and subpopulations of RG. A, B, Ex utero electroporation was used to co-transfect constitutively active stable CMVtdTomato construct, and a mouse Hes5 promoter driving destabilized GFP [t1/2 = 2 h, Hes5d2GFP, Ohtsuka et al. (2006)] reporter construct into wild-type E14.5 neocortices, which were sliced, cultured (1.5DIV), fixed briefly and imaged via high resolution 2-color MPM. Co-transfected INP progeny in the overlying SVZ exhibited lower overall Notch-dependent Hes5 activity compared withprogenitors in the VZ (A) and RGosvz (B, Hes5d2GFP+, apical−basal+ process and attachment; asterisks denote Hes5d2GFP-low INPsvz). C, D, Live-cell MPM imaging of Hes5d2GFP+ RGosvz division (D) and contacts with neighboring RGvz basal processes (C, reconstruction, hook-like displacement in RGvz basal process, arrow). E, F, Contextual Notch effectors Hes1 and Hes5 are differentially expressed in the RG pool. E, Hes1 immunolabeling (red) of sections from E14.5 Hes5GFP transgenic mice (green, endogenous GFP) reveal Hes1+/Hes5GFP− RGvz clusters (asterisks) nearby Hes5GFP+ RGvz clusters (bars), which contain Hes1+/Hes5GFP+ (arrows), and Hes1+/Hes5GFP− and Hes1−/Hes5GFP+ (arrowheads) RGvz progenitor subtypes (boxed region, single channel views): also note a few Hes1+ and/or Hes5GFP+ cells in the SVZ (RGosvz, large arrows). F, Live-cell LSCM image of wild-type E14.5 neocortex transfected with H1YH5C confirms differential output of Notch signaling in RGvz: higher Hes1 activity (yellow) or Hes5 activity (cyan), arrowheads; high dual Hes1+/Hes5+ activity (white), arrows. G, H, Although the embryonic niche generates higher canonical Notch activity in the total RG pool (A–F), acute inhibition of this canonical Notch signal synchronizes these contextually distinct RGvz Hes1/Hes5 subpopulations to uniformly fate switch into INPs. E14.5 acute neocortical slices from Tbr2GFP mice treated with DAPT (H) or DMSO(−) control (G) (n = 3/3 slices): note in G the extensive INP processes (Tbr2GFP+, arrowheads), INP basal division, and hook-like apical process (G, large and small arrows, respectively) interacting with underlying RGvz (Tbr2GFP−) (G, maximum intensity Z-projection 10-slices, 30 μm), which convert en masse to Tbr2GFP+ INPs with acute Notch activity inhibition (H, single optical section). Relative fluorescent intensity scale in B also refers to A. LSCM, Laser scanning confocal microscopy.
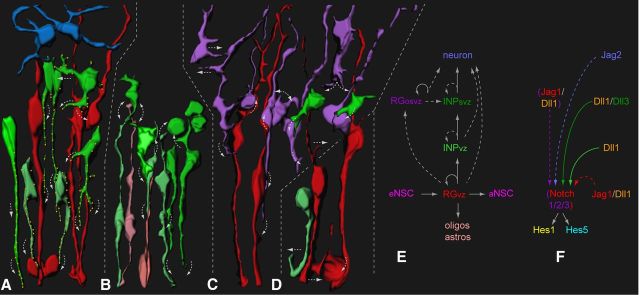
Altogether, these results suggest that the neocortical eNSC niche is an extremely dynamic “social network,” involving a multitude of transient cell–cell interactions between diverse INP and RG neocortical progenitor subpopulations. In particular, there is extensive cell–cell contact from basal progenitors to apical progenitors within radial columns, in a radial-relay type of cell–cell interaction, in addition to tangential cell–cell interactions across radial columns. These characteristic interactions constitute a potential cellular basis for contact-dependent signaling, including Notch signaling.
INPs and subpopulations of RG display different levels and output of Notch signaling
To better understand the molecular underpinnings of Notch signaling by different progenitor types, we used an unbiased gene expression profiling strategy to enrich for genes expressed in proliferating INPs versus proliferating RG. Replicating progenitors (>2N DNA content) were isolated from dissociated neocortices of E14.5 Tbr2GFP transgenic mice (Kwon and Hadjantonakis, 2007; Kowalczyk et al., 2009) via FACS sorting for DNA content and then GFP fluorescence. This sequential strategy enabled us to sort progenitors (>2N DNA) into RG (Tbr2GFP−) and INPs (Tbr2GFP+) (Fig. 4A). The sorted cells were used as RNA sources for comparative transcriptomic profiling of each progenitor type using microarrays.
Figure 4.
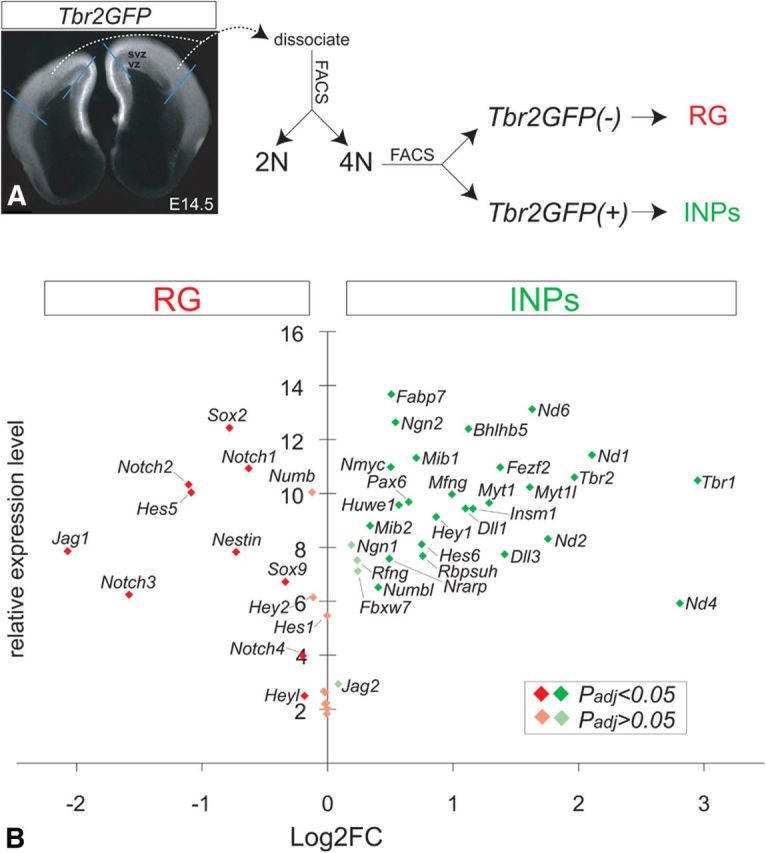
Gene expression profiling reveals molecular differences in Notch pathway components between INPs and RG. A, We used a FACS-based strategy to isolate 4N dissociated neocortical progenitors isolated from E14.5 Tbr2GFP embryos, which were further sorted into RG and INP subpopulations based on GFP fluorescence, and global gene expression profiles of sorted RG and INPs were obtained using an Affymetrix platform. B, Scatter plot comparing relative levels and enrichments of known RG and INP markers and neurogenic genes to selected Notch pathway components (receptors, ligands, effectors/targets, accessory molecules) in the RG and INP sorted progenitors, consistent with their endogenous mRNA expression patterns in the VZ and SVZ (Allen Brain Atlas, http://developingmouse.brain-map.org/: GenePaint, www.genepaint.org/: Brain Gene Expression Map BGEM, http://www.stjudebgem.org).
Evaluation of known RG and INP markers via GO and open-access gene expression pattern analysis of VZ, SVZ, or cortical plate confirmed efficient enrichment into respective pools (Allen Brain Atlas, http://developingmouse.brain-map.org/: GenePaint, www.genepaint.org/: Brain Gene Expression Map BGEM, http://www.stjudebgem.org). Although a complete analysis of this dataset will be described separately, we noticed initially that many components of the Notch signaling pathway including receptors, effectors, ligands, and accessory molecules showed differential enrichment in RG and INP pools (Fig. 4B). This is of key interest since recent studies point to a major role for Notch signaling in generating neocortical progenitor diversity. For example, Notch1–3, Hes5, HeyL, and Jag1 were enriched in RG, while Dll1, Dll3, and Hey1 were enriched in INPs; Hes1 and Hey2 expression showed nonsignificant trends toward enrichment in RG over INPs; Jag2 was not enriched in RG or INPs, but was predominantly expressed in postmitotic neurons (Fig. 4B, BGEM). The molecular enrichments described above were consistent with endogenous mRNA expression patterns in the VZ, SVZ, and cortical plate at E14.5 (Allen Brain Atlas, GenePaint, BGEM). Interestingly, some Notch components showed graded expression along the mediolateral and rostrocaudal axes, suggesting a potential role for Notch signaling in areal patterning of the neocortex, although this possibility was not explored further in the present study.
