Abstract
Competition through cross-reacting host immune responses, a form of apparent competition, is a major driver of pathogen evolution and diversity. Most models of pathogens have focused on intraspecific interactions to explain observed patterns. Two recent experiments suggested that Haemophilus influenzae, a common nasopharyngeal colonizer of humans, might alter the immune environment in a way that favors otherwise less fit serotypes of another common pathogen, pneumococcus. Using a computational model, we demonstrate that H. influenzae, if it consistently raises the fitness of the less fit serotypes, can strongly promote pneumococcal diversity. However, the effects of H. influenzae are so sensitive to the prevalence of H. influenzae that this species is unlikely to be the main driver of serotype coexistence. Interactions that significantly affect diversity could furthermore be extremely difficult to detect through co-occurrence analysis alone. These results suggest that small differences in strains’ adaptations to different immunological regimes, which are shaped by coinfections with other pathogens, can have dramatic effects on strain dynamics and patterns of phenotypic variation. Studies of microbial communities might therefore benefit from the use of varied approaches to infer the presence of indirect interactions.
Keywords: apparent competition, pneumococcus, Haemophilus influenzae, microbiome, immunity, pathogen
Introduction
Apparent competition—indirect competition between two or more species through a shared enemy (Holt 1977)—can be a major force shaping ecological communities (Bonsall and Hassell 1997; Chaneton and Bonsall 2000; Holt and Lawton 1994). In some cases, apparent competition may have stronger effects than interference or exploitation competition on species coexistence (Holt 1984; Holt and Lawton 1993; Price et al. 1986). Its role might be especially large in systems where consumers (predators or parasites) and resources (prey or hosts) have comparable generation times, allowing consumers to respond rapidly to changes in the population size of resources. Extinction of one or more resource species is a common outcome (Holt 1977; Holt 1984; Holt and Lawton 1993), although apparent competition can also enable coexistence (e.g., Bonsall and Hassell 1997).
In host-pathogen interactions, coexistence of species, or of genetic types within a species, can depend on apparent competition (Fenton and Perkins 2010; Holt and Dobson 2006; Mideo 2009; Pedersen and Fenton 2007). From one perspective, pathogens are consumers and hosts resources (Bowers and Turner 1997; Holt and Pickering 1985; Price et al. 1986; Tompkins et al. 2000). Host populations sharing a common pathogen can indirectly compete and be regulated by the pathogen through, for example, pathogen-induced mortality or a reduction in host fecundity (Anderson and May 1978; May and Anderson 1978). Especially when transmitted by a vector or environmental reservoir, a pathogen shared by multiple hosts can depress host populations enough that the hosts effectively do not compete for resources (Holt and Lawton 1993; Price et al. 1986). On another scale, the populations of cells and factors that comprise the host immune response might be viewed as predators of pathogenic prey (Fenton and Perkins 2010; Mideo 2009). Immune responses may be particularly powerful because they are not limited by the traditional energetic constraints of classical predators. From this perspective, the question is how shared immune responses affect the outcome of competition among pathogens within an individual host or host population. Competition for non-immune hosts can be thought of as competition for enemy-free space (Holt and Lawton 1993).
Theoretical models (e.g., Andreasen 2003; Bowers and Turner 1997; Ferguson et al. 2003; Gog and Grenfell 2002; Gupta et al. 1998; Koelle et al. 2006) have demonstrated that this second kind of apparent competition can shape pathogen diversity. In the absence of other effects such as spatial structure, the intrinsic reproductive numbers of different strains (R0) and their degree of antigenic relatedness, often referred to as cross-immunity, have been the focus of models that examine strain diversity. The majority of such models incorporate cross-immunity that arises from acquired immunity, such as T cells and B cells, which are at least somewhat specific to particular antigens expressed by pathogens and can persist in the host’s immune memory. In general, the specificity of these responses promotes negative frequency dependent selection, so that strains that have been seen more often before (by an individual host or a host population) are more easily targeted, promoting the growth of rarer, less cross-reactive strains. These dynamics can generate strain coexistence over long time periods. However, if strains share common immunological “predators” but differ substantially in their R0, coexistence is less certain (Omori et al. 2010).
Most studies have focused on mechanisms of competition between strains of the same species or genus [e.g., Plasmodium spp. (Buckee and Gupta 2010; Gupta et al. 1998), influenza viruses (Andreasen 2003; Ferguson et al. 2003; Koelle et al. 2006), and dengue viruses (Cummings et al. 2009; Nagao and Koelle 2008; Recker et al. 2009)] to explain strain dynamics. The presence of other pathogen species can add complexity by changing the immunological environment in which pathogens interact. Indirect positive interactions between species are one possible outcome. By reducing the CD4+ T cell population, for example, HIV facilitates coinfection with malaria; malaria also increases HIV viral loads (Abu-Raddad et al. 2006). Infection with helminths may predispose human hosts to acquisition of bacterial and viral pathogens via a tradeoff between the Th1 and Th2 responses (Ezenwa and Jolles 2011). Indirect competition can also result through shared inflammatory responses (Brown et al. 2008; Graham 2008). These examples demonstrate that pathogen abundance can be determined partly by indirect interactions, either positive or negative, with other species.
An open question is the extent to which the presence of one pathogen species can change the outcome of intraspecific competition in another pathogen species by modifying the immunological environment. In this scenario, the presence of one species alters the type of apparent competition experienced by strains of another species. If different outcomes are possible, then attempts to understand or manage the dynamics of any single pathogen species may need to consider the broader within-host community.
Recently, it has been suggested that colonization with Haemophilus influenzae might affect competition among individual serotypes of Streptococcus pneumoniae in humans (Lysenko et al. 2010; Margolis et al. 2010). Both H. influenzae and S. pneumoniae are extremely common nasopharyngeal colonizers (fig. B1, table A1). The >90 serotypes of S. pneumoniae, also called pneumococcus, have a stable rank order: the same serotypes tend to be common everywhere and over time (Cobey and Lipsitch 2012). Pneumococcus is typically found in the human nasopharynx, where it is said to be carried. The most common serotypes have relative long durations of carriage (Hogberg et al. 2007; Lipsitch et al. 2012). The most common serotypes are also better at preventing co-colonization with other serotypes (Lipsitch et al. 2012) and resisting phagocytosis by neutrophils (Weinberger et al. 2009). These observations raise the question of how the rarer and less obviously fit serotypes persist. Elsewhere, we proposed that coexistence can result from a combination of acquired non-serotype-specific immunity, which disproportionately reduces the fitness of the commonest serotypes, and a small amount of acquired serotype-specific (anticapsular) immunity (e.g., a ~30% reduction in susceptibility to a serotype if a host has carried it before) (Cobey and Lipsitch 2012). This amount of serotype-specific immunity is consistent with the few available observations (Goldblatt et al. 2005; Hill et al. 2008; McCool and Weiser 2004; Weinberger et al. 2008). An alternative or additional hypothesis is that H. influenzae indirectly promotes diversity by changing the immunological environment in which pneumococcal serotypes compete. Two limited experiments in mice have suggested that in the presence of H. influenzae, the outcome of competition between a common (presumably more fit) and rare (presumably less fit) serotype might be reversed: the common serotype outcompetes the rare serotype when H. influenzae is absent, and the rare serotype outcompetes the common serotype when H. influenzae is present (Lysenko et al. 2010; Margolis et al. 2010). This reversal is thought to arise from a tradeoff between a serotype’s ability to resist opsonic and non-opsonic phagocytosis. In non-opsonic phagocytosis, phagocytes such as neutrophils consume pneumococci directly. When phagocytosis is enhanced by opsonization, immune effectors such as complement protein or antibodies bind to the surface of a pathogen and make it easier to destroy. H. influenzae increases the rate of opsonization in a complement-dependent manner, which appears to harm the common serotypes disproportionately (Hyams et al. 2010; Lysenko et al. 2010; Lysenko et al. 2005). Thus, H. influenzae might change the kind of apparent competition experienced by serotypes.
In this study, we first consider whether species might coexist due to shifts in apparent competition induced by another species. Specifically, we ask how a change in apparent competition between pneumococcal serotypes would affect serotype diversity, assuming the impact of H. influenzae observed for the three serotypes examined in animal models were fully general across a range of pneumococcal serotypes. We computationally investigate whether tradeoffs in the survival of pneumococcal serotypes in hosts colonized or uncolonized with H. influenzae might explain the coexistence of pneumococcal serotypes.
