Abstract
Cisplatin is a widely used anticancer drug that acts by binding DNA and causing the formation of intrastrand and interstrand (ICL) cross-links, but the precise downstream effects of the latter damage are not well understood. In this study, we investigated the influence of cisplatin ICLs on synthetic nucleosomes that were platinated in a site-specific manner in vitro and on gene transcription in live mammalian cells. Nucleosome core particles (NCPs) that we constructed contained site-specific cisplatin 5′-d(G*pC)/5′-d(G*pC) ICLs, where the asterisk denotes the platinated nucleoside, to examine the influence of platinum lesions on the dynamic behavior of nucleosomes in solution. A cisplatin ICL, but not a 1,2-d(GpG) cross-link, significantly inhibited ATP-independent histone octamer-DNA sliding. We also used a novel linearization-recircularization strategy described here to synthesize mammalian expression vectors containing site-specific cisplatin ICLs. Plasmid vectors were tested in live mammalian cellsto study the transcription inhibition effects of cisplatin ICLs in the context of two different repair backgrounds. Cisplatin ICLs inhibit transcription as effectively as 1,2-d(GpG) cross-links. We determined that nucleotide excision repair plays a key role in the removal of cisplatin ICLs, acting in a replication-independent fashion. We also found that loss of mismatch repair function dramatically attenuatesthe transcription inhibition effects by cisplatin ICLs but not 1,2-d(GpG) intrastrand cross-links. Our results revealed the unique properties of cisplatin ICLs on nucleosome mobility and on transcription, and they defined how these adducts act in a manner completely different from that used for cisplatin 1,2-d(GpG) cross-links. These new findings provide direct support for a role of ICLs in the pharmacological activities of cisplatin, despite the lower frequency of their formation.
Introduction
Platinum-based chemotherapy has been successfully used to treat various types of cancer (1–3). FDA-approved platinum drugs, including cisplatin [cis-diamminedichloroplatinum(II)], oxaliplatin [(R,R-1,2-diaminocyclohexane)oxalateplatinum(II)], and carboplatin [cis-diammine(1,1′-cyclobutanedicarboxylato)platinum(II)] (Figure 1), bind DNA and form adducts that block transcription and ultimately induce apoptosis (4–6). The major DNA adducts formed by cisplatin are cis-{Pt(NH3)2}2+ 1,2-d(GpG) and –d(ApG) intrastrand cross-links, and minor adducts include 1,3-d(GpNpG) intrastrand cross-links and 5′-d(GpC)/5′-d(GpC) interstrand cross-links (ICLs). How the cells process cisplatin-DNA intrastrand cross-links has been extensively studied (7).
Figure 1.

Chemical structures of platinum (II) anticancer drugs: cisplatin, carboplatin, and oxaliplatin.
In a eukaryotes, DNA wraps around histone octamers forming nucleosomes, the fundamental building blocks of chromatin (8). Cisplatin binds both free and nucleosomal DNA (9). Determining the consequences of platinum-DNA adduct formation at the nucleosome level is an important step in understanding the details of cellular recognition of platinum drugsand of transcription stalling. With the use of DNA probes containing site-specific cisplatin lesions, hydroxyl radical footprinting experiments revealed that a cisplatin 1,2-d(GpG) or a 1,3-d(GpTpG) intrastrand cross-link overrides the rotational setting predefined in a nucleosome positioning sequence. The lesions face inward, toward the histone core, and affect the translational positioning of the DNA (10–12).
Cisplatin ICLs induce profound structural changes in free DNA duplexes and display a profile of binding to cellular proteins and a repair mechanism that are distinct from those of intrastrand cross-links (13–17). Relatively little is known about the effects of ICLs at the nucleosome level. Cisplatin forms ICLs on nucleosomal DNA, with the level of cross-linking decreasing near the nucleosome dyad (18). Other ICL-forming chemotherapeutic agents, such as nitrogen mustards and mitomycin C, also cross-link DNAatlevels that diminish near the dyad (19). Minor groove DNA-binding agents such as pyrrole-imidazole polyamides block temperature-induced histone octamer repositioning and inhibit transcription by bacteriophage T7 RNA polymerase (20, 21).
The transcription inhibition effects of cisplatin intrastrand cross-links have been extensively explored in both in vitro reconstituted systems and live mammalian cells (22). To obtain site-specifically platinated mammalian expression vectors for the latter studies, a “gapping” strategy was employed to incorporate cisplatin intrastrand cross-links within the multiple cloning site of a Gaussia luciferase plasmid, pGLuc (23). The plasmid was used to investigate transcription inhibition at cisplatin intrastr and cross-links in live mammalian cells (24). Subsequently, pGLuc containing a site-specific Pt-DNA lesion by a monofunctional platinum compound, pyriplatin [cis-diammine(pyridine)chloroplatinum(II)], was synthesized and its transcription inhibition effects and repair mechanism were evaluated (25). The“gapping” strategy is, however, not applicable for the synthesis of plasmids containing site-specific cisplatin ICLs.
Although their frequency is relatively low, cisplatin ICLs have long been a topic of interest (26). For instance, despite their limited occurrence, ICLs have the potential to be the most critical cisplatin lesion. ICLs have been associated with cisplatin resistance arising from gene-specific repair (17). Despite efforts to understand the cellular recognition and processing of cisplatin ICLs, their repair mechanism(s) in mammalian systems are not well established. Transcription inhibition studies in vitro reveal that cisplatin ICLs significantly stall both prokaryotic and eukaryotic RNA polymerases at the site of damage (27).
In order to probe the effects of cisplatin ICLsat the level of the nucleosome we posed the following questions. What differences, if any, are there in the properties of nucleosomes containing DNA havinga site-specific cisplatin intrastrand versusan interstrand cross-link? To what extentdo cisplatin ICLs inhibit transcription in live mammalian cells? Do cisplat inintrastr and versus interstrand DNA cross-links differ in their abilities to disrupt transcription? Is transcription-coupled repair the same for the two types of adduct?To address these questions, we constructed site-specifically modified nucleosomes containing a cisplatin ICL or a 1,2-d(GpG) intrastrand cross-link. Nucleosomeslacking platinum damage were utilized as a control. We compared the mobility of nucleosomes containing the different cisplatin-DNA cross-links. In addition, we established a novel linearization-recircularization strategy to synthesize plasmids containing site-specific cisplatin ICLs, and the plasmids were applied in transcription assays to reveal the transcription inhibition effects of ICLs in a variety of live mammalian cells. Unique inhibitory effects of cisplatin ICLs on nucleosome mobility and transcription were discovered, revealing that, although cisplatin ICLs represent a small portion of Pt-DNA cross-links, their cellular processing is completely different from that of the more abundant cisplatin 1,2-intrastrand cross-links.