Although RG are known to have overall higher levels of canonical Notch activity compared with INPs (Mizutani et al., 2007), differences at the level of actual Notch output on target effector genes were apparent. For example, in our dataset Hes1 and Hes5 showed different gene enrichment profiles and overall expression in RG (Fig. 4B, Allen Brain Atlas, GenePaint, BGEM). To confirm this output diversity of Notch signaling between RG subpopulations and INPs, we used ex utero electroporation to co-transfect Hes5d2GFP (mouse Hes5 promoter driving destabilized 2 h t1/2 GFP) (Ohtsuka et al., 2006) and tdTom (CMV promoter driving stable tdTomato) plasmids into E14.5 mouse neocortex, which was prepared for imaging as above. MPM revealed that many INPsvz (tdTom+, multipolar morphology, no apical/basal attachments, located in SVZ) exhibited decreased Hes5d2GFP fluorescence compared with RGvz (apical and basal processes and attachments) (Fig. 5A), confirming that Hes5 expression and activity are lower in INPs than in RG. However, some cells in the SVZ/OSVZ maintained high Hes5 reporter activity with a basal process connected to the pial surface, consistent with their identification as RGosvz (Fig. 5A,B). Live MPM of Hes5d2GFP transfected slices further revealed that RGosvz with high Hes5 activity contacted basal processes of neighboring RGvz that formed hook-like structures in the SVZ upon contact at the RGosvz soma (reconstructed RGvz/osvz pair, Fig. 5C), and that Hes5 active RGosvz underwent basal mitotic somal translocation (Fig. 5D), a cellular behavior consistent with RGosvz (Fig. 3) (Hansen et al., 2010; Shitamukai et al., 2011; Wang et al., 2011).
To further examine the apparent diversity of Notch output in RG, we analyzed sections from E14.5 transgenic Hes5GFP reporter mice, which exhibit mosaic levels and clusters of transgenically labeled RGvz (Fig. 5E) (Basak and Taylor, 2007). Immunolabeling Hes5GFP sections with an anti-Hes1 antibody revealed that RGvz are divided into Hes1+Hes5GFP−, Hes1+/Hes5GFP+, and Hes1−/Hes5GFP+ RGvz subpopulations: ∼33% of Hes1+ cells are Hes5GFP+ (85/261 Hes1 RGvz), and ∼59% of Hes5GFP+ cells are Hes1+ (85/145 RGvz). Although RGosvz were less easily distinguished with the Hes5GFP transgenic label, we did observe Hes1+ cells in the OSVZ (Fig. 5E).
To confirm that Notch signaling output can vary within RGvz subpopulations, we constructed a novel dual Hes1d2YFP:Hes5d2CFP (H1YH5C) reporter plasmid: mouse Hes1 promoter driving destabilized 2 h t1/2 YFP reporter; mouse Hes5 promoter driving destabilized 2 h t1/2 CFP reporter (Ohtsuka et al., 2006). This single construct contains separate expression cassettes to simultaneously monitor Hes1 versus Hes5 activity, and any differences in reporting should reflect promoter activity, not differences in reporter biosynthesis or turnover. We used ex utero electroporation to transfect the H1YH5C dual reporter into E13.5 wild-type neocortex. Live LSCM imaging revealed RGvz were in fact divided into high Hes1 (yellow), equal Hes1/Hes5 (white), and high Hes5 (cyan) RGvz subpopulations (Fig. 5F). These data confirmed that RGvz exhibit diverse output of two different canonical Notch signaling activities, suggestive of combinatorial Hes1 and Hes5 signaling within the RGvz pool.
Despite such variations, blocking canonical signaling from all Notch receptors pharmacologically with acute DAPT treatment uniformly synchronized the differentiation of the RGvz pool (Tbr2GFP− cells in the VZ) into Tbr2GFP+ INPs, compared with control DMSO-treated cultures (Fig. 5G,H, n = 3/3) (Nelson et al., 2007; Kawaguchi et al., 2008a; Ochiai et al., 2009). Thus, these results confirm that niche dynamics maintain a higher overall Notch activity in RG compared with INPs (Mizutani et al., 2007; Yoon et al., 2008; Ochiai et al., 2009), and furthermore demonstrate diverse Notch output into at least Hes1/Hes5 combinatorial subpopulations of RG resulting in a molecularly heterogeneous RG progenitor pool. Notwithstanding these differences in diverse Notch output within subpopulations of the RGvz pool, a major role of higher Notch signaling levels in RG overall is still to prevent their cell-fate transition into INPs.
INPs and RG are distinct sources for multiple Notch ligands
INPs are known to be an important molecular source of Dll1, a key activating ligand for maintaining high Notch activity in the RG pool (Kawaguchi et al., 2008b). Interestingly, it appears that multiple Notch ligands may also show differential enrichment in INPs or RG. For example, as mentioned above, in addition to Dll1, Dll3 also appears enriched in INPs, while Jag1 appears enriched in RG (Fig. 4B). Histological gene expression analysis by in situ hybridization demonstrated a “salt-and-pepper” pattern of Dll1 expression in the VZ and SVZ, while Dll3 appeared localized to the SVZ, and Jag1 was detected in the lateral neocortical VZ (Campos et al., 2001; Allen Brain Atlas, http://developingmouse.brain-map.org). However, in light of our new knowledge of cellular dynamics and interactions, it is key to understand where the Dll1, Dll3, and Jag1 proteins are localized in the VZ and SVZ, especially with regard to progenitor cell bodies and processes.
To this end, we used antibodies for immunofluorescence detection of Dll1, Dll3, and Jag1 proteins. Double immunofluorescence for Dll1 and Dll3 (Fig. 6A,B, E14.5; D, E12.5) revealed punctate Dll1 protein in the VZ and SVZ, while Dll3 protein was restricted to the SVZ and appeared to be localized mainly in cell bodies. Also, Dll1 and Dll3 protein levels were overall highest in dorsomedial cortex, and lower in ventrolateral regions. By contrast, Jag1 protein was expressed in the neocortical VZ with highest levels in ventrolateral regions, especially near the pallial-subpallial boundary (PSPB) (Fig. 6C). Higher expression of Jag1 was also noted in the GE on E14.5 (Fig. 6C), and was even more prominent on E16.5 (Fig. 6E). Comparison with cell-type specific markers revealed abundant Dll1 puncta surrounding Sox9-, Pax6-, and Hes1-positive nuclei in the VZ (Fig. 6F,H). Some punctate Dll1 protein also appeared to localize in or near Tbr2-positive INPvz/svz cell bodies (Fig. 6F,H). By contrast, Dll3 immunolabeling localized strictly around cell bodies of later-stage NeuroD1-expressing INPsvz (Fig. 6G), or in INPsvz with low-to-none Hes5GFP expression (Fig. 6I).
Figure 6.