We next examine whether it might be theoretically possible to detect an effect of H. influenzae on pneumococcus using available observations. In other systems, the presence of apparent competition has been demonstrated by direct manipulation of predator and prey densities (Balmer et al. 2009; Bonsall and Hassell 1997; Chaneton and Bonsall 2000; Graham 2008; Grosholz 1992; Karvonen et al. 2009), by fitting models to time series (Bonsall and Hassell 1998; Lello et al. 2004; Telfer et al. 2008; Telfer et al. 2010), and by analyzing correlations in nature (Behnke et al. 2005; Bottomley et al. 2005; Byrne et al. 2003; Chaneton and Bonsall 2000; Fenton et al. 2010; Karvonen et al. 2009; Lello et al. 2004; Leung and Poulin 2007; Tompkins et al. 2000). A persistent challenge is to distinguish the effects of apparent competition from those of resource and interference competition. Many of the traditional approaches are not amenable to the study of interactions between bacterial pathogens of humans. In mice, a standard animal model of pneumococcus, population sizes of the immune system and concentrations of chemical immune effectors are difficult to manipulate (apart from complete abrogation in genetic knockout mice) and to measure at the site of infection. Even if accurate measurement were possible, it is unclear if numerical responses are preserved across host species. At the population level, extensive time series of the co-colonization rates of H. influenzae and individual pneumococcal serotypes have not been gathered. We were thus interested in whether cross-sectional epidemiological studies might be used to detect an interaction between the species. If H. influenzae affects the colonization abilities of pneumococcal serotypes differently, we might expect to see lower rates of co-colonization with serotypes that are more sensitive to its effects.
We find that by altering the type of apparent competition experienced by pneumococcus, H. influenzae could strongly promote serotype diversity, but it is not a sufficient explanation for observed coexistence. Additionally, its role would be extremely difficult to infer from cross-sectional observations alone. These results demonstrate that small differences in the competitive ability of pathogen strains under different immunological states can strongly promote diversity, but additional lines of evidence are necessary to identify and quantify these effects. We briefly discuss implications for understanding the complex communities of the human microbiome from cross-sectional observations.
Model
We developed an agent-based model to simulate the transmission of 25 arbitrary pneumococcal serotypes and one strain of H. influenzae. The model of pneumococcal transmission has been described elsewhere (Cobey and Lipsitch 2012), but the major features are summarized here, followed by the modeled interactions with H. influenzae that are specific to this study. The simulated serotypes differed in their intrinsic durations of carriage and their ability, when already colonizing a host, to prevent colonization by an invading serotype. We assumed that the intrinsic duration of carriage and exclusion ability were positively correlated, so that the serotype with the longest duration of carriage was also best at inhibiting colonization by other serotypes (Lipsitch et al. 2012; Weinberger et al. 2008). The maximum and minimum intrinsic durations of carriage were taken from observations (Gray et al. 1980; Hogberg et al. 2007), and the durations of carriage of the other serotypes were evenly and linearly interpolated between these points. The exclusion abilities (Lipsitch et al. 2012) were also linearly interpolated, with the least fit serotype (which also has the shortest intrinsic duration of carriage) having no ability to exclude invaders. In the absence of evidence to the contrary, all serotypes were assumed to have identical transmission rates (Erasto et al. 2010).
Hosts colonized with pneumococcus developed two kinds of immunity. Serotype-specific immunity reduced susceptibility by a fraction σ if a host had ever been previously colonized with a serotype; for a given simulation, σ was the same for all serotypes. Initially, we assumed that non-serotype-specific (henceforth, “nonspecific”) immunity exponentially reduced the duration of carriage to a minimum level (here, 25 days) depending on the number of times a host had carried pneumococcus before. We also tested a linear decline to zero. For each functional form, we varied the slope of the initial decline. For each of these different parameterizations (form and slope) of nonspecific immunity, we also varied the strength of serotype-specific immunity from none (σ = 0) to strong but imperfect (σ = 0.9).
The model also simulated the dynamics of a single strain of H. influenzae. Although diverse types of H. influenzae circulate in nature, we know of no evidence or reason why these differences should affect interactions with pneumococcus. The acquisition of immunity to H. influenzae colonization following exposure to the organism has not been quantified, and we parsimoniously assumed it was similar to that of a pneumococcal serotype. Hosts that had previously cleared H. influenzae were less susceptible to future carriage with H. influenzae (σH = 0.3), and the duration of carriage declined nonlinearly with past exposure. Consistent with experiment (Lysenko et al. 2005; Margolis et al. 2010), the dynamics of H. influenzae were modeled to be independent of the dynamics of pneumococcus: current or past colonization with pneumococcus affected neither the risk of colonization nor the duration of colonization with H. influenzae. However, carriage of pneumococcus was sensitive to concurrent carriage of H. influenzae. When a host carrying pneumococcus acquired H. influenzae, each carried strain of serotype z cleared instantaneously with some probability h(z). When a host carrying H. influenzae became colonized with pneumococcus of serotype z, that pneumococcal colonization immediately cleared with the same probability. This rapid clearance is consistent with the observed speed of clearance in hosts who have received the pneumococcal conjugate vaccine, which induces a strong antibody response. For simplicity, we assumed a linear relationship between the probability that a strain of pneumococcus is cleared by colonization with H. influenzae, h(z), and the fitness rank (in the absence of H. influenzae) of that pneumococcal serotype, f(z). The serotype with the longest duration of carriage, which is always the most fit serotype in the absence of H. influenzae, has rank f(z) = 1, and the serotype with the shortest duration of carriage has rank f(z) = Z, where Z equals the number of serotypes. The probability of clearance of serotype z is given by
| (1) |
The parameter c equals the probability of clearing the otherwise fittest serotype (the one with the longest intrinsic duration of carriage and best exclusion abilities) and scales the sensitivity of all serotypes to clearance by H. influenzae. The otherwise least fit serotype (with the shortest intrinsic duration of carriage and no exclusion abilities) always has a positive probability (equal to c/Z) of being cleared.
We simulated the model for different carriage prevalences of H. influenzae (10%, 25%, or 50% in children <5 y old), maximum clearance probabilities c (c = 0, 0.05, 0.1, and 0.2), and levels of serotype-specific immunity σ (σ ∈ [0, 0.9]). For each set of parameters, we refitted pneumococcal transmission rates with H. influenzae present so that the total carriage prevalence of pneumococcus in children <5 y old was 40%. This prevalence is approximately the midpoint of the broad range of observed carriage rates in young children (table A1, fig. B1). At pneumococcal carriage rates of 37%-41%, the prevalence of H. influenzae varied between 11% and 70% and had a mean of 38% (table A1, fig. B1).
Results
Effects of H. influenzae are Sensitive to its Prevalence
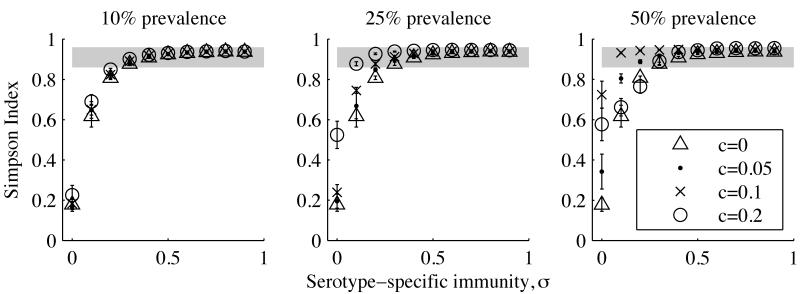
Depending on its prevalence, H. influenzae could exert large or inconsequential effects on the dynamics of pneumococcal serotypes. When colonizing only 10% of young children, H. influenzae had no significant effect on pneumococcal serotype diversity for the values of c tested (fig. 1A). This diversity is measured by the Simpson Index, which equals the probability of randomly picking two different serotypes from the same population with replacement; it was calculated as 1 – D, where , and pi denotes the frequency of the ith serotype. At H. influenzae carriage rates of 25%, maximal clearance rates of c = 0.1 and c = 0.2 were associated with a large increase in the Simpson Index at low levels of serotype-specific immunity, σ (fig. 1B). At carriage rates of 50%, a 10% clearance probability of the fittest serotype in the presence of H. influenzae (c = 0.1) could produce realistic values of the Simpson Index starting at very low serotype-specific immunity (σ = 0.1; fig. 1C). However, a larger maximal clearance probability (c = 0.2) at the same prevalence dramatically altered the outcome of competition. Rather than predominating, the serotypes with the longest durations of carriage went extinct, and the most frequent serotypes were the ones best able to resist clearance in the presence of H. influenzae (fig. B2). This reversal in rank order has not, to our knowledge, been reported in natural populations (Cobey and Lipsitch 2012).