Materials and Methods
Enzymes were obtained from New England Biolabs (Ipswich, MA) and chemicals and solvents from commercial sources unless specified otherwise. UV-vis spectra were obtained on a HP 8453 spectrometer. Platinum analysis was performed by flameless atomic absorption spectroscopy on a Perkin-Elmer AAnalyst 600 system. Analytical and preparative HPLC were run on an Agilent 1200 HPLC system. SequaGel concentrate used in the preparation of the denaturing polyacrylamide gels was purchased from National Diagnostics. Oligonucleotide properties were calculated using the online site http://www.basic.northwestern.edu/biotools/-oligocalc.html. Polyacrylamide gel electrophoresis (PAGE) was performed on either a Life Technologies S2 sequencing gel electrophoresis apparatus or a Protean II xi cell from Biorad. Electrophoresis gels with radioactive samples were dried and documented on a Storm 840 Phosphorimager system from Amersham/GE Healthcare or exposed wet to a Kodak Biomax MS film. Radioactive samples were quantified on a Beckman LS 6500 scintillation counter. Agarose gel electrophoreses containing 0.5 μg/mL ethidium bromide were imaged using a BioRad Fluor-S MultiImager. A QIAquick PCR Purification Kit was purchased from Qiagen (Valencia, CA). HeLa cells were purchased from the American Type Culture Collection. U2OS-MOCK and XPF-1128 cells were supplied by Dr. Nora Graf in the Department of Chemistry at the Massachusetts Institute of Technology. HEC59 and HEC59+Chr2 cells were obtained from Dr. Thomas Kunkel from the National Institutes of Health.
Synthesis of Platinated Oligonucleotides for the Construction of 146-bp DNA Duplexes
The small DNA duplexes containing a cisplatin ICL or1,2-d(GpG) cross-link were synthesized and purified by HPLC following the protocol reported previously (14, 28). The platinated 21-mer strands were characterized by UV-vis spectroscopy, atomic absorption spectroscopy, and a nuclease S1 digestion assay (data not shown). The cisplatin ICL was characterized by 12% polyacrylamide gel electrophoresis (Figure S2) and stored in 0.1 M NaClO4 at −80°C.
Preparation of 146-bp DNA Duplexes
63-Mer, 57-mer, 62-mer, and 56-mer oligonucleotides were chemically synthesized and purified by 8% preparative denaturing PAGE. The purified strands were quantified by UV-vis spectroscopy. The strands were also radiolabeled with [γ-32P]-ATP by T4 polynucleotide kinase (PNK) and revealed on an 8% denaturing gel to confirm their purity (Figure S3). Sequences are provided in Figure S4. A 300 pmol quantity of each oligonucleotide was first phosphorylated with ATP or [γ-32P]-ATP by T4 PNK (20 U) at 37 °C for 1.5 h. The enzyme was deactivated by heat and removed by phenol extraction. The oligonucleotides were then ethanol precipitated. The large duplexes were prepared (Figure S5) by annealing 63-mer and 57-mer, 62-mer and 56-mer, separately, in 50 μL of annealing buffer containing 100 mMNaCl, 50 mMTris-HCl, 10 mM MgCl2, pH 7.4 in a temperature gradient of 95 °C to 4 °C over 8 h. A 20 pmol quantity of each of the 21-mer/33-mer duplexes was then annealed with 40 pmol of the two large duplexes (Figure S5) in annealing buffer using a temperature gradient of 37 °C to 4 °C over 2 h. The buffer was adjusted to 100 mMNaCl, 100 mMTris-HCl, 20 mM MgCl2, and 1 mM ATPpH 7.4 in a total volume of 800 μL supplemented with 4 U of T4 DNA ligase. The ligation was carried out at 16 °C overnight and the enzyme was deactivated by heat and removed by phenol extraction. The volume of the mixture was reduced by ethanol precipitation and the ligation products were purified and characterized by a 5% denaturing PAGE (Figure S6).
Purification of Native Oligonucleosomes
High molecular weight chromatin from HeLa cells was isolated and oligonucleosomes with linker histone depletion were prepared as reported previously (11). The purity of the oligonucleosomes was confirmed by a 1.5% native agarose gel and a 12% SDS-PAGE (Figure S7).
Nucleosome Reconstitution
Nucleosome reconstitution experiments were carried out as reported previously (11). Details are reported in the Supplementary Information section.
Vector Construction and Preparation
An unreplicable mammalian vector expressing Gaussia luciferase, pGLuc, was derived from pCMV-GLuc as reported previously (23). The backbone for construction of Gaussia luciferase plasmids containing site-specific platinum-DNA 1,2-d(GpG) cross-links, pGLuc4temGG, was derived from pGLuc plasmid as described previously (23). Another Gaussia luciferase plasmid for subsequent incorporation of a site-specific cisplatin ICL, designated as pGLuc-Bsm2, was prepared by insertion of a heteroduplex oligonucleotide, 5′-AGCTTGAGCGGTACCAGGGAGAGACG/5′-GATCCGTCTCTCCCTGG-TACCGCTCA, between the HindIII and BamHI sites of pGLuc. This modified plasmid contains unique HindIII and BsmBI restriction sites for the incorporation of acisplatin ICL between the CMV promoter and Gaussia luciferase gene (Figure 4).
Figure 4.

Sequence design of pGLuc9temG from pGLuc plasmid for the incorporation of site-specific cis-{Pt(NH3)2}2+ ICL.
Preparation of Insertion Strands for Site-Specifically Modified Plasmids
A 15-bp DNA duplex containing a site-specific cisplatin ICL, 5′-AGCTGGAGAAGAG*CAAGAG/5′-CCCTCTCTTG*CTCTTCTCC, where the asterisks denote the platinated bases, was synthesized as reported previously (14) (Figure 4).