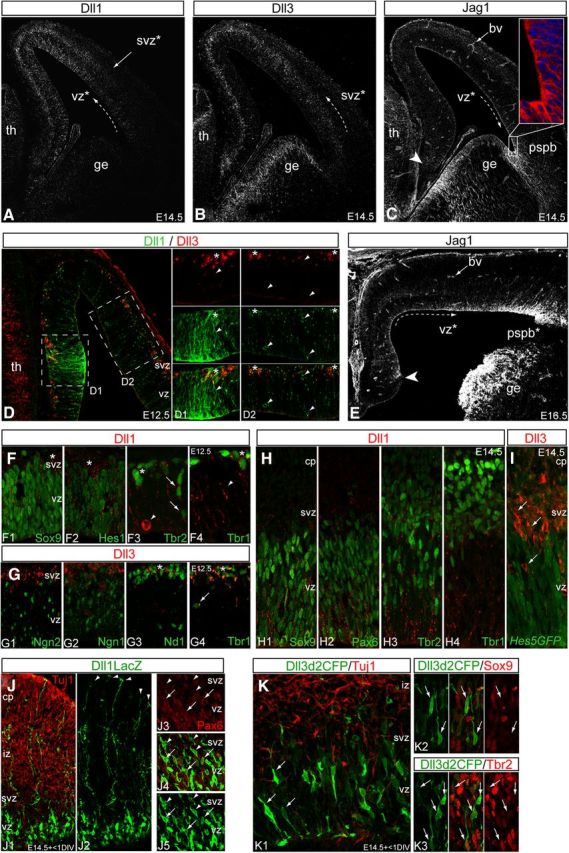
INPs and RG constitute multiple sources of Notch-ligands with graded protein distributions across the developing neocortex. A–C, E14.5 coronal sections were immunolabeled with anti-Dll1 (A), anti-Dll3 (B), and anti-Jag1 (C) antibodies, and analyzed by LSCM, which revealed in low-power grayscale views, counter gradients of highest levels of Dll1 and Dll3 proteins distributed medially, by contrast to the highest levels of Jag1 protein distributed laterally, particularly in the PSPB (C, inset Jag1 red, nuclei blue DAPI counterstain). Also note Dll1 protein in the VZ and SVZ (A) by contrast to Dll3 protein localized primarily to the SVZ (B). D, E12.5 section co-immunolabeled with anti-Dll1 (green) and anti-Dll3 antibodies (red) reveal highest expression of Dll1 medially (D1, arrowhead) transitioning into more discrete puncta laterally within the VZ and SVZ (D2, arrowheads), while Dll3 localizes to clusters of cells in the SVZ (D1, D2, asterisks). E, Jag1 immunolabeling in E16.5 neocortex reveals its maintained mediolateral gradient of expression in the VZ (arrowhead, extent of medial expression). F–G, Comparison of Dll1 (F) and Dll3 (G) immunolabeling with RG and INP transcription factor markers reveals extensive Dll1 puncta surrounding Sox9+ (F1) and Hes1+ (F2) RGvz nuclei, as well as around Tbr2+ INPvz (arrows), and around clusters of Tbr2+ (F3) and Tbr1+ (F4) INPsvz nuclei (asterisks): note that occasional Dll1+ cells are found at the ventricular surface (F3, arrowhead). Dll3 immunolabeling correlates best with later stage TFs such as NeuroD1 (Nd1, G3) and Tbr1 (G4), compared with upstream TFs such as Neurog2 (Ngn2, G1) or Neurog1 (Ngn1, G2). H, I, Similarly, at E14.5, extensive Dll1 puncta was observed around Sox9+ (H1), Pax6+ (H2) RGvz nuclei, especially associated with Tbr2+ nuclei in the VZ and SVZ (H3), but less so with Tbrb1+ nuclei (INPsvz) (H4). Dll3 immunolabeling at E14.5 in sections from Hes5GFP transgenic mice confirm Dll3 protein in Hes5GFP− INPsvz cells (I, arrows). J, K, Dll1LacZ and Dll3d2CFP reporter constructs were transfected into E14.5 wild-type neocortex, cultured as slices for <1DIV, fixed, sectioned and immunolabeled with anti-βgal or anti-GFP antibodies to detect Dll1LacZ (J) and Dll3d2CFP (K) reporters, respectively. Co-immunolabeling with anti-Tuj1 reveals Dll1LacZ+ cell bodies in the VZ, with long, basal processes extending through the Tuj1+ IZ and CP, all the way to the pial surface (J1, J2, arrowheads), while Dll3d2CFP expressing cells at this time were Tuj1−, localized within the VZ/SVZ, and had INPvz/svz morphologies (K1, arrows). Co-immunolabeling for Dll1LacZ and Pax6 revealed that some Dll1LacZ cells were Pax6+ RGvz (arrows), while others were Pax6− INPvz/svz(arrowheads) (J3-5). Co-immunolabeling for Dll3d2CFP revealed that cells in the VZ were Sox9− (K2, arrows) and Tbr2+ (arrows, K3). Abbreviations: th, thalamus; ge, ganglionic eminence; *, graded distribution; PSPB, pallial-subpallial boundary; bv, blood vessel; LSCM, laser scanning confocal microscopy; DIV, days in vitro.
As a complementary approach, we used ex utero electroporation to transfect Dll1LacZ and Dll3d2CFP reporter plasmids (Beckers et al., 2000; Nelson et al., 2009) into E14.5 neocortex. Slices were prepared and cultured for <24 h, fixed, and sections immunolabeled with anti-β -galactosidase or -GFP antibodies (Dll1LacZ, Dll3d2CFP, respectively) in combination with cell-type specific markers. Under these conditions of short duration slice culture, Dll1LacZ was active in Pax6+/Tuj1− RGvz, with many Dll1LacZ+ processes extending to the pial surface, and in some Pax6- cells (presumptive INPsvz, Fig. 6J). By contrast, Dll3d2CFP was active in Sox9−/Tuj1−/Tbr2+ INPvz (Fig. 6K). These results indicated that Dll1 was expressed in not only INPsvz, but also INPvz and likely RGvz subpopulations (see below), while Dll3 reporter activity was specifically upregulated in INPvz, and subsequent Dll3 protein was localized in or near INPsvz cell bodies.
INPsvz extend long-range processes containing punctate Dll1 protein contact mitotic progenitors at the ventricular surface
Even with LSCM, punctate Dll1 immunolabeling was difficult to clearly localize to a specific cell type (labeled with transcription factor antibodies) in the VZ. To test whether punctate Dll1 protein observed in the VZ was actually due to its presence in INPsvz processes extending into the VZ (Figs. 1–3), we analyzed both Dll1 and Dll3 protein localization in sections from Tbr2GFP embryos. Co-immunolabeling for Dll1 (gold) and Tbr2 (magenta) in E12.5 Tbr2GFP (endogenous GFP, green) sections revealed that much of the punctate Dll1 labeling in the VZ was present within Tbr2+/Tbr2GFP+ INPvz/svz cells, and especially within their long processes extending into the VZ (Fig. 7A). Co-immunolabeling for Dll1 (gold) and Tbr1 (magenta) labeling further confirmed extensive Dll1 puncta within Tbr1-/Tbr2GFP+ INPvz, within long radial processes of Tbr1−/Tbr2GFP+ INPsvz, and within the cell bodies of some Tbr1+/Tbr2GFP+ late-stage INPsvz and/or newly postmitotic neurons (Fig. 7B). By contrast, co-immunolabeling for Dll3 (gold) and Tbr1 (magenta) revealed that Dll3 was mainly localized in the cell bodies of Tbr1−/Tbr2GFP+ INPsvz progenitors, and in some Tbr1+/Tbr2GFP+ late-stage INPsvz or newly postmitotic neurons (Fig. 7C). Co-immunolabeling for Dll1 (gold) and Dll3 (magenta) antibodies, as well as endogenous Tbr2GFP (green) and DAPI (blue), revealed that long-range Dll1+/Dll3−/Tbr2GFP+ processes extended from INPsvz into the VZ and contacted mitotic RG (DAPI M-phase nuclei at the ventricular surface) (Fig. 7D–F), exactly as seen by live-cell MPM imaging (Figs. 2–3). Moreover, higher magnifications revealed some overlap of Dll1/Dll3 proteins in INPsvz cells (Fig. 7G). However, Dll1 protein consistently appeared more punctate and separate from Dll3 protein, whereas Dll3 protein appeared more diffuse and localized near INPsvz cell bodies, particularly at contact points within clustered Dll3+/Tbr2GFP+ INPsvz cells (Fig. 7G).
Figure 7.