Figure 1.
Pneumococcal serotype diversity, as measured by the Simpson Index, for different levels of serotype-specific immunity σ, prevalences of H. influenzae, and maximal clearance rates c. Measures were obtained by averaging annual estimates of the Simpson Index for each of the last 20 y of each simulation. Gray areas denote the range observed in carriage studies (Cobey and Lipsitch 2012). Error bars show SD from 10 simulations.
The rank-frequency distributions provide additional evidence that H. influenzae can profoundly alter the outcome of pneumococcal competition, but not in a way consistent with observations (figs. B3-B5). For many combinations of c and σ, simulations generate realistic values of the Simpson Index (figs. 1, B3, B4), but the rank-frequency distributions are significantly flatter than observed. There is no combination of c and σ at which H. influenzae can significantly increase diversity and maintain a realistic rank-frequency distribution across a wide range of prevalences. Because the maximal rate of clearance c and strength of serotype-specific immunity σ should not vary between populations, the rank-frequency distributions suggest that H. influenzae is not the primary driver of serotype coexistence.
The presence of H. influenzae can nonetheless promote the diversity of serotypes that would otherwise be unable to coexist without higher levels of serotype-specific immunity, σ. A relevant question is whether, in a population where H. influenzae did in fact play such a role, a significant interaction would be detectable in a cross-sectional, observational study of reasonable size.
Detection from Cross-sectional Data may be Impractical
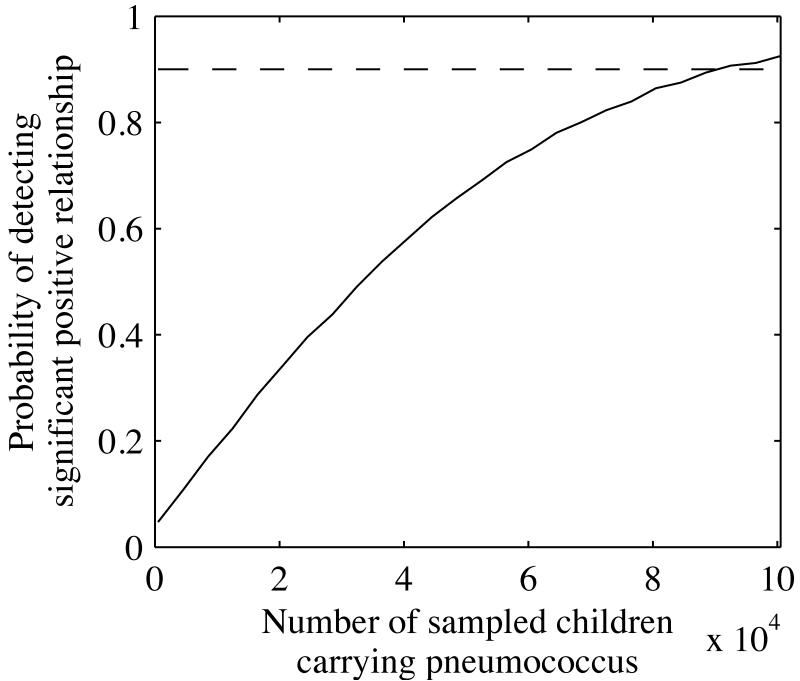
Through repeated simulations, we calculated that even when H. influenzae exerts substantial selective pressure on serotypes (c = 0.2, σ = 0.1, and 25% prevalence of H. influenzae), the probability that a child <5 years old who was colonized with serotype z also carried H. influenzae ranged only 2% between the serotypes: the probability of cocolonization with H. influenzae was ~24.5% for the commonest serotype and ~26.5% for the rarest. We used these probabilities to simulate binomial random draws for different sample sizes and calculated the probability of inferring, via logistic regression, a significant positive relationship between serotype rank (which correlates linearly with resistance to clearance in the presence of H. influenzae) and the probability of being co-colonized with H. influenzae (fig. 2). To have a 90% chance of detecting a significant positive relationship would require a sample size of ~90,000 children; sample sizes of several thousand pneumococcus-positive children, exceeding the size of published studies, would provide less than a 10% chance of detecting a true trend. This analysis suggests that epidemiological studies of co-colonization might well find null results, even if another species such as H. influenzae is a significant force promoting serotype diversity.
Figure 2.
Probability of detecting a significant positive trend between serotype rank and the rate of co-colonization with H. influenzae for different sample sizes when H. influenzae significantly increases pneumococcal diversity (c = 0.2, H. influenzae prevalence = 25%, σ = 0.1). Serotypes are ranked in descending order of their frequency (or intrinsic duration of carriage), with rank 1 denoting the most common serotype and 25 the rarest. The sample size refers to the number of children <5 y old colonized with pneumococcus. The dashed line shows a 90% probability of detecting a significant positive trend by logistic regression.
Discussion
We previously found that some serotype-specific immunity (σ ~0.3 or higher) was needed to match observed levels of pneumococcal diversity (Cobey and Lipsitch 2012). However, adaptation to different immune environments might explain pneumococcal diversity without the need to invoke serotype-specific immunity. This study was designed to determine whether a tradeoff in the ability of pneumococcal serotypes to persist in the presence versus the absence of H. influenzae might promote the coexistence of serotypes and if such an interaction might be epidemiologically detectable from cross-sectional studies. We found that H. influenzae could be a force that strongly promotes coexistence, but that it is not a plausible driver of observed pneumococcal serotype diversity. The extent to which H. influenzae increased diversity was sensitive to its prevalence and the maximal clearance rate c: there was no situation (combination of c and σ) in which H. influenzae could substantially increase diversity while maintaining realistic rank-frequency distributions and rank orders over a range of prevalences (10%-50%). Therefore, when c is high, variations in the prevalence of H. influenzae have large consequences for serotype coexistence. If H. influenzae is an important mediator of pneumococcal diversity in nature, the observed range in prevalence of H. influenzae (fig. B1) thus implies there should be more variability in pneumococcal rank-frequency distributions and rank orders than what is actually observed.
This study’s second finding is that even if the range of carriage rates of H. influenzae for a given prevalence of pneumococcus were narrower in real populations and the hypothesis thereby plausible, an interaction would be hard to detect epidemiologically by studying patterns of association between H. influenzae and individual pneumococcal serotypes in carriage. The patterns are subtle and require very large sample sizes to assess statistically.
While arguing against a primary role for an H. influenzae-induced shift in apparent competition for explaining pneumococcal serotype coexistence, our results more generally suggest a need to consider hypotheses invoking shifts in apparent competition, potentially induced by other species, to explain intraspecific pathogen diversity. They also point to the need for multiple lines of evidence to detect what might be small differences in pathogens’ (or other microbial commensals’) adaptations to different immunological environments.
This analysis underscores the challenges of relying on cross-sectional observations of many pathogens to infer what amount to subtle differences between them. Here, the question was not whether pneumococcal serotypes compete intraspecifically but whether the terms of this competition might vary in different environments. A system containing more competing species will generally suffer from smaller sample sizes (the number of hosts carrying each kind of parasite), hindering quantitative inference. This may partly explain why studies reporting correlations have involved fewer species (Behnke 2008; Lello et al. 2004). Pathogen association studies in general can be limited by the presence of hidden confounding factors, such as shared risks for pathogen acquisition, and spurious associations generated by the system’s nonlinear dynamics (Behnke et al. 2005; Fenton et al. 2010; Karvonen et al. 2009; Lello et al. 2008). In our simulations, the age-specific hazards were extremely similar for all serotypes and H. influenzae (results not shown), interspecific interactions were instantaneous (i.e., occurred only during co-colonization), and the dynamics reached apparently stable point equilibria, but such assumptions might not be easily extended to other multi-pathogen systems. An alternative approach to measuring the interaction here might involve looking across populations for a positive correlation between serotype evenness and the prevalence of H. influenzae at a given carriage rate of pneumococcus. However, this analysis would require many replicates and adjustment for temporal trends, such as pneumococcal vaccination rates, and could be more difficult than a large cross-sectional study. Approaches based on fitting models to time series data (e.g., Shrestha et al. 2011), especially when they contain perturbations and parallel measures of immune function, might be more revealing. Our findings also emphasize the utility of controlled experiments to identify pairwise interactions.