Preparation of Site-Specifically Platinated pGLuc Probes
Synthesis of the Gaussia luciferase expression vector containing a site-specific cisplatin ICL, pGLuc9temG+ICL, was carried out using a linearization-recircularization strategy (Figure 5). A 700 μg quantity of pGLuc-Bsm2 was digested with 700 units of HindIII at 37 °C for 2 h. The resulting linear plasmid was purified by isopycnic centrifugation at 58,000 rpm, 20 °C for 24 h with a CsCl gradient containing 120 μg/mL ethidium bromide to separate the linearized plasmid from its circular form. The fraction containing the linearized plasmid was extracted under UV shadowing, and ethidium bromide was removed by three n-butanol extraction steps. An ethanol precipitation step was performed to reduce the final volume, and the linearized pGLuc-Bsm2 was stored at −20 °C. Next, an 80 μg quantity of purified linearized plasmid was ligated with 100 equiv of phosphorylated 15-bp duplex containing a site-specific cisplatin ICL in ligation buffer containing 50 mMTris-HCl, pH 7.6, 10 mM MgCl2, and 1 mM ATP. This buffer was prepared without DTT, which could potentially interact with the platinum atom. The mixture was heated at 37 °C for 10 min and kept at 4 °C for 10 min. T4 DNA Ligase (320 units) was added to the final mixture, which was incubated at 16 °C overnight. The excess amount of 15-bp ICL insertion strands was subsequently removed with a QIAquick PCR purification kit. The plasmid was further digested with 200 units of BsmBI at 55 °C for 1 h, andthe resulting small duplex DNA in the solution was removed by the QIAquick PCR purification kit. The recircularization step was carried out in the presence of 320 units of T4 DNA ligase in a buffer containing 50 mMTris-HCl, pH 7.6, 10 mM MgCl2, and 1 mM ATP at room temperature for 3 h in a total volume of 6.0 mL. The final product was purified using isopycnic centrifugation. The fraction containing the closed circular form of plasmid was visualized by UV shadowing and extracted. Ethidium bromide was removed by n-butanol extraction, and the plasmids were ethanol precipitated and stored in TE buffer (10 mMTris-HCl, 1 mM EDTA, pH 7.4). The plasmid containing a site-specific ICL is designated as pGLuc9temG+ICL. A control plasmid containing unplatinated small duplex insertion strand, designated as pGLuc9temG+IS, was prepared as well. The DNA concentrations were determined by a Quant-iT PicoGreen dsDNA kit (Invitrogen). The yields from linearized plasmids range from 2–5%. The pGLuc plasmid containing site-specific cisplatin 1,2-d(GpG) cross-links and the control plasmid with no platinum were synthesized according to a published protocol (23).
Figure 5.

Linearization-recircularization strategy for the synthesis of pGLuc plasmid containing a site-specific cis-{Pt(NH3)2}2+ ICL.
Transient Transfection of Cells and GLuc Reporter Transcription Assays
Transient transfection of different mammalian cells and GLuc reporter transcription assays were carried out following protocols published previously (25).
Results
Synthesis of Site-Specifically Platinated Nucleosomes
To build site-specifically modified nucleosomes containing different platinum lesions, we adopted the sequence used previously in our laboratory to study the structure of nucleosomal DNA containing a cisplatin 1,3-d(GpTpG) cross-link (10). This sequence has a moderate affinity for histone octamers and does not form a well-positioned nucleosome (10). In order to synthesize 146-bp DNAs containing site-specific platinum lesions, small 21-mer/33-mer duplexes containing a cisplatin ICLor a cisplatin intrastrand cross-link were first prepared and purified (Figures S1 and S2). The larger strands, 63-mer, 57-mer, 62-mer, and 56-mer, were synthesized, purified, and characterized by a 12% denaturing PAGE to confirm their purity (Figure S3; sequences shown in Figure S4). The 63-mer and 57-mer, and the 62-mer and 56-mer, were annealed in a pairwise fashion to obtain the larger duplexes, which were then annealed and ligated with the small duplexes to yield site-specific 146-bp strands (Figure S5). During this process, [γ-32P]-ATP was used to label the 5′-end of the 63-mer so that, in the final products, the duplexes were radiolabeled on the top strand. The synthesized site-specific 146-bp strands were characterized on a 5% denaturing gel (Figure S6). The duplex containing a cisplatin ICL had reduced migration, confirming the presence of a site-specific ICL. When the ICL was treated with NaCN, the platinum link was removed, converting the duplex into 146-mer strands having the same migration as duplexes containing no platinum (Figure S6). Native oligonucleosomes containing core histones with linker histone depletion from HeLa cells were purified (Figure S7) (11). First, we transferred the labeled duplexes with or without a site-specific cisplatin ICL to donor chromatin having > 3 nucleosomes, 2–3 nucleosomes, or 1–2 nucleosomes. The transfer efficiency for the centered, radiolabeled forms of the nucleosome core particle (NCP) assembly and off-centered NCP assembly was similar when different sizes of chromatin were utilized (Figure S8). We therefore used donor chromatin having 2–3 nucleosomes in the following assays.
Effects of Cisplatin ICLs on Nucleosome Repositioning
When the NCP samples were heated at 45 °C for 2 h, most of the off-centered NCPs containing no platinum shifted to form centered NCPs. NCPs containing a site-specific cisplatin ICL were, however, unable to be heat shifted to form centered NCPs, indicating that a cisplatin ICL alters nucleosome mobility (Figure S8). We next examined, in much greater detail, the influence of a cisplatin ICL on the dynamic behavior of nucleosome in solution by testing its effect on nucleosome repositioning induced by heating. NCPs containing a site-specific cisplatin ICL, a cisplatin 1,2-d(GpG) cross-link, or no platinum were compared. The samples were heat shifted at 37 °C or 45 °C for 0, 0.5, 1, 2, and 3 h. The same amount of sample was loaded in each lane of a native polyacrylamide gel to compare the percent of centered and off-centered NCPs. At 37 °C, the majority of the NCPs were in the centered, thermodynamically stable form after 1 h when the core particle contained either no platinum or a site-specific cisplatin 1,2-d(GpG) cross-link, and most of the off-centered NCPs were transformed back to centered NCPs after 3 h (Figure 2). The shift process was faster at 45 °C, where the majority of the NCPs containing a 1,2-d(GpG) cross-link or no platinum were in the centered form after 0.5 h (Figure S9). The nucleosome containing a site-specific cisplatin ICL, however, displayed a dramatically different nucleosome mobility profile. Firstly, upon the assembly of nucleosome containing an ICL, there are a greater number off-centered NCPs (from 55% to 60%) compared to nucleosomes containing a 1,2-d(GpG) cross-link or no cross-link (from 40% to 50%). Secondly, heat shiftingconverted only a small portion of off-centered to centered nucleosomes, even after 3 h at both 37 °C and 45 °C. The transfer speed was higher at the higher temperature, but still a large portion of the nucleosomes remained in the off-centered form (Figure 2 and Figure S9). A quantitative analysis of the influence of different platinum adducts on nucleosome positional preference is shown in Figure 3.