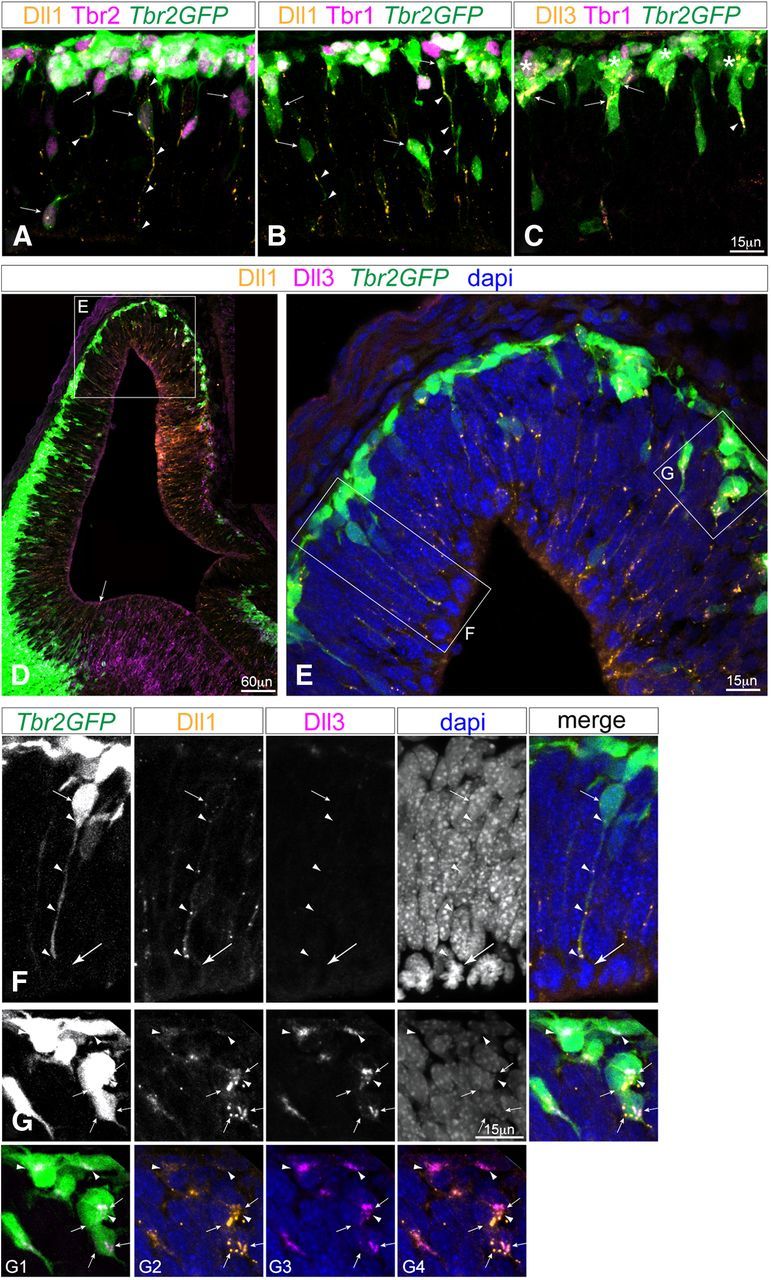
INPs extend long-range processes containing punctate Dll1 protein that contact mitotic progenitors at the apical surface. A–C, E12.5 sections from Tbr2GFP transgenic mice co-immunolabeled with anti-Dll1 and anti-Tbr2 (A, Dll1, orange; Tbr2, magenta; endogenous GFP, green; dapi not shown) or anti-Dll1 and anti-Tbr1 (B, Dll1, orange; Tbr1, magenta; endogenous GFP, green; dapi not shown) visualized with LSCM confirm that punctate Dll1 proteins localize to Tbr2+/Tbr2GFP+ INPvz and INPsvz (arrows) and their processes (arrowheads), compared withTbr1+/Tbr2GFP+ later-stage INPsvz or postmitotic neurons. C, Co-immunolabeling with anti-Dll3 and anti-Tbr1 (Dll3, orange; Tbr1, magenta; endogenous GFP, green; DAPI not shown) reveals Dll3 is primarily located at contact points between Tbr1−/Tbr2GFP+ INPsvz cells (arrows), clusters (asterisks), and occasional short Tbr1−/Tbr2GFP+ INPsvz processes (C, arrowhead). D–G, Co-immunolabeling with anti-Dll1 and anti-Dll3 (Dll1, orange; Dll3, magenta; endogenous GFP, green; dapi, blue) reveals INPsvz cells with punctate Dll1 protein in apical processes contacting mitotic RG at the ventricular surface, whereas Dll3 protein is concentrated at contact regions between INPsvz. D, Low-power view of E12.5 Tbr2GFP section co-immunolabeled with anti-Dll1 and ant-Dll3 antibodies show strong Dll1 and Dll3 protein distribution medially, as well as the upregulation of Dll3 in the GE, juxtaposed to the ventral Tbr2GFP expression limit at the pallial-subpallial boundary (arrow). E, Higher power view of boxed region in D with DAPI channel (blue). F, Higher-power gray-scale single channel views from region indicated in E showing individual Tbr2GFP, Dll1, Dll3, and DAPI labels and merged image reveals specific punctate Dll1 protein in Tbr2GFP+ INPsvz process (arrowheads) contacting a mitotic cell (dapi) at the ventricular surface (big arrow). G, Higher-power gray-scale single channel views from region indicated in E showing individual labels and merged image as in F, reveals Dll3 protein enriched at the contact between adjacent cell bodies of clustering Tbr2GFP+ INPsvz cells (arrowheads), and that while Dll3 and Dll1 immunolabeling shows some overlap within individual INPsvz cells, the two ligands show enrichment in discrete cellular compartments: G1, Tbr2GFP merged with Dll3; G2, Dll1 merged with dapi; G3, Dll3 merged with dapi; G4, Dll1, Dll3, merged with DAPI. Scale bars: A–C, F–G, 15 μm; D, 60 μm.
Altogether, these data not only confirm that INPs are a major source of the Dll1, but also show that INPs and RG subpopulations are each sources of diverse Notch-ligands including Dll1, Dll3, and Jag1. These data further suggest a cellular mechanism for INP-mediated Dll1 activation of Notch signaling, whereby INPs extend long-range dynamic processes containing punctate Dll1 protein to contact and activate Notch signaling in RG.
Diverse Dll1+ progenitor cells interact through dynamic and transient filopodia-like processes
Although we noticed that much of the punctate Dll1 protein in the VZ was localized to INP processes, it was clear that Dll1 protein and promoter activity were also expressed in subpopulations of RG (Figs. 6, 7). To directly observe Dll1 activity in RG as well as INPs, we subcloned the mouse Dll1 promoter from Dll1LacZ (Fig. 2) (Beckers et al., 2000) into a destabilized fluorescent YFP reporter (t1/2 2 h, Dll1d2YFP), and used ex utero electroporation to transfect VZ progenitors in E12.5–13.5 wild-type neocortices that were then prepared for live-cell MPM imaging. Dll1d2YFP activity was detected in multiple neocortical progenitor subtypes (RGvz, RGosvz, INPvz, and INPsvz) (Fig. 8) that exhibited diverse cellular network interactions matching those shown above (Figs. 2–3). For example, in the VZ, Dll1d2YFP+ RGvz and INPvz apical processes formed hook-like structures around other Dll1d2YFP+ mitotic cells at the ventricular surface; both of which maintained specific contact with the most apical daughter cell (Fig. 8A1,A2). In experiments with more sparse and discrete labeling, isolated Dll1d2YFP+ RGvz and INPvz displayed short dynamic filopodia-like structures extending tangentially from the main apical process, presumably making lateral contacts with neighboring VZ progenitors (Fig. 8B,B1–B2). In the SVZ, Dll1d2YFP+ dividing progenitors included RGosvz (Fig. 8C) and INPsvz. The latter exhibited multipolar morphologies with dynamic long- and short-range apical processes, some with smaller branched filopodia, contacting radially aligned cells in the SVZ and VZ (Fig. 8D). For example, within the extensive network of Dll1 active cells at E13.5 (Fig. 8E), a Dll1d2YFP active INPsvz with a long radial apical process (∼60–70 μm) interacted specifically with an underlying Dll1d2YFP+ INPvz, which underwent apical interkinetic nuclear migration (INM) to divide at the ventricular surface after the INPsvz retracted its apical process and migrated basally (Fig. 8F). This behavior was similar to the INPvz in Figure 8G1. By contrast, in another Dll1d2YFP+/+ INPsvz/vz interaction, after the INPsvz retracted its dynamic apical process, rather than dividing apically, the INPvz withdrew its apical process and divided basally (Fig. 8G2). Furthermore, Dll1d2YFP+ cells not only included actively dividing INPvz/svz (Fig. 8A,E–H), but also apparently some symmetrically dividing RGvz at the ventricular surface (Fig. 8H; cleavage plane angle = 90°), which also extended tangential filopodia-like processes (Fig. 8I). Thus, these live MPM imaging data reveal that the VZ/SVZ niche includes diverse Dll1 active neocortical progenitors interacting through dynamic radial and tangential processes. These types of cellular interactions presumably represent the cellular basis for Dll1 signals to reactivate/maintain Notch activity, providing a cellular basis for progenitor diversification.