Like other ecological models, ours makes simplifying assumptions in the presence of incomplete information, and these assumptions should be revisited as more is learned. H. influenzae might interact with pneumococci in ways not examined here. The model assumes a smooth tradeoff, i.e., h(z) changes linearly with the intrinsic duration of carriage. First, resistance to opsonic phagocytosis might be further mediated by factors other than the capsule, which could make this relationship noisy and even harder to detect (Hyams et al. 2011). Second, some serotypes might be particularly well or poorly adapted to cocolonization with H. influenzae. For such serotypes, it might be possible to detect sharper effects in smaller sample sizes. A mechanistic understanding of serotypes’ interactions with the immune system could lead the way toward simpler yet more predictive models. For example, is there a positive correlation between a serotype’s propensity for opsonic phagocytosis (h(z)) and its susceptibility to acquired specific immunity (σ)? Such a relationship might exist if antibody and complement responses were both driven by capsular surface area. It is also possible that strains of H. influenzae differ in their effects on opsonic phagocytosis. Ignoring these differences would make a tradeoff even more difficult to detect. Our claim that certain regimes of parameter space yielded dynamics that were unrealistically sensitive to H. influenzae warrants careful testing, as suggested above: are increases in carriage of H. influenzae ever associated with a transient rise in serotype evenness? Further simultaneous surveillance of both H. influenzae and pneumococcal serotypes over multiple locations and time points would be helpful to this end.
The challenges posed by these two species suggest hazards for more complex models of commensal microbial communities that interact with host immunity, as occurs in the gut and respiratory tract. There is a shortage of basic information about the biology of these systems, including which direct interactions are present and on what aspects of phenotype they are based. These uncertainties limit the scope of hypotheses that can be tested. For example, the tradeoff described here might be a general phenomenon of Gram-negative bacteria, not just H. influenzae (Lysenko et al. 2010). Another Gram-negative species, Moraxella catarrhalis, is also a frequent colonizer of the upper respiratory tract, but few epidemiological studies have examined its co-occurrence with H. influenzae and individual pneumococcal serotypes. We know of no experiments investigating the interaction of M. catarrhalis and individual pneumococcal serotypes in mice or experiments that compare the immune environments induced by each Gram-negative species. This uncertain phenotypic resolution (e.g., whether increased opsonic phagocytosis comes from H. influenzae in particular or Gram-negative bacteria in general) pervades research on commensal microbes. Many studies of the human microbiome identify taxonomic units based on 16S RNA, ignoring differences in antigenic phenotype, and effectively omit the consumer (immune) populations. These kinds of data are necessary to develop and test model structure at a rudimentary level, i.e., to infer who is interacting directly with whom and how.
A more accurate picture of these communities can be acquired incrementally and iteratively through a combination of experiments, observation, and modeling, but our results support the idea that there may be practical limits to the measurement of indirect interactions in unmanipulated, species-rich communities. Traditional experiments to demonstrate the presence of indirect effects have focused on small motifs of a few species (Wootton 1994). We show that cross-sectional, correlational analyses may be underpowered in detecting differences in the strength of apparent competition in different environments, even when the interaction of interest has a large effect on diversity.
Host-symbiont communities, including the subset of interactions between hosts and pathogens, provide a challenging context for understanding species coexistence. The typical immune response includes components ranging from the nonspecific to the highly specific, and many of these components are unobserved. The presence of less specific components presents an opportunity for apparent competition between different species, and the amount of competition should be influenced by the persistence and functional responses of their immunological predators. Much of the variation observed in pathogens might result from adaptation to different immunological environments, which are continuously shaped by the dynamics of other symbionts (Seppala et al. 2009). Although this study suggests that H. influenzae does not play a major role in indirectly regulating the coexistence of pneumococcal serotypes, we have shown that pathogen diversity can in theory be strongly affected by a weak tradeoff in escaping different kinds of immunity. Detecting this tradeoff only from correlations in natural carriage might be extremely difficult and potentially misleading (Fenton et al. 2010; Holt 1984; Holt and Lawton 1993). Experiments in animal models, longitudinal coinfection studies augmented by immunological data, and interventions such as vaccines that target specific pathogens or strains could help to identify and, critically, quantify the strengths of direct and indirect effects (Behnke 2008; Bradley and Jackson 2008; Telfer et al. 2010).
Supplementary Material
Acknowledgements
We thank Bill Hanage, Jennie Lavine, Justin O’Hagan, and an anonymous reviewer for helpful comments. The computations in this paper were run on the Odyssey cluster supported by the FAS Science Division Research Computing Group at Harvard University. The project described was supported by award numbers 5R01AI048935 from the National Institute of Allergy and Infectious Diseases (NIAID) and 1F32GM97997 and U54GM088558 from the National Institute of General Medical Sciences (NIGMS). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIAID, NIGMS, or the National Institutes of Health. M.L. has received consulting fees or honoraria from Pfizer, Novartis, AIR Worldwide, and the Avian/Pandemic Flu Registry (Outcome Sciences), supported by Roche.
Appendix A
Table A1.
Studies were cross-sectional or obtained isolates over a period of less than one year. Studies were excluded if they did not use nasopharyngeal swabs or if the sampled population was chosen based on a suspected or confirmed preexisting condition (e.g., acute otitis media, respiratory illness, or hospitalization). DCC = day-care center; Abor. = Aboriginal population; orph. = orphanage.
| Study | Age range | Prevalence or percent H. influenzae |
positive samples S. pneumoniae |
|---|---|---|---|
| Abdullahi et al. 2008 | 0-4 y | 26 | 57 |
| Almeida Vde et al. 2011 | 2-59 mos | 32 | 58 |
| Aniansson et al. 1992 | 2 mos | 5 | 12 |
| 6 mos | 6 | 30 | |
| 10 mos | 13 | 32 | |
| 18 mos | 24 | 32 | |
| Bakir et al. 2001 | 0-10 y | 22 | 8 |
| Capeding et al. 1995 | 6-8 wk | 21 | 28 |
| 10-12 wk | 30 | 40 | |
| 14-17 wk | 39 | 47 | |
| 18-22 wk | 34 | 54 | |
| 32-39 wk | 50 | 60 | |
| 46-65 wk | 41 | 54 | |
| Chavanet et al. 2011 | 3 mos – 3 y | 55 | 48 |
| Christenson et al. 1997 | ≤ 2 y | 45 | 60 |
| (estimated from fig. 2) | 3 y | 41 | 40 |
| 4 y | 30 | 40 | |
| 5 y | 30 | 25 | |
| 6 y | 15 | 24 | |
| 7 y | 6 | 21 | |
| Dalle et al. 2000 | 3 mos – 2 y | 33 | 48 |
| De Lencastre et al. 1999 | 6 mos – 6 y | 72 | 46 |
| Dunais et al. 2003 | 6-36 mos | 24 (non-DCC) 37 (DCC) |
34 (non-DCC) 55 (DCC) |