Figure 2.

Representative autoradiograph of NCPs containing cis-{Pt(NH3)2}2+ ICLs, cis-{Pt(NH3)2}2+ 1,2-d(GpG) cross-links, or No Pt. NCPs were heat shifted at 37 °C. A 4.5% native polyacrylamide gel was used.
Figure 3.

Influence of platinum adducts on nucleosome positional preference at (A) 37 °C or (B) 45 °C. Blue diamond, cis-{Pt(NH3)2}2+ ICL; red square, cis-{Pt(NH3)2}2+ 1,2-d(GpG) cross-link; green triangle, No Pt. The error bars came from at least two independent experiments from different batches of the 146-bp duplexes.
Construction of pGLuc Containing Site-Specific ICLs
We used pGLuc as the vector for the incorporation of an ICL between the CMV promoter and GLuc expression gene to investigate the transcription inhibition effects of ICLs in live mammalian cells (Figure 4). HindIII and BsmBI restriction sites were cloned into pGLuc plasmids, designated as pGLuc-Bsm2. A 15-bp heteroduplex oligonucleotide containing a site-specific ICL with 5′-HindIII and 5′-BsmBI overhangs was inserted into the pGLuc-Bsm2 plasmid between the HindIII and BsmBI sites. The resulting plasmids were designated as pGLuc9temG+IS and pGLuc9temG+ICL, for unplatinated control and site-specifically platinated ICL plasmids, respectively.
The overall strategy for constructing a site-specific ICL plasmid is given in Figure 5. The acceptor plasmid pGLuc-Bsm2 was first linearized by digestion with HindIII and then purified (Figure 5, a and b). A 100-fold molar excess of the phosphorylated heteroduplex oligonucleotide containing a site-specific ICL, as shown in Figure 4, was annealed to the linearized pGLuc-Bsm2 in the presence of T4 DNA ligase. Because these oligos contain a 5′-AGCT overhang complementary to the HindIII-generated ends, they could be annealed to the linearized pGLuc-Bsm2 in a specific direction. This is a dead-end reaction because the HindIII ligation site was eliminated after the addition of the heteroduplex, with the resulting linearized plasmid containing two 5′-CCCT overhangs that could not be annealed andligated to form a circularized plasmid. The excess amount of small oligos was eliminated. The next step required removal of the platinated heteroduplex oligonucleotide from one end of the plasmid to generate a ligation-competent plasmid end suitable for recircularization (Figure 5, c). In the current design, the heteroduplex contains a 5′-CCCT overhang complementary to that generated by BsmBI digestion. We used size exclusion chromatography to remove the smaller DNA fragments, because they could potentially affect the subsequent annealing and ligation steps (Figure 5, d). Thispurified linear plasmid contains a 5′-CCCT overhang from the inserted small oligo and a 5′-AGGG overhang from the BsmBI digestion, and it could therefore beclosed by T4 DNA ligase. The ligation was carried out at low plasmid concentrations to favor formation of circular products by intramolecular rather than intermolecular reactions, thereby avoiding the formation of multimers (Figure 5, e). The final site-specific circular plasmid, pGLuc9temG+ICL, was purified by isopycnic ultracentrifugation (Figure 5, f). With this strategy, aheteroduplex oligonucleotide containing a site-specific cisplatin ICL could be incorporated into the pGLuc plasmid in a specific direction. In parallel we synthesized a control plasmid incorporating a heteroduplex oligonucleotide containing no platinum, designated pGLuc9temG+IS. Restrictiondigestion analysis of the synthesized and purified plasmids (Figure S10) confirmed their purity and the successful incorporation of an ICL.
Transcription Inhibition at Cisplatin ICLs in NER-Proficient and -Deficient Cells
We next studied the transcription inhibition at cisplatin ICLs in live mammalian cells. The pGLuc9temG+ICL plasmid was employed in a transcription assay using osteosarcoma cancer cells with down regulation of XPF gene expression (XPF1128) and with control cells (U2OS-mock). The detailed protocol for this transcription assay has been described previously (25). In brief, the cells were transfected with pGLuc9temG+ICL and pGLuc9temG+IS plasmids, as well as witha site-specific plasmid carrying acisplatin 1,2-d(GpG) intrastrand cross-link and an unplatinated control plasmid, and the secreted Gaussia luciferase in the culture media was collected at 24, 48, and 72 h. The amount of luciferase was quantitated. Transcription in the presence of site-specific platinum cross-links was normalized to that of unplatinated controls at different time points in U2OS-mock and XPF1128 cells. Dataassembled from three independent experiments are presented in Figure 6. A cisplatin ICL strongly inhibited transcription in both cell lines, at a level comparable to that of the cisplatin 1,2-d(GpG) cross-link. Transcription recovery was observed for all the plasmids over 72 h in both cell lines, indicating repair of the platinum cross-links during this time period. Measurably greater transcription inhibition was observed for both a cisplatin 1,2-d(GpG) cross-link and an ICL in XPF-knockdown cells. The transcription levels in the presence of a cisplatin ICL were 44.6%, 66.3%, and 68.3% in U2OS-mock cells, and the levels were 36.3%, 54.7%, and 58.9% in XPF-1128 cells at 24, 48, and 72 h, respectively. The present data confirm that cisplatin ICLs strongly inhibit transcription in live mammalian cells and that XPF has a role in the repair of cisplatin ICLs.
Figure 6.