Discussion
Our studies suggest that the embryonic neocortical progenitor niche constitutes an extremely dynamic social network at the cellular level, incorporating an abundance of transient cell–cell interactions between diverse INP and RG subpopulations. Dynamic and transient long-range processes are a fundamental cellular property of INPs throughout neurogenesis, with extensive long-range interactions from basally located progenitors to more apical progenitors within radial columns, in addition to tangential interactions across radial columns. Notch signaling activity is overall higher in RG than in INPs, but exhibits diverse output resulting in a molecularly heterogeneic RG pool. While a major role of Notch signaling in RG is to prevent their cell-fate transition into INPs, heterogeneous Notch signaling may also contribute to progenitor diversification. Furthermore, our results have also shown that INPs and RG are distinct sources for multiple Notch ligands, and that gradients of Notch signaling components are present within the developing neocortex. INPs extend long-range processes containing punctate Dll1 protein that contact mitotic RG at the ventricular surface, and Dll1-active neocortical progenitors interact through radial and tangential processes. The cell–cell interactions mediated by these short- and long-range processes likely transmit Dll1 signals to reactivate/maintain Notch activity in neighboring progenitors, and coordinate progenitor cell division and differentiation across radial and zonal boundaries (Fig. 9).
Figure 9.
Dynamic cell–cell interactions as a mechanism for Notch signaling in the embryonic neocortical NSC niche. A–D, A series of composite images of RGvz (red), RGosvz (purple), and INPvz/svz (green) progenitors from live-cell MPM imaging experiments summarizes the types of dynamic intercellular interactions in the embryonic neocortical NSC niche. A, INPs use dynamic filopodia containing punctate Dll1 protein (orange dots) to reactivate/maintain high canonical Notch activity in the RG progenitor pool. Note that while INPs are depicted as a major source of Dll1 (Fig. 1-3, 6), subsets of RGvz and RGosvz were also Dll1 active or contained Dll1 punctate protein (data not shown for simplicity, see Fig. 2, 3, 6). Also, identified postmitotic neurons in the overlying intermediate zone/cortical plate (blue) were included for reference and completeness of cell types in the embryonic neocortical NSC niche. B, INPsvz also dynamically interact with underlying INPvz (light green) and recently divided RGvz (light red) through radial relay cell interactions. C, Similar to INPsvz, RGosvz also exhibited varied morphologies and interactions, including extending/retracting radial filopodia into the VZ, and interacting with neighboring INPsvz and RGvz basal processes. D, Both RGvz and INPvz were also observed to extend/retract shorter tangential filopodia from their respective apical processes that could interact directly with neighboring apically dividing progenitors. Finally, progenitor divisions themselves induced hook-like displacements in apical processes of adjacent RGvz and INPvz, as well as basal processes of RGvz (A–D). E, F, Model of the production of cell types within the embryonic neocortical neural stem cells (eNSC) niche with respect to molecular feedback from the sequential and reiterative Notch ligand distribution, which generates diverse contextual canonical Notch signaling output in the RG pool: solid lines indicate the cellular mechanism of INP/Dll1 feedback to RG, parentheses/dashed lines indicate less well understood molecular/cellular input.
Contacts between neocortical progenitors are thought to constitute cell–cell signaling mechanisms (Marthiens et al., 2010; Zhang et al., 2010) necessary for establishing the in vivo architecture required to create functional circuitries in the adult cortex (Johansson et al., 2010). Live-cell imaging has been key to understanding how diverse INPs and RG progenitor pools interact. Initial single-cell imaging studies suggested that INPs undergo four different stages or modes of migration with unique morphologies: (1), rapid movement to the SVZ; (2), migratory arrest in the SVZ; (3), retrograde migration toward the ventricle; (4), migration to the cortex (Noctor et al., 2004). However, the possibility that INP processes mediate not only migration but also cell–cell signaling was not explored, and it remained unclear what types of interactions occur at the population level over shorter time periods. We took advantage of the high spatial and temporal resolution of MPM to determine how INPs in the neocortex behave at the population level in live organotypic slices prepared fromTbr2GFP transgenic embryos (Kwon and Hadjantonakis, 2007; Kowalczyk et al., 2009). We also targeted electroporation of constitutively active tdTom or mTom to densely label mosaics of progenitors, their progeny, and especially their processes in the Tbr2GFP transgenic background to determine how diverse neocortical progenitors interact in their niche. These approaches confirmed many findings from previous studies of single INPs, and further revealed additional INP population dynamics, including: (1) INPsvz extend transient long-range processes into the apical VZ and to the ventricular surface, where they exhibit complex filopodial branching and morphological changes such as hook-like structures around dividing apical progenitors; and (2) INPvz interact with overlying INPsvz via short basal processes, and dynamic tangential filopodia-like protrusions emanating from their apical processes and from their attachments (Figs. 1, 9A–D). Our two-color MPM also allowed us to visualize for the first time novel types of potential RG and INP process-based radial interactions within and between radial columns (discussed below).
INPs have been identified as an important molecular source of Dll1 to maintain Notch activity in neighboring RG (Kawaguchi et al., 2008c; Yoon et al., 2008). We hypothesized that, at the cellular level, Delta-Notch signaling may occur through the ensemble of INP dynamic processes and filopodia. Our gene expression profiling of INPs, detailed imaging of punctate Dll1 protein in Tbr2GFP+ INP processes contacting mitotic apical progenitors, and live-cell imaging of Dll1-active progenitors in the eNSC niche indeed suggest that during specific phases of migration, INPs also use their dynamic filopodia to transmit Dll1 signals (and perhaps other types of cell–cell contact-dependent signaling pathways as well).
Canonical Notch signaling is inherently transient (Fortini, 2009; Kopan and Ilagan, 2009). In embryonic neocortex, Notch levels within individual RG are known to oscillate over short time periods (2 h), and progress through higher and lower states during the cell cycle, reaching lowest levels near the ventricular surface during mitosis (Tokunaga et al., 2004; Mizutani et al., 2007; Shimojo et al., 2008; Ochiai et al., 2009). Thus, during increasingly longer G1/S transition as the cell cycle lengthens during neurogenesis and RG integrate intrinsic and extrinsic cues (Takahashi et al., 1995; Caviness et al., 1999; Miyata, 2007), Notch must be continuously reactivated to prevent RG-to-INP differentiation (Kawaguchi et al., 2008b; Ochiai et al., 2009). A similar phenomenon has been observed in the timing of Notch signaling in embryonic retinal NSCs (Nelson et al., 2007). Since Notch activation requires biophysical force from ligand binding (Fortini, 2009; Kopan and Ilagan, 2009), and Dll1 functions as a major Notch ligand in the neocortex (Kawaguchi et al., 2008c), the presence of Dll1 in long-range processes and filopodia around apical RG mitoses can be inferred to provide an important source for initial Notch reactivation in newly divided RGvz. These types of interactions in the mouse neocortex are reminiscent of similar functions of neuroblasts in the developing Drosophila ectoderm, which extend long-range dynamic filopodia containing punctate Delta protein to activate Notch throughout the entire cluster of cells (De Joussineau et al., 2003; Rajan et al., 2009; Axelrod, 2010; Cohen et al., 2010; Milán and Cohen, 2010). Such long-range signaling provides robustness to the process of cellular diversification in tissue, necessary for correct numbers and patterns of sensory organs (De Joussineau et al., 2003; Rajan et al., 2009; Axelrod, 2010; Cohen et al., 2010; Milán and Cohen, 2010).
Some Notch activation force may also be generated by INM of VZ progenitors (Del Bene et al., 2008; Taverna and Huttner, 2010). We observed punctate Dll1 protein/activity in RGvz and INPvz anchored apical processes, and Jag1 protein in apical RG (Figs. 6, 8). Since INM and apical divisions induce hook-like formations in adjacent apical process (Figs. 2, 8, 9) (Miyata, 2008), these hook-like contacts could serve as another source for Notch activation, although it may also involve non-canonical Notch signals and potential cross talk with other pathways (Minaki et al., 2005; Mizuhara et al., 2005; Zhang et al., 2010; Ohata et al., 2011). INM is not continuous, and in basal VZ regions dynamic contacts from overlying INPsvz and/or basal processes of INPvz may be important for maintaining Notch activity in RGvz progenitors during later cell cycle stages. For example, we found that in the SVZ, multiple types of dynamic interactions occur between overlying (more basal) progenitors (INPsvz/RGosvz) and underlying (more apical) progenitors (INPsvz, INPvz, RGvz, and translocating RGosvz). Many of these interactions appear to be mediated by dynamic apical processes in a cell-to-cell-to-cell radial-relay fashion (Figs. 2, 3, 9). The basal niche is also composed of different spatially restricted sources of Dll1 and Dll3 in INPsvz, and Dll1 in RGosvz (Figs. 6–8, 9E,F). RGvz and RGosvz basal processes might also receive different Notch input, since Jag2 is expressed in the cortical plate (BGEM). Additionally, INPvz and RGvz also generated transient dynamic tangential filopodia from their anchored apical processes and attachments (Figs. 1, 2, 8, 9). Interestingly, transgenic Hes5GFP showed Notch activation in clusters of RGvz subpopulations (Basak and Taylor, 2007). We found that Hes5GFP+ clusters were separated from each other by neighboring Hes1+/Hes5GFP− RGvz clusters (Fig. 5). Within Hes5GFP clusters, further differences in contextual Notch signaling were also observed on the basis of Hes1 signaling (Fig. 5). Our novel Hes1d2YFP:Hes5d2CFP double reporter also revealed mixed Hes1/Hes5 subpopulations of RGvz (Fig. 5), suggestive of a hierarchical or combinatorial Notch signaling state in RGvz subpopulations.