| Factor et al. 2005 | 2-5 mos | 45 | 49 |
| 6-11 mos | 59 | 64 | |
| 12-23 mos | 60 | 66 | |
| 24-35 mos | 60 | 62 | |
| 36-47 mos | 58 | 58 | |
| 48-69 mos | 54 | 51 | |
| Gessner et al. 1998, | <2 y | 32 | 48 |
| Soewignjo et al. 2001 | |||
| Greenberg et al. 2006 | 1-59 mos | 71 | 67 |
| Gunnarsson and Ekdahl 1998 | 0-7 y | 13 | 19 |
| Homoe et al. 1996 | 0-1 y | 77 | 38 |
| 3-4 y | 93 | 27 | |
| 5-8 y | 70 | 23 | |
| Huang et al. 2005 | 0-7 y | 17 (in 2001) 15 (in 2004) |
26 (in 2001) 23 (in 2004) |
| Ito et al. 2002 | DCC and 18- month-olds |
56 | 58 |
| Jacoby et al. 2011 | <2 y | 41 (Abor.) 11 (non-Abor.) |
49 (Abor.) 25 (non-Abor.) |
| Jain et al. 2005 | 5-10 y | 42 | 53 |
| Janapatla et al. 2011 | ≤1 y | 3 | 19 |
| 2 y | 6 | 28 | |
| 3 y | 7 | 29 | |
| 4 y | 2 | 16 | |
| 5 y | 12 | 20 | |
| overall | 5 | 23 | |
| Jourdain et al. 2011 | 3-6 y | 61 | 43 |
| Kwambana et al. 2011 | <1 y | 70 | 78 |
| Lo et al. 2003, Wang et al. 2008 | ≤ 5 y | 5 | 27 |
| Mackenzie et al. 2010 | 2-15 y | 61 (in 2002) 53 (in 2004) |
70 (in 2002) 65 (in 2004) |
| Madhi et al. 2007 | 5.7 y +/− 0.7 y | 52 | 53 |
| Marchisio et al. 2001 | 1-7 y | 13 (fall) 18 (spring) |
4 (fall) 5 (spring) |
| Marcus and van Dyk 1996 | 0-8 y | 20 | 41 |
| Mastro et al. 1993 | 2-59 mos | 37 | 62 |
| Masuda et al. 2002 | 1 mo. – 5 y | 53 | 60 |
| Moulin et al. 1999 | 6-30 mos | 45 | 57 |
| Naaber et al. 2000 | 2-7 y | 17 | 46 |
| Peerbooms et al. 2002 | 3-36 mos | 37 (DCC) 11 (control) |
58 (DCC) 37 (control) |
| Prymula et al. 2009 | 15-18 mos | 18 | 22 |
| Sa-Leao et al. 2008 | 14-37 mos | 87 (mean) | 61 (mean) |
| Stubbs et al. 2005 | 3-7 y (Abor.) | 80 | 90 |
| ≤4 y (non- Abor. DCC) |
41 | 43 | |
| Sulikowska et al. 2004 | 6 mos – 5 y | 24 (DCC, winter) 58 (orph., winter) 11 (home, winter) 50 (DCC, spring) 70 (orph., spring) 3 (home, spring) |
57 (DCC, winter) 63 (orph., winter) 26 (home, winter) 54 (DCC, spring) 51 (orph., spring) 19 (home, spring) |
| Sung et al. 1995 | 2 mos – 5 y | 6 (Chinese) 67 (Vietnamese) |
10 (Chinese) 56 (Vietnamese) |
| Torun et al. 2009 | 6-10 y | 32 | 29 |
| van Gils et al. 2011 | 6 wks | 15 | 16 |
| estimated from Figure 1 | 6 mos | 35 | 48 |
| 12 mos | 43 | 66 | |
| 18 mos | 57 | 66 | |
| 24 mos | 52 | 64 | |
| Villasusa Paez et al. 2006 | 0-6 y | 55 | 78 |
| Vives et al. 1997 | 1 mo. | 4 | 3 |
| 12 mos. | 10 | 19 | |
| Wolf et al. 1999 | 4 mos – 5 y | 41 (Brazil) 42 (Angola) 70 (Netherlands) |
19 (Brazil) 35 (Angola) 41 (Netherlands) |
| Zemlickova et al. 2006 | 3-6 y | 25 | 38 |
Footnotes
Online manuscript elements: Appendix B (figures B1-B5)
Literature cited
- Abdullahi O, Nyiro J, Lewa P, Slack M, Scott JA. The descriptive epidemiology of Streptococcus pneumoniae and Haemophilus influenzae nasopharyngeal carriage in children and adults in Kilifi district, Kenya. Pediatr Infect Dis J. 2008;27:59–64. doi: 10.1097/INF.0b013e31814da70c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Raddad LJ, Patnaik P, Kublin JG. Dual infection with HIV and malaria fuels the spread of both diseases in sub-Saharan Africa. Science. 2006;314:1603–1606. doi: 10.1126/science.1132338. [DOI] [PubMed] [Google Scholar]
- Almeida Vde C, Negrini BV, Cervi MC, Isaac Mde L, Mussi-Pinhata MM. Pneumococcal nasopharyngeal carriage among infants born to human immunodeficiency virus-infected mothers immunized with pneumococcal polysaccharide vaccine during gestation. Pediatr Infect Dis J. 2011;30:466–470. doi: 10.1097/INF.0b013e31820a1ec6. [DOI] [PubMed] [Google Scholar]
- Anderson RM, May RM. Regulation and stability of host-parasite population interactions. I. Regulatory processes. Journal of Animal Ecology. 1978;47 [Google Scholar]
- Andreasen V. Dynamics of annual influenza A epidemics with immuno-selection. Journal of Mathematical Biology. 2003;46:504–536. doi: 10.1007/s00285-002-0186-2. [DOI] [PubMed] [Google Scholar]
- Aniansson G, Alm B, Andersson B, Larsson P, Nylen O, Peterson H, Rigner P, et al. Nasopharyngeal colonization during the first year of life. J Infect Dis. 1992;165(Suppl 1):S38–42. doi: 10.1093/infdis/165-supplement_1-s38. [DOI] [PubMed] [Google Scholar]
- Bakir M, Yagci A, Ulger N, Akbenlioglu C, Ilki A, Soyletir G. Asymtomatic carriage of Neisseria meningitidis and Neisseria lactamica in relation to Streptococcus pneumoniae and Haemophilus influenzae colonization in healthy children: apropos of 1400 children sampled. Eur J Epidemiol. 2001;17:1015–1018. doi: 10.1023/a:1020021109462. [DOI] [PubMed] [Google Scholar]
- Balmer O, Stearns SC, Schotzau A, Brun R. Intraspecific competition between co-infecting parasite strains enhances host survival in African trypanosomes. Ecology. 2009;90:3367–3378. doi: 10.1890/08-2291.1. [DOI] [PubMed] [Google Scholar]
- Behnke JM. Structure in parasite component communities in wild rodents: predictability, stability, associations and interactions … or pure randomness? Parasitology. 2008;135:751–766. doi: 10.1017/S0031182008000334. [DOI] [PubMed] [Google Scholar]
- Behnke JM, Gilbert FS, Abu-madi MA, Lewis JW. Do the helminth parasites of wood mice interact? Journal of Animal Ecology. 2005;74:982–993. [Google Scholar]
- Bonsall MB, Hassell MP. Apparent competition structures ecological assemblages. Nature. 1997;3888:371–373. [Google Scholar]
- Bonsall MB, Hassell MP. The population dynamics of apparent competition in a host-parasitoid assemblage. Journal of Animal Ecology. 1998;67:918–929. doi: 10.1046/j.1365-2656.1998.6760918.x. [DOI] [PubMed] [Google Scholar]
- Bottomley C, Isham V, Basanez MG. Population biology of multispecies helminth infection: interspecific interactions and parasite distribution. Parasitology. 2005;131:417–433. doi: 10.1017/s0031182005007791. [DOI] [PubMed] [Google Scholar]
- Bowers RG, Turner J. Community structure and the interplay between interspecific infection and competition. J Theor Biol. 1997;187:95–109. doi: 10.1006/jtbi.1997.0418. [DOI] [PubMed] [Google Scholar]
- Bradley JE, Jackson JA. Measuring immune system variation to help understand host-pathogen community dynamics. Parasitology. 2008;135:807–823. doi: 10.1017/S0031182008000322. [DOI] [PubMed] [Google Scholar]
- Brown SP, Le Chat L, Taddei F. Evolution of virulence: triggering host inflammation allows invading pathogens to exclude competitors. Ecol Lett. 2008;11:44–51. doi: 10.1111/j.1461-0248.2007.01125.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckee CO, Gupta S. Modelling malaria population structure and its implications for control. Adv Exp Med Biol. 2010;673:112–126. doi: 10.1007/978-1-4419-6064-1_8. [DOI] [PubMed] [Google Scholar]
- Byrne CJ, Holland CV, Kennedy CR, Poole WR. Interspecific interactions between Acanthocephala in the intestine of brown trout: are they more frequent in Ireland? Parasitology. 2003;127:399–409. doi: 10.1017/s0031182003003846. [DOI] [PubMed] [Google Scholar]
- Capeding MR, Nohynek H, Sombrero LT, Pascual LG, Sunico ES, Esparar GA, Esko E, et al. Evaluation of sampling sites for detection of upper respiratory tract carriage of Streptococcus pneumoniae and Haemophilus influenzae among healthy Filipino infants. J Clin Microbiol. 1995;33:3077–3079. doi: 10.1128/jcm.33.11.3077-3079.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaneton EJ, Bonsall MB. Enemy-mediated apparent competition: empirical patterns and the evidence. Oikos. 2000;88:380–394. [Google Scholar]
- Chavanet P, Atale A, Mahy S, Neuwirth C, Varon E, Dabernat H, Portier H. [Nasopharyngeal carriage, antibiotic susceptibility and serotyping of Streptococcus pneumoniae and Haemophilus influenzae in children attending day care centers] Med Mal Infect. 2011;41:307–317. doi: 10.1016/j.medmal.2011.02.001. [DOI] [PubMed] [Google Scholar]