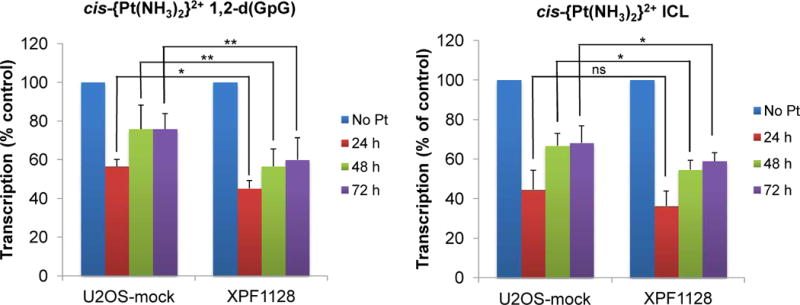
Transcription profiles of site-specifically platinated probes containing a cis-{Pt(NH3)2}2+ 1,2-d(GpG) cross-link or a cis-{Pt(NH3)2}2+ ICL in U2OS-mock and XPF1128 cells. The error bars came from three independent experiments using different batches of probes. ns, not significant; *, P < 0.05; **, P < 0.01.
Transcription Inhibition Effects at Cisplatin ICLs in MMR-Proficient and -Deficient Cells
Site-specifically platinated pGLuc plasmids containing cisplatin 1,2-d(GpG) or ICL were utilized in a transcription assay with HEC59 (MMR-deficient) and HEC59+Chr2 (MMR-proficient) human endometrial cancer cellcells as indicated in Figure 7. Cisplatin ICLs strongly inhibited transcription in the MMR-proficient endometrial cancer cells to an extent that was significantly higher than that for cisplatin 1,2-d(GpG) cross-links. No significant difference of transcription was observed between the two cell lines when a site-specific cisplatin 1,2-d(GpG) cross-link was presented in the pGLuc plasmid. Transcription inhibition by the cisplatin ICL was clearly stronger in MMR-proficient compared to MMR-deficient cells. Transcription levels in the presence of a cisplatin ICL were 74.4%, 79.4%, 79.7% in HEC59 cells, and the levels were 26.4%, 35.2%, and 39.4% in HEC59+Chr2 cells at 24, 48, and 72 h, respectively. These data indicate that MMR has a role in processing cisplatin ICLs but not 1,2-d(GpG) cross-links in replication-independent pathways.
Figure 7.

Transcription profiles of site-specifically platinated probes containing a cis-{Pt(NH3)2}2+ 1,2-d(GpG) cross-link or a cis-{Pt(NH3)2}2+ ICL in HEC59 and HEC59+Chr2 cells. The error bars came from three independent experiments using different batches of probes. ns, not significant; **, P < 0.01.
Discussion
Influence of Cisplatin ICLs on Nucleosome Mobility
Chromatin is highly dynamic during transcription, with nucleosome displacement that involves histone modification, ATP-dependent chromatin remodeling, and histone chaperones (29). Several mechanisms including transient site-exposure, ATP-independent histone octamer sliding, re-modeling of chromatin and nucleosomes, and changes in chromatin higher-order structure regulate DNA accessibility (30). Damage can be exposed through translocation of histone octamer along the DNA, allowing full access of damage-response proteins to the lesion (31). Studies in vivo revealed that cisplatin forms DNA adducts in the promoter region, inhibiting hormone receptor-dependent chromatin remodeling and subsequent binding of transcription factors (32). Our results further shed light on the influence of platinum adducts at the nucleosomal level.
A recent paper revealed that global platination by cisplatin or oxaliplatinon 146-bp nucleosomal DNA inhibits histone octamer-DNA sliding, although the authors claimed that the platinum adducts in the nucleosome core did not alter positioning (33). When there was one platinum-DNA adduct per NCP, either that of cisplatin or oxaliplatin, it did not significantly inhibit DNA-histone sliding. Our results are in accord with these conclusions if we consider the following. When a 146-bp DNA duplex is globally platinated, cisplatin intrastrand cross-linkswill dominate. We find here that a cisplatin 1,2-d(GpG) cross-link only slightlyaffects nucleosome mobility. In results previously published by our laboratory, a site-specific cisplatin 1,3-d(GpTpG) cross-link in nucleosomal DNA did notinhibit histone octamer-DNA sliding at all (34). Here we find that heat shifting of nucleosomal DNA containing a cisplatin ICL does not convertan off-centered NCP product to a centered product. Thus, it is cisplatin ICLs rather than intrastrand cross-links that most significantly inhibit ATP-independent histone octamer-DNA sliding.
A cisplatin ICL induces larger distortions in native DNA than those caused by intrastrand cross-links. Structural studies reveal that the cis-{Pt(NH3)2}2+unit in an ICL lies in the minor groove and that the double helix is locally reversed, forming a left-handed, Z-DNA structure. Cytosines in the central 5′-GC/5′-GC motif are no longer paired to guanines but instead are extruded from the double helix (35, 36). Our results here are consistent with other findings that the minor-groove binding polyamides suppress histone octamer-DNA sliding in the nucleosome (21). A cisplatin ICL may lock the histoneoctamer in a translational position by blocking rotation of the DNA on the surface of the core particle. This result has implications for the repair of different cisplatin adducts at the nucleosomal level. The cisplatin ICL may block its own repair by inhibiting dynamic site exposure. Because histone-octamer association inhibits the repair of platinum adducts (37), a cisplatin ICL on the nucleosome may present greater challenges to the cellular repair machinery.
Synthesis of Plasmid Containing Site-Specific Cisplatin ICLs
To further elucidate the transcription inhibition effects of cisplatin ICLs in live mammalian cells, we established a linearization-recircularization strategy to synthesize plasmid containing site-specific cisplatin ICLs. There are advantages and disadvantages of such an approach, which introduces a novel design for the direction-specific incorporation of aheteroduplex oligonucleotide containing DNA damage. The same host vector, linearized pGLuc-Bsm2 in this case, can be utilized toinsert not only site-specific cisplatin ICLs, but site-specific intrastrand cross-links or any other type of damage as well. Different numbers of platinum adducts can be incorporated into the same backbone. For the “gapping” strategy that we published previously (23), different plasmid backbones have to be cloned and manipulatedto incorporate different platinum-DNA cross-links, and the “gapping” strategy is not applicable for the construction of ICL-containing plasmids. The linearization-recircularization strategy has a relatively low yield, ranging from 2–5% in final yield from the linearized plasmid. The “gapping” strategy can provide yield up to 10%.