While it is clear that canonical Notch signaling is key for maintaining neocortical RG, the precise roles of individual and combinatorial Notch signaling components within RG subpopulations remain largely unclear. Combined conditional knock-outs of Notch receptors (Notch1/Notch3) or Hes genes (Hes1/Hes3/Hes5) cannot recapitulate the neocortical phenotype of conditional ablation of Rbpsuh alone (Hatakeyama et al., 2004; Mason et al., 2005; Imayoshi et al., 2008; Imayoshi et al., 2010), indicative of a high degree of molecular redundancy in neocortical NSCs compared with other regions of the nervous system. However, it also appears that specific modes of Hes1 activity (oscillatory vs sustained) determine the biological outcome of Notch signaling (Kageyama et al., 2008; Shimojo et al., 2008), and that Hes5 may exhibit specific noncompensated roles in Fezf1/Fezf2-depleted neocortical NSCs (Shimizu et al., 2010). Notch ligands in the neocortex seem to exhibit less redundancy. Conditional knock-out of Dll1 (or Mindbomb1, a key cofactor for Dll1 activity) in neocortical progenitors recapitulates much of the phenotype observed in the Rbpsuh knock-out (Kawaguchi et al., 2008c; Yoon et al., 2008), indicating that Dll1 is a major non-compensated Notch ligand in the neocortex. By contrast, while Dll3 exhibits specific functions during somitogenesis and T-cell development (Dunwoodie, 2009; Hoyne et al., 2011), there is no reported phenotype in the neocortex, similar to the cochlea and retina (Hartman et al., 2007; Nelson et al., 2009), indicating possible compensatory mechanisms for Dll3 functions in the nervous system. These unique roles are further supported by evidence that Dll1-specific functions in the nervous system cannot be rescued by genetic replacement of Dll3 (Geffers et al., 2007). However, misregulation of Dll3 in Huwe1-depleted neocortical NSCs generates excessive Notch activation and proliferation, a major pathway leading to malignant brain tumor formation (Zhao et al., 2009), although the mechanism of Dll3-Notch signaling remains controversial (Ladi et al., 2005; Geffers et al., 2007). Our gene expression profiling revealed that multiple Notch receptors are present in RG (Fig. 4), which can exhibit differential target selectivity for Hes1/5 (Ong et al., 2006). Hence, Dll1 on dynamic processes (INPs especially) may have a general role in activating Notch receptors, but this activity may be composed of differential combinations of Notch1/2/3/(4), contributing to diverse Hes output in RG subpopulations. This molecular capability coupled to highly dynamic radially and tangentially oriented cellular processes and additional discrete sources of Notch ligands may provide differential Notch feedback within radial units to better control temporal aspects of neurogenic output, and between radial units to better coordinate neocortical patterning.
Our results support the notion that extensive contacts between both INP and RG progenitor cells generate diversity and maintain appropriate numbers of RG stem-like progenitors throughout stages of neurogenesis. Cell contact-dependent signals in vivo are communicated through a highly interactive social network of diverse progenitors with dynamic processes that function, at least in part, to convey differential Notch signaling. Our results also suggest that general manipulations aimed at perturbing specific cellular dynamics, such as INM, may also inadvertently affect related but distinct dynamics such as Delta-Notch signaling. Thus, new tools are required to specifically address molecular and cellular complexities observed in mammalian neocortical Notch signaling (Pierfelice et al., 2011), such as the ability to inactivate specific ligands and/or manipulate process dynamics in discrete cell types, and determine where specific activation of different Notch receptors occur. Nevertheless, our results provide a foundation for defining the cellular basis of Notch signaling and cellular diversification in neocortex and other regions of the mammalian nervous system. Since dynamic niches regulate Notch activity, it becomes apparent how even transient alterations in dynamics may affect subsequent neural output and connections, contributing to complex neurological disorders.
Notes
Supplemental material for this article is available at http://depts.washington.edu/cibr/?page_id=1239; host site for companion live-cell multiphoton movies and descriptions for the present study. This material has not been peer reviewed.
Footnotes
This work was supported by NIH Grants R21 MH087070 and RO1 MH080766-S to R.F.H., and an American Heart Association Research Fellowship 10POST2610067 to R.D.H. We thank R.O. Wong, V. Taylor, R. Kageyama, Y. Gotoh, A. Gossler, N. Brown, J. Breunig, and P. Rakic for sharing reagents and thoughtful discussions, and R. Daza for technical assistance. We also thank J. Breunig and P. Rakic for sharing data prior to publication.
The authors declare no competing financial interests.
References
- Ables JL, Breunig JJ, Eisch AJ, Rakic P. Not(ch) just development: Notch signalling in the adult brain. Nat Rev Neurosci. 2011;12:269–283. doi: 10.1038/nrn3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold SJ, Huang GJ, Cheung AF, Era T, Nishikawa SI, Bikoff EK, Molnár Z, Robertson EJ, Groszer M. The T-box transcription factor Eomes/Tbr2 regulates neurogenesis in the cortical subventricular zone. Genes Dev. 2008;22:2479–2484. doi: 10.1101/gad.475408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attardo A, Calegari F, Haubensak W, Wilsch-Bräuninger M, Huttner WB, Reh TA. Live imaging at the onset of cortical neurogenesis reveals differential appearance of the neuronal phenotype in apical versus basal progenitor progeny. PLoS ONE. 2008;3:e2388. doi: 10.1371/journal.pone.0002388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelrod JD. Delivering the lateral inhibition punchline: it's all about the timing. Sci Signal. 2010;3:pe38. doi: 10.1126/scisignal.3145pe38. [DOI] [PubMed] [Google Scholar]
- Basak O, Taylor V. Identification of self-replicating multipotent progenitors in the embryonic nervous system by high Notch activity and Hes5 expression. Eur J Neurosci. 2007;25:1006–1022. doi: 10.1111/j.1460-9568.2007.05370.x. [DOI] [PubMed] [Google Scholar]
- Beckers J, Caron A, Hrab é de Angelis M, Hans S, Campos-Ortega JA, Gossler A. Distinct regulatory elements direct delta1 expression in the nervous system and paraxial mesoderm of transgenic mice. Mech Dev. 2000;95:23–34. doi: 10.1016/S0925-4773(00)00322-1. [DOI] [PubMed] [Google Scholar]
- Bedogni F, Hodge RD, Elsen GE, Nelson BR, Daza RA, Beyer RP, Bammler TK, Rubenstein JL, Hevner RF. Tbr1 regulates regional and laminar identity of postmitotic neurons in developing neocortex. Proc Natl Acad Sci U S A. 2010;107:13129–13134. doi: 10.1073/pnas.1002285107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos LS, Duarte AJ, Branco T, Henrique D. mDll1 and mDll3 expression in the developing mouse brain: role in the establishment of the early cortex. J Neurosci Res. 2001;64:590–598. doi: 10.1002/jnr.1111. [DOI] [PubMed] [Google Scholar]
- Caviness VS, Jr, Takahashi T, Nowakowski RS. The G1 restriction point as critical regulator of neocortical neuronogenesis. Neurochem Res. 1999;24:497–506. doi: 10.1023/A:1022579712262. [DOI] [PubMed] [Google Scholar]
- Cohen M, Georgiou M, Stevenson NL, Miodownik M, Baum B. Dynamic filopodia transmit intermittent Delta-Notch signaling to drive pattern refinement during lateral inhibition. Dev Cell. 2010;19:78–89. doi: 10.1016/j.devcel.2010.06.006. [DOI] [PubMed] [Google Scholar]