- Christenson B, Sylvan SP, Noreen B. Carriage of multiresistant Streptococcus pneumoniae among children attending day-care centres in the Stockholm area. Scand J Infect Dis. 1997;29:555–558. doi: 10.3109/00365549709035893. [DOI] [PubMed] [Google Scholar]
- Cobey S, Lipsitch M. Niche and neutral effects of acquired immunity permit coexistence of pneumococcal serotypes. Science. 2012 doi: 10.1126/science.1215947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings DA, Iamsirithaworn S, Lessler JT, McDermott A, Prasanthong R, Nisalak A, Jarman RG, et al. The impact of the demographic transition on dengue in Thailand: insights from a statistical analysis and mathematical modeling. PLoS Med. 2009;6:e1000139. doi: 10.1371/journal.pmed.1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalle F, Leroy J, Pechinot A, Neuwirth C, Dupont MJ, Plesiat P, Estavoyer JM, et al. Nasopharyngeal carriage of Streptococcus pneumoniae and Haemophilus influenzae, and antibiotic susceptibilities in children attending day-care centers. Medecine et Maladies Infectieuses. 2000;30:510–514. [Google Scholar]
- De Lencastre H, Kristinsson KG, Brito-Avo A, Sanches IS, Sa-Leao R, Saldanha J, Sigvaldadottir E, et al. Carriage of respiratory tract pathogens and molecular epidemiology of Streptococcus pneumoniae colonization in healthy children attending day care centers in Lisbon, Portugal. Microb Drug Resist. 1999;5:19–29. doi: 10.1089/mdr.1999.5.19. [DOI] [PubMed] [Google Scholar]
- Dunais B, Pradier C, Carsenti H, Sabah M, Mancini G, Fontas E, Dellamonica P. Influence of child care on nasopharyngeal carriage of Streptococcus pneumoniae and Haemophilus influenzae. Pediatr Infect Dis J. 2003;22:589–592. doi: 10.1097/01.inf.0000073203.88387.eb. [DOI] [PubMed] [Google Scholar]
- Erasto P, Hoti F, Granat SM, Mia Z, Makela PH, Auranen K. Modelling multi-type transmission of pneumococcal carriage in Bangladeshi families. Epidemiol Infect. 2010;138:861–872. doi: 10.1017/S0950268809991415. [DOI] [PubMed] [Google Scholar]
- Ezenwa VO, Jolles AE. From host immunity to pathogen invasion: the effects of helminth coinfection on the dynamics of microparasites. Integr Comp Biol. 2011;51:540–551. doi: 10.1093/icb/icr058. [DOI] [PubMed] [Google Scholar]
- Factor SH, LaClaire L, Bronsdon M, Suleymanova F, Altynbaeva G, Kadirov BA, Shamieva U, et al. Streptococcus pneumoniae and Haemophilus influenzae type B Carriage, Central Asia. Emerg Infect Dis. 2005;11:1476–1479. doi: 10.3201/eid1109.040798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton A, S E. Perkins. Applying predator-prey theory to modelling immune-mediated, within-host interspecific parasite interactions. Parasitology. 2010;137:1027–1038. doi: 10.1017/S0031182009991788. [DOI] [PubMed] [Google Scholar]
- Fenton A, Viney ME, J Lello. Detecting interspecific macroparasite interactions from ecological data: patterns and process. Ecol Lett. 2010;13:606–615. doi: 10.1111/j.1461-0248.2010.01458.x. [DOI] [PubMed] [Google Scholar]
- Ferguson NM, Galvani AP, Bush RM. Ecological and immunological determinants of influenza evolution. Nature. 2003;422:428–433. doi: 10.1038/nature01509. [DOI] [PubMed] [Google Scholar]
- Gessner BD, Sutanto A, Steinhoff M, Soewignjo S, Widjaya A, Nelson C, Arjoso S. A population-based survey of Haemophilus influenzae type b nasopharyngeal carriage prevalence in Lombok Island, Indonesia. Pediatr Infect Dis J. 1998;17:S179–182. doi: 10.1097/00006454-199809001-00018. [DOI] [PubMed] [Google Scholar]
- Gog JR, Grenfell BT. Dynamics and selection of many-strain pathogens. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:17209–17214. doi: 10.1073/pnas.252512799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldblatt D, Hussain M, Andrews N, Ashton L, Virta C, Melegaro A, Pebody R, et al. Antibody responses to nasopharyngeal carriage of Streptococcus pneumoniae in adults: a longitudinal household study. J Infect Dis. 2005;192:387–393. doi: 10.1086/431524. [DOI] [PubMed] [Google Scholar]
- Graham AL. Ecological rules governing helminth-microparasite coinfection. Proc Natl Acad Sci U S A. 2008;105:566–570. doi: 10.1073/pnas.0707221105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray BM, Converse GM, 3rd, Dillon HC., Jr. Epidemiologic studies of Streptococcus pneumoniae in infants: acquisition, carriage, and infection during the first 24 months of life. J Infect Dis. 1980;142:923–933. doi: 10.1093/infdis/142.6.923. [DOI] [PubMed] [Google Scholar]
- Greenberg D, Givon-Lavi N, Broides A, Blancovich I, Peled N, Dagan R. The contribution of smoking and exposure to tobacco smoke to Streptococcus pneumoniae and Haemophilus influenzae carriage in children and their mothers. Clin Infect Dis. 2006;42:897–903. doi: 10.1086/500935. [DOI] [PubMed] [Google Scholar]
- Grosholz ED. Interactions of intraspecific, interspecific, and apparent competition with host-pathogen population dynamics. Ecology. 1992;73:507–514. [Google Scholar]
- Gunnarsson O, K Ekdahl. Previous respiratory tract infections and antibiotic consumption in children with long- and short-term carriage of penicillin-resistant Streptococcus pneumoniae. Epidemiol Infect. 1998;121:523–528. doi: 10.1017/s0950268898001599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Ferguson NM, Anderson RM. Chaos, persistence, and evolution of strain structure in antigenically diverse infectious agents. Science. 1998;280:912–915. doi: 10.1126/science.280.5365.912. [DOI] [PubMed] [Google Scholar]
- Hill PC, Cheung YB, Akisanya A, Sankareh K, Lahai G, Greenwood BM, Adegbola RA. Nasopharyngeal carriage of Streptococcus pneumoniae in Gambian infants: a longitudinal study. Clin Infect Dis. 2008;46:807–814. doi: 10.1086/528688. [DOI] [PubMed] [Google Scholar]
- Hogberg L, Geli P, Ringberg H, Melander E, Lipsitch M, Ekdahl K. Age- and serogroup-related differences in observed durations of nasopharyngeal carriage of penicillin-resistant pneumococci. J Clin Microbiol. 2007;45:948–952. doi: 10.1128/JCM.01913-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt R, Pickering J. Infectious disease and species coexistence: a model of Lotka-Volterra form. American Naturalist. 1985;126:196–211. [Google Scholar]
- Holt RD. Predation, apparent competition, and the structure of prey communities. Theor Popul Biol. 1977;12:197–129. doi: 10.1016/0040-5809(77)90042-9. [DOI] [PubMed] [Google Scholar]
- Holt RD. Spatial heterogeneity, indirect interactions, and the coexistence of prey species. The American Naturalist. 1984;124:377–406. doi: 10.1086/284280. [DOI] [PubMed] [Google Scholar]
- Holt RD, Dobson AP. Extending the principles of community ecology to address the epidemiology of host-pathogen communities. In: Collinge SK, Ray C, editors. Disease Ecology: Community Structure and Pathogen Dynamics. Oxford University Press; Oxford: 2006. [Google Scholar]
- Holt RD, Lawton JH. Apparent competition and enemy-free space in insect host-parasitoid communities. Am Nat. 1993;142:623–645. doi: 10.1086/285561. [DOI] [PubMed] [Google Scholar]
- Holt RD, Lawton JH. The ecological consequences of shared natural enemies. Annual Review of Ecological Systems. 1994;25:495–520. [Google Scholar]
- Homoe P, Prag J, Farholt S, Henrichsen J, Hornsleth A, Kilian M, Jensen JS. High rate of nasopharyngeal carriage of potential pathogens among children in Greenland: results of a clinical survey of middle-ear disease. Clin Infect Dis. 1996;23:1081–1090. doi: 10.1093/clinids/23.5.1081. [DOI] [PubMed] [Google Scholar]
- Huang SS, Platt R, Rifas-Shiman SL, Pelton SI, Goldmann D, Finkelstein JA. Post-PCV7 changes in colonizing pneumococcal serotypes in 16 Massachusetts communities, 2001 and 2004. Pediatrics. 2005;116:e408–413. doi: 10.1542/peds.2004-2338. [DOI] [PubMed] [Google Scholar]