Role of NER in the Repair of Cisplatin-DNA Cross-Links
NER is the major pathway for removing cisplatin intrastrand cross-links. The role of NER in the repair of cisplatin 1,2-d(GpG) cross-links and pyriplatin-DNA lesions has been previously described (24, 25). A growing body of evidence has shown that NER may also function in the repair of ICLs. Several NER components, including XPF·ERCC1 and RPA, participate in ICL repair in vitro (38). NER makes dual incisions in E. coli and yeast, bracketing the ICL on one of the cross-linked strands to “unhook” the cross-link (39, 40). Other evidence reveals, however, that NER isdispensable in mammalian ICL repair (41). Unlike the NER-defective mutants in E. coli and yeast, the mammalian equivalents are only moderately sensitive to ICL-forming agents such as mitomycin C (42). Moreover, biochemical studies have demonstrated that the dual incisions by mammalian NER do not unhook ICLs (43). These lines of evidence support the current model in which, instead of playing a role in the initial steps of ICL repair, NER may help remove the cross-link following incision/translesion synthesis (44).
Using the linearization-recircularization strategy, we constructed plasmids containing a site-specific cisplatin 5′-d(G*pC)/5′-d(G*pC) ICL, where the asterisks denote the platinated deoxynucleosides. The ability of the ICL to inhibit transcription was studied in XPF-knockdown and XPF-normal cells. We observed stronger transcription inhibition in the XPF-knockdown cells compared to that in XPF-normal cells, indicating a role for NER in replication-independent repair of cisplatin ICLs. Our results are in good agreement with a very recent paper in which transcription-coupled, but not global-genome, NER appears to be involved in repair of cisplatin ICLs (45).
Role of MMR in the Repair of Cisplatin-DNA Cross-Links
Besides the direct role of MMR in DNA repair, ittriggers apoptosis signaling pathways following platinum-DNA damage (46). MMR, which contributes to cisplatin resistance (47, 48), is involved in eliminating ICLs formed by psoralenin human cells (49). The role of MMR proteins in processing cisplatin ICLs has also been investigated. In particular, we showed in an electrophoretic mobility shift assay that hMutSβ, a heterodimer of hMSH2 and hMSH3, binds to a cisplatin ICL (14). In addition, transcription assays with globally platinatedp GLuc plasmids reveal that the transcription inhibition following cisplatin and pyriplatin damage is greater in hMSH2-proficient cells (25). The results from site-specifically platinated plasmids containing ICLs are in agreement with these prior results using plasmids globally platinated by cisplatin, with stronger transcription inhibition occurring in MMR-proficient cells. The MMR apparatus, instead of facilitating removal of cisplatin cross-links, promotes the transcription inhibition effects of cisplatin ICLs but not 1,2-d(GpG) cross-links. For the first time, to the best of our knowledge, we have discovered in this study that loss of MMR functionalitysignificantly attenuates the transcription inhibition effects of cisplatin ICLs, suggesting that such activity may promote cisplatin resistance. Thus, cisplatin ICLs rather than 1,2-d(GpG) cross-links may contribute more to cisplatin resistance for cells lacking hMSH2. The role of hMSH2 in blocking transcription of plasmids containing a cisplatin ICL may arise from binding of the protein at the site of the cross-link, which then could interfere with Pol II transcription past the lesion.
Cellular Processing of Cisplatin ICLs and the Pharmacological Properties of Cisplatin
It is generally agreed that intrastrand cross-links contribute more to the pharmacological properties of cisplatin than ICLs owing to their greater abundance. The cellular processing of cisplatin intrastrand cross-linkshave been extensively investigated, revealing nuclear proteins that recognize the damage, repair pathways that remove it, and the influence of such cross-links on DNA metabolism and nucleosome properties. Compared to other DNA-damaging chemotherapeutics including nitrogen mustards, psoralen, and mitomycin-C, which mainly form ICLs, cisplatin appears to have a distinctive mechanism of action owing to the lower frequency of ICL formation (39). The present work reveals, however, that the cellular processing of cisplatin ICLs can significantly influence the pharmacological activity of cisplatin. A previous study reported greater gene-specific repair efficiency of cisplatininter- but not intrastrand cross-links that was linked to cisplatin resistance (17). Because of their unique structure, however, cisplatin ICLs are more challenging for the cellular repair machinery to eliminate. A detailed repair mechanism for ICLs is not fully worked out, and recent work has identified replication-dependent repair of ICLs in which the Fanconi anemia (FA) pathway plays a pivotal role (16, 50). Little is known about the replication-independent repair mechanism of ICLs and their influence on nucleosomal DNA. We find here that cisplatin ICLs inhibit DNA-histone sliding and therefore potentially shield themselves from cellular repair. Cisplatin ICLs strongly inhibit transcription to an extent not less than that of cisplatin 1,2-d(GpG) cross-links. NER plays a role, albeit not a significant one, in the replication-independent repair of cisplatin ICLs, and loss of MMR function greatly attenuates the transcription inhibition effects of cisplatin ICLs but not 1,2-d(GpG) intrastrand cross-links. These findings, together with our previously published work (14), confirm that cisplatin ICLs are uniquely recognized by nuclear proteins, strongly inhibit transcription in live mammalian cells, and have special pathways for removal. Owing to these unique properties, we now speculate that cisplatin ICLs may also contribute significantly to the cytotoxicity of cisplatin as well as to cellular resistance to the drug. The manner by which cells process cisplatin ICLs will therefore dramatically affect the pharmacological activity of cisplatin. More detailed studies of the cellular processing of cisplatin ICLs, such as an in-depth analysis of replication-dependent and –independent repair mechanisms at the nucleosome level, should provide a more complete picture of the mechanism of action of cisplatin, the “penicillin” of modern chemotherapeutics, and offer information regarding resistance to this drug. Such information will also guide the design of future generation platinum-based antineoplastic agents with improved pharmacological activities and reduced side effects.
Supplementary Material
Acknowledgments
The authors are most grateful to Dr. Thomas Kunkel (National Institutes of Health) for MMR-deficient cells and Dr. Nora Graf (Massachusetts Institute of Technology) for XPF-knockdown cells. L.S. acknowledges MIT’s John Reed (1961) Fund for summer research funding.
Grant Support
This work was supported by grant CA034992 from the National Cancer Institute to S.J.L. and Project 7000253 from the City University of Hong Kong to G.Z.