- Corbin JG, Gaiano N, Juliano SL, Poluch S, Stancik E, Haydar TF. Regulation of neural progenitor cell development in the nervous system. J Neurochem. 2008;106:2272–2287. doi: 10.1111/j.1471-4159.2008.05522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Joussineau C, Soul é J, Martin M, Anguille C, Montcourrier P, Alexandre D. Delta-promoted filopodia mediate long-range lateral inhibition in Drosophila. Nature. 2003;426:555–559. doi: 10.1038/nature02157. [DOI] [PubMed] [Google Scholar]
- Del Bene F, Wehman AM, Link BA, Baier H. Regulation of neurogenesis by interkinetic nuclear migration through an apical-basal notch gradient. Cell. 2008;134:1055–1065. doi: 10.1016/j.cell.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunwoodie SL. The role of Notch in patterning the human vertebral column. Current opinion in genetics and development. 2009;19:329–337. doi: 10.1016/j.gde.2009.06.005. [DOI] [PubMed] [Google Scholar]
- Elsen GE, Hodge RD, Bedogni F, Daza RA, Nelson BR, Shiba N, Reiner SL, Hevner RF. The protomap is propagated to cortical plate neurons through an Eomes-dependent intermediate map. Proc Natl Acad Sci U S A. 2013;110:4081–4086. doi: 10.1073/pnas.1209076110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund C, Fink A, Lau C, Pham D, Daza RA, Bulfone A, Kowalczyk T, Hevner RF. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fietz SA, Kelava I, Vogt J, Wilsch-Bräuninger M, Stenzel D, Fish JL, Corbeil D, Riehn A, Distler W, Nitsch R, Huttner WB. OSVZ progenitors of human and ferret neocortex are epithelial-like and expand by integrin signaling. Nat Neurosci. 2010;13:690–699. doi: 10.1038/nn.2553. [DOI] [PubMed] [Google Scholar]
- Fortini ME. Notch signaling: the core pathway and its posttranslational regulation. Dev Cell. 2009;16:633–647. doi: 10.1016/j.devcel.2009.03.010. [DOI] [PubMed] [Google Scholar]
- Franco SJ, Müller U. Shaping our minds: stem and progenitor cell diversity in the Mammalian neocortex. Neuron. 2013;77:19–34. doi: 10.1016/j.neuron.2012.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal JS, Morozov YM, Ayoub AE, Chatterjee M, Rakic P, Haydar TF. Molecular and morphological heterogeneity of neural precursors in the mouse neocortical proliferative zones. J Neurosci. 2006;26:1045–1056. doi: 10.1523/JNEUROSCI.4499-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geffers I, Serth K, Chapman G, Jaekel R, Schuster-Gossler K, Cordes R, Sparrow DB, Kremmer E, Dunwoodie SL, Klein T, Gossler A. Divergent functions and distinct localization of the Notch ligands DLL1 and DLL3 in vivo. J Cell Biol. 2007;178:465–476. doi: 10.1083/jcb.200702009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen DV, Lui JH, Parker PR, Kriegstein AR. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature. 2010;464:554–561. doi: 10.1038/nature08845. [DOI] [PubMed] [Google Scholar]
- Hartman BH, Hayashi T, Nelson BR, Bermingham-McDonogh O, Reh TA. Dll3 is expressed in developing hair cells in the mammalian cochlea. Dev Dyn. 2007;236:2875–2883. doi: 10.1002/dvdy.21307. [DOI] [PubMed] [Google Scholar]
- Hatakeyama J, Bessho Y, Katoh K, Ookawara S, Fujioka M, Guillemot F, Kageyama R. Hes genes regulate size, shape and histogenesis of the nervous system by control of the timing of neural stem cell differentiation. Development. 2004;131:5539–5550. doi: 10.1242/dev.01436. [DOI] [PubMed] [Google Scholar]
- Haubensak W, Attardo A, Denk W, Huttner WB. Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: a major site of neurogenesis. Proc Natl Acad Sci U S A. 2004;101:3196–3201. doi: 10.1073/pnas.0308600100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hevner RF, Haydar TF. The (not necessarily) convoluted role of basal radial glia in cortical neurogenesis. Cereb Cortex. 2012;22:465–468. doi: 10.1093/cercor/bhr336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyne GF, Chapman G, Sontani Y, Pursglove SE, Dunwoodie SL. A cell autonomous role for the Notch ligand Delta-like 3 in alphabeta T-cell development. Immunol Cell Biol. 2011;89:696–705. doi: 10.1038/icb.2010.154. [DOI] [PubMed] [Google Scholar]
- Imayoshi I, Shimogori T, Ohtsuka T, Kageyama R. Hes genes and neurogenin regulate non-neural versus neural fate specification in the dorsal telencephalic midline. Development. 2008;135:2531–2541. doi: 10.1242/dev.021535. [DOI] [PubMed] [Google Scholar]
- Imayoshi I, Sakamoto M, Yamaguchi M, Mori K, Kageyama R. Essential roles of Notch signaling in maintenance of neural stem cells in developing and adult brains. J Neurosci. 2010;30:3489–3498. doi: 10.1523/JNEUROSCI.4987-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson PA, Cappello S, Gotz M. Stem cells niches during development–lessons from the cerebral cortex. Curr Opin Neurobiol. 2010;20:400–407. doi: 10.1016/j.conb.2010.04.003. [DOI] [PubMed] [Google Scholar]
- Kageyama R, Ohtsuka T, Shimojo H, Imayoshi I. Dynamic Notch signaling in neural progenitor cells and a revised view of lateral inhibition. Nat Neurosci. 2008;11:1247–1251. doi: 10.1038/nn.2208. [DOI] [PubMed] [Google Scholar]
- Kawaguchi A, Ikawa T, Kasukawa T, Ueda HR, Kurimoto K, Saitou M, Matsuzaki F. Single-cell gene profiling defines differential progenitor subclasses in mammalian neurogenesis. Development. 2008a;135:3113–3124. doi: 10.1242/dev.022616. [DOI] [PubMed] [Google Scholar]
- Kawaguchi D, Yoshimatsu T, Hozumi K, Gotoh Y. Selection of differentiating cells by different levels of delta-like 1 among neural precursor cells in the developing mouse telencephalon. Development. 2008b;135:3849–3858. doi: 10.1242/dev.024570. [DOI] [PubMed] [Google Scholar]
- Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk T, Pontious A, Englund C, Daza RA, Bedogni F, Hodge R, Attardo A, Bell C, Huttner WB, Hevner RF. Intermediate neuronal progenitors (basal progenitors) produce pyramidal projection neurons for all layers of cerebral cortex. Cereb Cortex. 2009;19:2439–2450. doi: 10.1093/cercor/bhn260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegstein A, Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci. 2009;32:149–184. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon GS, Hadjantonakis AK. Eomes::GFP—a tool for live imaging cells of the trophoblast, primitive streak, and telencephalon in the mouse embryo. Genesis. 2007;45:208–217. doi: 10.1002/dvg.20293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladi E, Nichols JT, Ge W, Miyamoto A, Yao C, Yang LT, Boulter J, Sun YE, Kintner C, Weinmaster G. The divergent DSL ligand Dll3 does not activate Notch signaling but cell autonomously attenuates signaling induced by other DSL ligands. J Cell Biol. 2005;170:983–992. doi: 10.1083/jcb.200503113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marthiens V, Kazanis I, Moss L, Long K, Ffrench-Constant C. Adhesion molecules in the stem cell niche–more than just staying in shape? J Cell Sci. 2010;123:1613–1622. doi: 10.1242/jcs.054312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Cerdeño V, Cunningham CL, Camacho J, Antczak JL, Prakash AN, Cziep ME, Walker AI, Noctor SC. Comparative analysis of the subventricular zone in rat, ferret and macaque: evidence for an outer subventricular zone in rodents. PLoS ONE. 2012;7:e30178. doi: 10.1371/journal.pone.0030178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason HA, Rakowiecki SM, Raftopoulou M, Nery S, Huang Y, Gridley T, Fishell G. Notch signaling coordinates the patterning of striatal compartments. Development. 2005;132:4247–4258. doi: 10.1242/dev.02008. [DOI] [PubMed] [Google Scholar]
- Milán M, Cohen SM. Notch signaling: filopodia dynamics confer robustness. Curr Biol. 2010;20:R802–R804. doi: 10.1016/j.cub.2010.07.030. [DOI] [PubMed] [Google Scholar]
- Minaki Y, Mizuhara E, Morimoto K, Nakatani T, Sakamoto Y, Inoue Y, Satoh K, Imai T, Takai Y, Ono Y. Migrating postmitotic neural precursor cells in the ventricular zone extend apical processes and form adherens junctions near the ventricle in the developing spinal cord. Neurosci Res. 2005;52:250–262. doi: 10.1016/j.neures.2005.03.014. [DOI] [PubMed] [Google Scholar]