- Hyams C, Opel S, Hanage W, Yuste J, Bax K, Henriques-Normark B, Spratt BG, et al. Effects of Streptococcus pneumoniae strain background on complement resistance. PLoS One. 2011;6:e24581. doi: 10.1371/journal.pone.0024581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyams C, Yuste J, Bax K, Camberlein E, Weiser JN, Brown JS. Streptococcus pneumoniae resistance to complement-mediated immunity is dependent on the capsular serotype. Infect Immun. 2010;78:716–725. doi: 10.1128/IAI.01056-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Ito K, Yoshizaki T, Nishimura T, Miwa T, Furukawa M. Nasopharyngeal penicillin-resistant Streptococcus pneumoniae strains among young children in Japan. Otol Neurotol. 2002;23:349–352. doi: 10.1097/00129492-200205000-00021. [DOI] [PubMed] [Google Scholar]
- Jacoby P, Carville KS, Hall G, Riley TV, Bowman J, Leach AJ, Lehmann D. Crowding and other strong predictors of upper respiratory tract carriage of otitis media-related bacteria in Australian Aboriginal and non-Aboriginal children. Pediatr Infect Dis J. 2011;30:480–485. doi: 10.1097/INF.0b013e318217dc6e. [DOI] [PubMed] [Google Scholar]
- Jain A, Kumar P, Awasthi S. High nasopharyngeal carriage of drug resistant Streptococcus pneumoniae and Haemophilus influenzae in North Indian schoolchildren. Trop Med Int Health. 2005;10:234–239. doi: 10.1111/j.1365-3156.2004.01379.x. [DOI] [PubMed] [Google Scholar]
- Janapatla RP, Chang HJ, Hsu MH, Hsieh YC, Lin TY, Chiu CH. Nasopharyngeal carriage of Streptococcus pneumoniae, Haemophilus influenzae, Moraxella catarrhalis, and Alloiococcus otitidis in young children in the era of pneumococcal immunization, Taiwan. Scand J Infect Dis. 2011;43:937–942. doi: 10.3109/00365548.2011.601754. [DOI] [PubMed] [Google Scholar]
- Jourdain S, Smeesters PR, Denis O, Dramaix M, Sputael V, Malaviolle X, Van Melderen L, et al. Differences in nasopharyngeal bacterial carriage in preschool children from different socio-economic origins. Clin Microbiol Infect. 2011;17:907–914. doi: 10.1111/j.1469-0691.2010.03410.x. [DOI] [PubMed] [Google Scholar]
- Karvonen A, Seppala O, Tellervo E. Host immunization shapes interspecific associations in trematode parasites. J Anim Ecol. 2009;78:945–952. doi: 10.1111/j.1365-2656.2009.01562.x. [DOI] [PubMed] [Google Scholar]
- Koelle K, Cobey S, Grenfell B, Pascual M. Epochal evolution shapes the phylodynamics of interpandemic influenza. Science. 2006;314:1898–1903. doi: 10.1126/science.1132745. [DOI] [PubMed] [Google Scholar]
- Kwambana BA, Barer MR, Bottomley C, Adegbola RA, Antonio M. Early acquisition and high nasopharyngeal co-colonisation by Streptococcus pneumoniae and three respiratory pathogens amongst Gambian new-borns and infants. BMC Infect Dis. 2011;11:175. doi: 10.1186/1471-2334-11-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lello J, Boag B, Fenton A, Stevenson IR, Hudson PJ. Competition and mutualism among the gut helminths of a mammalian host. Nature. 2004;428:840–844. doi: 10.1038/nature02490. [DOI] [PubMed] [Google Scholar]
- Lello J, Norman RA, Boag B, Hudson PJ, Fenton A. Pathogen interactions, population cycles, and phase shifts. Am Nat. 2008;171:176–182. doi: 10.1086/525257. [DOI] [PubMed] [Google Scholar]
- Leung TL, Poulin R. Interactions between parasites of the cockle Austrovenus stutchburyi: hitch-hikers, resident-cleaners, and habitat-facilitators. Parasitology. 2007;134:247–255. doi: 10.1017/S0031182006001478. [DOI] [PubMed] [Google Scholar]
- Lipsitch M, Abdullahi O, D’Amour A, Xie W, Weinberger D, Tchetgen Tchetgen E, Scott JAG. Estimating rates of carriage acquisition and clearance and competitive ability for pneumococcal serotypes in Kenya with a Markov transition model. Epidemiology. 2012;23:510–519. doi: 10.1097/EDE.0b013e31824f2f32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo WT, Wang CC, Yu CM, Chu ML. Rate of nasopharyngeal carriage, antimicrobial resistance and serotype of Streptococcus pneumoniae among children in northern Taiwan. J Microbiol Immunol Infect. 2003;36:175–181. [PubMed] [Google Scholar]
- Lysenko ES, Lijek RS, Brown SP, Weiser JN. Within-host competition drives selection for the capsule virulence determinant of Streptococcus pneumoniae. Curr Biol. 2010;20:1222–1226. doi: 10.1016/j.cub.2010.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysenko ES, Ratner AJ, Nelson AL, Weiser JN. The role of innate immune responses in the outcome of interspecies competition for colonization of mucosal surfaces. PLoS Pathog. 2005;1 doi: 10.1371/journal.ppat.0010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie GA, Leach AJ, Carapetis JR, Fisher J, Morris PS. Epidemiology of nasopharyngeal carriage of respiratory bacterial pathogens in children and adults: cross-sectional surveys in a population with high rates of pneumococcal disease. BMC Infect Dis. 2010;10:304. doi: 10.1186/1471-2334-10-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhi SA, Adrian P, Kuwanda L, Jassat W, Jones S, Little T, Soininen A, et al. Long-term immunogenicity and efficacy of a 9-valent conjugate pneumococcal vaccine in human immunodeficient virus infected and non-infected children in the absence of a booster dose of vaccine. Vaccine. 2007;25:2451–2457. doi: 10.1016/j.vaccine.2006.09.019. [DOI] [PubMed] [Google Scholar]
- Marchisio P, Gironi S, Esposito S, Schito GC, Mannelli S, Principi N. Seasonal variations in nasopharyngeal carriage of respiratory pathogens in healthy Italian children attending day-care centres or schools. J Med Microbiol. 2001;50:1095–1099. doi: 10.1099/0022-1317-50-12-1095. [DOI] [PubMed] [Google Scholar]
- Marcus L, van Dyk JC. Incidence of asymptomatic carriage of potentially pathogenic respiratory organisms among preschool Pretoria children. S Afr Med J. 1996;86:1132–1134. [PubMed] [Google Scholar]
- Margolis E, Yates A, Levin BR. The ecology of nasal colonization of Streptococcus pneumoniae, Haemophilus influenzae and Staphylococcus aureus: the role of competition and interactions with host’s immune response. BMC Microbiol. 2010;10:59. doi: 10.1186/1471-2180-10-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastro TD, Nomani NK, Ishaq Z, Ghafoor A, Shaukat NF, Esko E, Leinonen M, et al. Use of nasopharyngeal isolates of Streptococcus pneumoniae and Haemophilus influenzae from children in Pakistan for surveillance for antimicrobial resistance. Pediatr Infect Dis J. 1993;12:824–830. doi: 10.1097/00006454-199310000-00006. [DOI] [PubMed] [Google Scholar]
- Masuda K, Masuda R, Nishi J, Tokuda K, Yoshinaga M, Miyata K. Incidences of nasopharyngeal colonization of respiratory bacterial pathogens in Japanese children attending day-care centers. Pediatr Int. 2002;44:376–380. doi: 10.1046/j.1442-200x.2002.01587.x. [DOI] [PubMed] [Google Scholar]
- May RM, Anderson RM. Regulation and stability of host-parasite population interactions. II. Destabilizing processes. Journal of Animal Ecology. 1978;47:249–267. [Google Scholar]
- McCool TL, Weiser JN. Limited role of antibody in clearance of Streptococcus pneumoniae in a murine model of colonization. Infect Immun. 2004;72:5807–5813. doi: 10.1128/IAI.72.10.5807-5813.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mideo N. Parasite adaptations to within-host competition. Trends Parasitol. 2009;25:261–268. doi: 10.1016/j.pt.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Moulin F, Armand-Lefevre L, Le Thomas I, Corneau A, Raymond J, Gendrel D. Epidemiology of nasopharyngeal Streptococcus pneumoniae and Haemophilus influenzae colonization in children living in a group home. Archives de Pediatrie. 1999;6:620S–624S. doi: 10.1016/s0929-693x(99)80379-0. [DOI] [PubMed] [Google Scholar]