Footnotes
Disclosure of Potential Conflicts of Interest
S.J. Lippard is a cofounder and serves as head of the Scientific Advisory Boardof Blend Therapeutics, a biopharmaceutical company which isdeveloping nanoparticle drug combinations that may include platinumcomplexes.
References
- 1.Rosenberg B, VanCamp L, Trosko JE, Mansour VH. Platinum compounds: a new class of potent antitumour agents. Nature. 1969;222:385–6. doi: 10.1038/222385a0. [DOI] [PubMed] [Google Scholar]
- 2.Einhorn LH, Williams SD. The role of cis-platinum in solid-tumor therapy. N Engl J Med. 1979;300:289–91. doi: 10.1056/NEJM197902083000605. [DOI] [PubMed] [Google Scholar]
- 3.Smith IE, Talbot DC. Cisplatin and its analogues in the treatment of advanced breast cancer: a review. Br J Cancer. 1992;65:787–93. doi: 10.1038/bjc.1992.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jung Y, Lippard SJ. Direct cellular responses to platinum-induced DNA damage. Chem Rev. 2007;107:1387–407. doi: 10.1021/cr068207j. [DOI] [PubMed] [Google Scholar]
- 5.Wang D, Lippard SJ. Cellular processing of platinum anticancer drugs. Nat Rev Drug Discov. 2005;4:307–20. doi: 10.1038/nrd1691. [DOI] [PubMed] [Google Scholar]
- 6.Kelland L. The resurgence of platinum-based cancer chemotherapy. Nat Rev Cancer. 2007;7:573–84. doi: 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]
- 7.Jamieson ER, Lippard SJ. Structure, recognition, and processing of cisplatin-DNA adducts. Chem Rev. 1999;99:2467–98. doi: 10.1021/cr980421n. [DOI] [PubMed] [Google Scholar]
- 8.Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–94. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 9.Lippard SJ, Hoeschele JD. Binding of cis- and trans-dichlorodiammineplatinum(II) to the nucleosome core. Proc Natl Acad Sci USA. 1979;76:6091–5. doi: 10.1073/pnas.76.12.6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Danford AJ, Wang D, Wang Q, Tullius TD, Lippard SJ. Platinum anticancer drug damage enforces a particular rotational setting of DNA in nucleosomes. Proc Natl Acad Sci USA. 2005;102:12311–6. doi: 10.1073/pnas.0506025102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ober M, Lippard SJ. Cisplatin damage overrides the predefined rotational setting of positioned nucleosomes. J Am Chem Soc. 2007;129:6278–86. doi: 10.1021/ja0706145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ober M, Lippard SJ. A 1,2-d(GpG) cisplatin intrastrand cross-link influences the rotational and translational setting of DNA in nucleosomes. J Am Chem Soc. 2008;130:2851–61. doi: 10.1021/ja710220x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang H, Zhu L, Reid BR, Drobny GP, Hopkins PB. Solution structure of a cisplatin-induced DNA interstrand cross-link. Science. 1995;270:1842–5. doi: 10.1126/science.270.5243.1842. [DOI] [PubMed] [Google Scholar]
- 14.Zhu G, Lippard SJ. Photoaffinity labeling reveals nuclear proteins that uniquely recognize cisplatin-DNA interstrand cross-links. Biochemistry. 2009;48:4916–25. doi: 10.1021/bi900389b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Räschle M, Knipscheer P, Enoiu M, Angelov T, Sun J, Griffith JD, et al. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell. 2008;134:969–80. doi: 10.1016/j.cell.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knipscheer P, Räschle M, Smogorzewska A, Enoiu M, Ho TV, Schärer OD, et al. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science. 2009;326:1698–701. doi: 10.1126/science.1182372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhen W, Link CJ, Jr, O’Connor PM, Reed E, Parker R, Howell SB, et al. Increased gene-specific repair of cisplatin interstrand cross-links in cisplatin-resistant human ovarian cancer cell lines. Mol Cell Biol. 1992;12:3689–98. doi: 10.1128/mcb.12.9.3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Millard JT, Wilkes EE. cis- and trans-Diamminedichloroplatinum(II) interstrand cross-linking of a defined sequence nucleosomal core particle. Biochemistry. 2000;39:16046–55. doi: 10.1021/bi0022285. [DOI] [PubMed] [Google Scholar]
- 19.Millard JT, Spencer RJ, Hopkins PB. Effect of nucleosome structure on DNA interstrand cross-linking reactions. Biochemistry. 1998;37:5211–9. doi: 10.1021/bi972862r. [DOI] [PubMed] [Google Scholar]
- 20.Gottesfeld JM, Belitsky JM, Melander C, Dervan PB, Luger K. Blocking transcription through a nucleosome with synthetic DNA ligands. J Mol Biol. 2002;321:249–63. doi: 10.1016/s0022-2836(02)00598-3. [DOI] [PubMed] [Google Scholar]
- 21.Suto RK, Edayathumangalam RS, White CL, Melander C, Gottesfeld JM, Dervan PB, et al. Crystal structures of nucleosome core particles in complex with minor groove DNA-binding ligands. J Mol Biol. 2003;326:371–80. doi: 10.1016/s0022-2836(02)01407-9. [DOI] [PubMed] [Google Scholar]
- 22.Todd RC, Lippard SJ. Inhibition of transcription by platinum antitumor compounds. Metallomics. 2009;1:280–91. doi: 10.1039/b907567d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ang WH, Brown WW, Lippard SJ. Preparation of mammalian expression vectors incorporating site-specifically platinated-DNA lesions. Bioconjug Chem. 2009;20:1058–63. doi: 10.1021/bc900031a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ang WH, Myint M, Lippard SJ. Transcription inhibition by platinum-DNA cross-links in live mammalian cells. J Am Chem Soc. 2010;132:7429–35. doi: 10.1021/ja101495v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu G, Myint M, Ang WH, Song L, Lippard SJ. Monofunctional platinum-DNA adducts are strong inhibitors of transcription and substrates for nucleotide excision repair in live mammalian cells. Cancer Res. 2012;72:790–800. doi: 10.1158/0008-5472.CAN-11-3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deans AJ, West SC. DNA interstrand crosslink repair and cancer. Nat Rev Cancer. 2011;11:467–80. doi: 10.1038/nrc3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corda Y, Job C, Anin M-F, Leng M, Job D. Spectrum of DNA-platinum adduct recognition by prokaryotic and eukaryotic DNA-dependent RNA polymerases. Biochemistry. 1993;32:8582–8. doi: 10.1021/bi00084a027. [DOI] [PubMed] [Google Scholar]