- Miyata T. Asymmetric cell division during brain morphogenesis. In: Macieira-Coelho A, editor. Progress in molecular and subcellular biology. Berlin Heidelberg: Springer; 2007. pp. 122–142. [DOI] [PubMed] [Google Scholar]
- Miyata T. Development of three-dimensional architecture of the neuroepithelium: role of pseudostratification and cellular ‘community’. Dev Growth Differ. 2008;50(Suppl 1):S105–S112. doi: 10.1111/j.1440-169X.2007.00980.x. [DOI] [PubMed] [Google Scholar]
- Miyata T, Kawaguchi A, Saito K, Kawano M, Muto T, Ogawa M. Asymmetric production of surface-dividing and non-surface-dividing cortical progenitor cells. Development. 2004;131:3133–3145. doi: 10.1242/dev.01173. [DOI] [PubMed] [Google Scholar]
- Mizuhara E, Nakatani T, Minaki Y, Sakamoto Y, Ono Y, Takai Y. MAGI1 recruits Dll1 to cadherin-based adherens junctions and stabilizes it on the cell surface. J Biol Chem. 2005;280:26499–26507. doi: 10.1074/jbc.M500375200. [DOI] [PubMed] [Google Scholar]
- Mizutani K, Yoon K, Dang L, Tokunaga A, Gaiano N. Differential Notch signalling distinguishes neural stem cells from intermediate progenitors. Nature. 2007;449:351–355. doi: 10.1038/nature06090. [DOI] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- Nelson BR, Hartman BH, Georgi SA, Lan MS, Reh TA. Transient inactivation of Notch signaling synchronizes differentiation of neural progenitor cells. Dev Biol. 2007;304:479–498. doi: 10.1016/j.ydbio.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BR, Hartman BH, Ray CA, Hayashi T, Bermingham-McDonogh O, Reh TA. Acheate-scute like 1 (Ascl1) is required for normal delta-like (Dll) gene expression and notch signaling during retinal development. Dev Dyn. 2009;238:2163–2178. doi: 10.1002/dvdy.21848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor SC, Martínez-Cerdeño V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Martínez-Cerdeño V, Kriegstein AR. Distinct behaviors of neural stem and progenitor cells underlie cortical neurogenesis. J Comp Neurol. 2008;508:28–44. doi: 10.1002/cne.21669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochiai W, Nakatani S, Takahara T, Kainuma M, Masaoka M, Minobe S, Namihira M, Nakashima K, Sakakibara A, Ogawa M, Miyata T. Periventricular notch activation and asymmetric Ngn2 and Tbr2 expression in pair-generated neocortical daughter cells. Mol Cell Neurosci. 2009;40:225–233. doi: 10.1016/j.mcn.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Ohata S, Aoki R, Kinoshita S, Yamaguchi M, Tsuruoka-Kinoshita S, Tanaka H, Wada H, Watabe S, Tsuboi T, Masai I, Okamoto H. Dual roles of Notch in regulation of apically restricted mitosis and apicobasal polarity of neuroepithelial cells. Neuron. 2011;69:215–230. doi: 10.1016/j.neuron.2010.12.026. [DOI] [PubMed] [Google Scholar]
- Ohtsuka T, Imayoshi I, Shimojo H, Nishi E, Kageyama R, McConnell SK. Visualization of embryonic neural stem cells using Hes promoters in transgenic mice. Mol Cell Neurosci. 2006;31:109–122. doi: 10.1016/j.mcn.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Ong CT, Cheng HT, Chang LW, Ohtsuka T, Kageyama R, Stormo GD, Kopan R. Target selectivity of vertebrate notch proteins. Collaboration between discrete domains and CSL-binding site architecture determines activation probability. J Biol Chem. 2006;281:5106–5119. doi: 10.1074/jbc.M506108200. [DOI] [PubMed] [Google Scholar]
- Pierfelice T, Alberi L, Gaiano N. Notch in the vertebrate nervous system: an old dog with new tricks. Neuron. 2011;69:840–855. doi: 10.1016/j.neuron.2011.02.031. [DOI] [PubMed] [Google Scholar]
- Pontious A, Kowalczyk T, Englund C, Hevner RF. Role of Intermediate Progenitor Cells in Cereb Cortex Development. Dev Neurosci. 2008;30:24–32. doi: 10.1159/000109848. [DOI] [PubMed] [Google Scholar]
- Rajan A, Tien AC, Haueter CM, Schulze KL, Bellen HJ. The Arp2/3 complex and WASp are required for apical trafficking of Delta into microvilli during cell fate specification of sensory organ precursors. Nat Cell Biol. 2009;11:815–824. doi: 10.1038/ncb1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P. Mode of cell migration to the superficial layers of fetal monkey neocortex. J Comp Neurol. 1972;145:61–83. doi: 10.1002/cne.901450105. [DOI] [PubMed] [Google Scholar]
- Sessa A, Mao CA, Hadjantonakis AK, Klein WH, Broccoli V. Tbr2 directs conversion of radial glia into basal precursors and guides neuronal amplification by indirect neurogenesis in the developing neocortex. Neuron. 2008;60:56–69. doi: 10.1016/j.neuron.2008.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Nakazawa M, Kani S, Bae YK, Shimizu T, Kageyama R, Hibi M. Zinc finger genes Fezf1 and Fezf2 control neuronal differentiation by repressing Hes5 expression in the forebrain. Development. 2010;137:1875–1885. doi: 10.1242/dev.047167. [DOI] [PubMed] [Google Scholar]
- Shimojo H, Ohtsuka T, Kageyama R. Oscillations in Notch signaling regulate maintenance of neural progenitors. Neuron. 2008;58:52–64. doi: 10.1016/j.neuron.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Shitamukai A, Konno D, Matsuzaki F. Oblique radial glial divisions in the developing mouse neocortex induce self-renewing progenitors outside the germinal zone that resemble primate outer subventricular zone progenitors. J Neurosci. 2011;31:3683–3695. doi: 10.1523/JNEUROSCI.4773-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stancik EK, Navarro-Quiroga I, Sellke R, Haydar TF. Heterogeneity in ventricular zone neural precursors contributes to neuronal fate diversity in the postnatal neocortex. J Neurosci. 2010;30:7028–7036. doi: 10.1523/JNEUROSCI.6131-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Nowakowski RS, Caviness VS., Jr The cell cycle of the pseudostratified ventricular epithelium of the embryonic murine cerebral wall. J Neurosci. 1995;15:6046–6057. doi: 10.1523/JNEUROSCI.15-09-06046.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna E, Huttner WB. Neural progenitor nuclei IN motion. Neuron. 2010;67:906–914. doi: 10.1016/j.neuron.2010.08.027. [DOI] [PubMed] [Google Scholar]
- Tokunaga A, Kohyama J, Yoshida T, Nakao K, Sawamoto K, Okano H. Mapping spatio-temporal activation of Notch signaling during neurogenesis and gliogenesis in the developing mouse brain. J Neurochem. 2004;90:142–154. doi: 10.1111/j.1471-4159.2004.02470.x. [DOI] [PubMed] [Google Scholar]
- Wang X, Tsai JW, LaMonica B, Kriegstein AR. A new subtype of progenitor cell in the mouse embryonic neocortex. Nat Neurosci. 2011;14:555–561. doi: 10.1038/nn.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon KJ, Koo BK, Im SK, Jeong HW, Ghim J, Kwon MC, Moon JS, Miyata T, Kong YY. Mind bomb 1-expressing intermediate progenitors generate notch signaling to maintain radial glial cells. Neuron. 2008;58:519–531. doi: 10.1016/j.neuron.2008.03.018. [DOI] [PubMed] [Google Scholar]
- Zhang J, Woodhead GJ, Swaminathan SK, Noles SR, McQuinn ER, Pisarek AJ, Stocker AM, Mutch CA, Funatsu N, Chenn A. Cortical neural precursors inhibit their own differentiation via N-cadherin maintenance of beta-catenin signaling. Dev Cell. 2010;18:472–479. doi: 10.1016/j.devcel.2009.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Teng EM, Summers RG, Ming G-L, Gage FH. Distinct morphological stages of dentate granule neuron maturation in the adult mouse hippocampus. J Neurosci. 2006;26:3–11. doi: 10.1523/JNEUROSCI.3648-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, D'Arca D, Lim WK, Brahmachary M, Carro MS, Ludwig T, Cardo CC, Guillemot F, Aldape K, Califano A, Iavarone A, Lasorella A. The N-Myc-DLL3 cascade is suppressed by the ubiquitin ligase Huwe1 to inhibit proliferation and promote neurogenesis in the developing brain. Dev Cell. 2009;17:210–221. doi: 10.1016/j.devcel.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]