- 28.Zhu G, Chang P, Lippard SJ. Recognition of platinum-DNA damage by poly(ADP-ribose) polymerase-1. Biochemistry. 2010;49:6177–83. doi: 10.1021/bi100775t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Workman JL. Nucleosome displacement in transcription. Genes Dev. 2006;20:2009–17. doi: 10.1101/gad.1435706. [DOI] [PubMed] [Google Scholar]
- 30.Luger K. Dynamic nucleosomes. Chromosome Res. 2006;14:5–16. doi: 10.1007/s10577-005-1026-1. [DOI] [PubMed] [Google Scholar]
- 31.Saha A, Wittmeyer J, Cairns BR. Chromatin remodelling: the industrial revolution of DNA around histones. Nat Rev Mol Cell Biol. 2006;7:437–47. doi: 10.1038/nrm1945. [DOI] [PubMed] [Google Scholar]
- 32.Mymryk JS, Zaniewski E, Archer TK. Cisplatin inhibits chromatin remodeling, transcription factor binding, and transcription from the mouse mammary tumor virus promoter in vivo. Proc Natl Acad Sci USA. 1995;92:2076–80. doi: 10.1073/pnas.92.6.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu B, Davey CA. Platinum drug adduct formation in the nucleosome core alters nucleosome mobility but not positioning. Chem Biol. 2008;15:1023–8. doi: 10.1016/j.chembiol.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 34.Todd RC, Lippard SJ. Consequences of cisplatin binding on nucleosome structure and dynamics. Chem Biol. 2010;17:1334–43. doi: 10.1016/j.chembiol.2010.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paquet F, Pérez C, Leng M, Lancelot G, Malinge J-M. NMR solution structure of a DNA decamer containing an interstrand cross-link of the antitumor drug cis-diamminedichloroplatinum (II) J Biomol Struct Dyn. 1996;14:67–77. doi: 10.1080/07391102.1996.10508930. [DOI] [PubMed] [Google Scholar]
- 36.Coste F, Malinge J-M, Serre L, Shepard W, Roth M, Leng M, et al. Crystal structure of a double-stranded DNA containing a cisplatin interstrand cross-link at 1.63 Å resolution: hydration at the platinated site. Nucleic Acids Res. 1999;27:1837–46. doi: 10.1093/nar/27.8.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang D, Hara R, Singh G, Sancar A, Lippard SJ. Nucleotide excision repair from site-specifically platinum-modified nucleosomes. Biochemistry. 2003;42:6747–53. doi: 10.1021/bi034264k. [DOI] [PubMed] [Google Scholar]
- 38.Zhang N, Lu X, Legerski RJ. Partial reconstitution of human interstrand cross-link repair in vitro: characterization of the roles of RPA and PCNA. Biochem Biophys Res Commun. 2003;309:71–8. doi: 10.1016/s0006-291x(03)01535-3. [DOI] [PubMed] [Google Scholar]
- 39.Noll DM, Mason TM, Miller PS. Formation and repair of interstrand cross-links in DNA. Chem Rev. 2006;106:277–301. doi: 10.1021/cr040478b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muniandy PA, Liu J, Majumdar A, Liu S-T, Seidman MM. DNA interstrand crosslink repair in mammalian cells: step by step. Crit Rev Biochem Mol Biol. 2010;45:23–49. doi: 10.3109/10409230903501819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De Silva IU, McHugh PJ, Clingen PH, Hartley JA. Defining the roles of nucleotide excision repair and recombination in the repair of DNA interstrand cross-links in mammalian cells. Mol Cell Biol. 2000;20:7980–90. doi: 10.1128/mcb.20.21.7980-7990.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thompson LH. Evidence that mammalian cells possess homologous recombinational repair pathways. Mutat Res. 1996;363:77–88. doi: 10.1016/0921-8777(96)00008-0. [DOI] [PubMed] [Google Scholar]
- 43.Bessho T, Mu D, Sancar A. Initiation of DNA interstrand cross-link repair in humans: the nucleotide excision repair system makes dual incisions 5′ to the cross-linked base and removes a 22- to 28-nucleotide-long damage-free strand. Mol Cell Biol. 1997;17:6822–30. doi: 10.1128/mcb.17.12.6822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McHugh PJ, Spanswick VJ, Hartley JA. Repair of DNA interstrand crosslinks: molecular mechanisms and clinical relevance. Lancet Oncol. 2001;2:483–90. doi: 10.1016/S1470-2045(01)00454-5. [DOI] [PubMed] [Google Scholar]
- 45.Enoiu M, Jiricny J, Schärer OD. Repair of cisplatin-induced DNA interstrand crosslinks by a replication-independent pathway involving transcription-coupled repair and translesion synthesis. Nucl Acids Res. 2012;40:8953–64. doi: 10.1093/nar/gks670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Topping RP, Wilkinson JC, Scarpinato KD. Mismatch repair protein deficiency compromises cisplatin-induced apoptotic signaling. J Biol Chem. 2009;284:14029–39. doi: 10.1074/jbc.M809303200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fink D, Nebel S, Aebi S, Zheng H, Cenni B, Nehmé A, et al. The role of DNA mismatch repair in platinum drug resistance. Cancer Res. 1996;56:4881–6. [PubMed] [Google Scholar]
- 48.Lin X, Howell SB. DNA mismatch repair and p53 function are major determinants of the rate of development of cisplatin resistance. Mol Cancer Ther. 2006;5:1239–47. doi: 10.1158/1535-7163.MCT-05-0491. [DOI] [PubMed] [Google Scholar]
- 49.Wu Q, Christensen LA, Legerski RJ, Vasquez KM. Mismatch repair participates in error-free processing of DNA interstrand crosslinks in human cells. EMBO Rep. 2005;6:551–7. doi: 10.1038/sj.embor.7400418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu T, Ghosal G, Yuan J, Chen J, Huang J. FAN1 acts with FANCI-FANCD2 to promote DNA interstrand cross-link repair. Science. 2010;329:693–6. doi: 10.1126/science.1192656. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