- Naaber P, Tamm E, Putsepp A, Koljalg S, Maimets M. Nasopharyngeal carriage and antibacterial susceptibility of Streptococcus pneumoniae, Haemophilus influenzae and Moraxella catarrhalis in Estonian children. Clin Microbiol Infect. 2000;6:675–677. doi: 10.1046/j.1469-0691.2000.00184.x. [DOI] [PubMed] [Google Scholar]
- Nagao Y, Koelle K. Decreases in dengue transmission may act to increase the incidence of dengue hemorrhagic fever. Proc Natl Acad Sci U S A. 2008;105:2238–2243. doi: 10.1073/pnas.0709029105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omori R, Adams B, Sasaki A. Coexistence conditions for strains of influenza with immune cross-reaction. J Theor Biol. 2010;262:48–57. doi: 10.1016/j.jtbi.2009.09.015. [DOI] [PubMed] [Google Scholar]
- Pedersen AB, Fenton A. Emphasizing the ecology in parasite community ecology. Trends Ecol Evol. 2007;22:133–139. doi: 10.1016/j.tree.2006.11.005. [DOI] [PubMed] [Google Scholar]
- Peerbooms PG, Engelen MN, Stokman DA, van Benthem BH, van Weert ML, Bruisten SM, van Belkum A, et al. Nasopharyngeal carriage of potential bacterial pathogens related to day care attendance, with special reference to the molecular epidemiology of Haemophilus influenzae. J Clin Microbiol. 2002;40:2832–2836. doi: 10.1128/JCM.40.8.2832-2836.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price PW, Westoby M, Rice B, Atsatt PR, Fritz RS, Thompson JN, Mobley K. Parasite mediation in ecological interactions. Annual Review of Ecology and Systematics. 1986;17:487–505. [Google Scholar]
- Prymula R, Kriz P, Kaliskova E, Pascal T, Poolman J, Schuerman L. Effect of vaccination with pneumococcal capsular polysaccharides conjugated to Haemophilus influenzae-derived protein D on nasopharyngeal carriage of Streptococcus pneumoniae and H. influenzae in children under 2 years of age. Vaccine. 2009;28:71–78. doi: 10.1016/j.vaccine.2009.09.113. [DOI] [PubMed] [Google Scholar]
- Recker M, Blyuss KB, Simmons CP, Hien TT, Wills B, Farrar J, Gupta S. Immunological serotype interactions and their effect on the epidemiological pattern of dengue. Proc Biol Sci. 2009;276:2541–2548. doi: 10.1098/rspb.2009.0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sa-Leao R, Nunes S, Brito-Avo A, Alves CR, Carrico JA, Saldanha J, Almeida JS, et al. High rates of transmission of and colonization by Streptococcus pneumoniae and Haemophilus influenzae within a day care center revealed in a longitudinal study. J Clin Microbiol. 2008;46:225–234. doi: 10.1128/JCM.01551-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seppala O, Karvonen A, Valtonen ET, Jokela J. Interactions among co-infecting parasite species: a mechanism maintaining genetic variation in parasites? Proc Biol Sci. 2009;276:691–697. doi: 10.1098/rspb.2008.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha S, King AA, Rohani P. Statistical inference for multi-pathogen systems. PLoS Comput Biol. 2011;7:e1002135. doi: 10.1371/journal.pcbi.1002135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soewignjo S, Gessner BD, Sutanto A, Steinhoff M, Prijanto M, Nelson C, Widjaya A, et al. Streptococcus pneumoniae nasopharyngeal carriage prevalence, serotype distribution, and resistance patterns among children on Lombok Island, Indonesia. Clin Infect Dis. 2001;32:1039–1043. doi: 10.1086/319605. [DOI] [PubMed] [Google Scholar]
- Stubbs E, Hare K, Wilson C, Morris P, Leach AJ. Streptococcus pneumoniae and noncapsular Haemophilus influenzae nasal carriage and hand contamination in children: a comparison of two populations at risk of otitis media. Pediatr Infect Dis J. 2005;24:423–428. doi: 10.1097/01.inf.0000160945.87356.ca. [DOI] [PubMed] [Google Scholar]
- Sulikowska A, Grzesiowski P, Sadowy E, Fiett J, Hryniewicz W. Characteristics of Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis isolated from the nasopharynges of asymptomatic children and molecular analysis of S. pneumoniae and H. influenzae strain replacement in the nasopharynx. J Clin Microbiol. 2004;42:3942–3949. doi: 10.1128/JCM.42.9.3942-3949.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung RY, Ling JM, Fung SM, Oppenheimer SJ, Crook DW, Lau JT, Cheng AF. Carriage of Haemophilus influenzae and Streptococcus pneumoniae in healthy Chinese and Vietnamese children in Hong Kong. Acta Paediatr. 1995;84:1262–1267. doi: 10.1111/j.1651-2227.1995.tb13545.x. [DOI] [PubMed] [Google Scholar]
- Telfer S, Birtles R, Bennett M, Lambin X, Paterson S, Begon M. Parasite interactions in natural populations: insights from longitudinal data. Parasitology. 2008;135:767–781. doi: 10.1017/S0031182008000395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telfer S, Lambin X, Birtles R, Beldomenico P, Burthe S, Paterson S, Begon M. Species interactions in a parasite community drive infection risk in a wildlife population. Science. 2010;330:243–246. doi: 10.1126/science.1190333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompkins DM, Draycott RAH, Hudson PJ. Field evidence for apparent competition mediated via the shared parasites of two gamebird species. Ecology Letters. 2000;3:10–14. [Google Scholar]
- Torun MM, Namal N, Demirci M, Bahar H. Nasopharyngeal carriage and antibiotic resistance of Haemophilus influenzae, Streptococcus pneumoniae and Moraxella catarrhalis in healthy school children in Turkey. Indian J Med Microbiol. 2009;27:86–88. [PubMed] [Google Scholar]
- van Gils EJ, Veenhoven RH, Rodenburg GD, Hak E, Sanders EA. Effect of 7-valent pneumococcal conjugate vaccine on nasopharyngeal carriage with Haemophilus influenzae and Moraxella catarrhalis in a randomized controlled trial. Vaccine. 2011;29:7595–7598. doi: 10.1016/j.vaccine.2011.08.049. [DOI] [PubMed] [Google Scholar]
- Villasusa Paez I, Martinez Motas I, Alvarez Garcia N. Prevalence of potentially pathogenic bacteria in the nasopharynx of healthy children attending a day care center in Havana City. Revista Cubana de Medicina Tropical. 2006;58 [PubMed] [Google Scholar]
- Vives M, Garcia ME, Saenz P, Mora MA, Mata L, Sabharwal H, Svanborg C. Nasopharyngeal colonization in Costa Rican children during the first year of life. Pediatr Infect Dis J. 1997;16:852–858. doi: 10.1097/00006454-199709000-00007. [DOI] [PubMed] [Google Scholar]
- Wang SR, Lo WT, Chou CY, Chen YY, Tsai SY, Chu ML, Wang CC. Low rate of nasopharyngeal carriage and high rate of ampicillin resistance for Haemophilus influenzae among healthy children younger than 5 years old in northern Taiwan. J Microbiol Immunol Infect. 2008;41:32–40. [PubMed] [Google Scholar]
- Weinberger DM, Dagan R, Givon-Lavi N, Regev-Yochay G, Malley R, Lipsitch M. Epidemiologic evidence for serotype-specific acquired immunity to pneumococcal carriage. J Infect Dis. 2008;197:1511–1518. doi: 10.1086/587941. [DOI] [PubMed] [Google Scholar]
- Weinberger DM, Trzcinski K, Lu YJ, Bogaert D, Brandes A, Galagan J, Anderson PW, et al. Pneumococcal capsular polysaccharide structure predicts serotype prevalence. PLoS Pathog. 2009;5:e1000476. doi: 10.1371/journal.ppat.1000476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf B, Gama A, Rey L, Fonseca W, Roord J, Fleer A, Verhoef J. Striking differences in the nasopharyngeal flora of healthy Angolan, Brazilian and Dutch children less than 5 years old. Ann Trop Paediatr. 1999;19:287–292. doi: 10.1080/02724939992383. [DOI] [PubMed] [Google Scholar]
- Wootton JT. The nature and consequences of indirect effects in ecological communities. Annu. Rev. Ecol. Syst. 1994;25:443–466. [Google Scholar]
- Zemlickova H, Urbaskova P, Adamkova V, Motlova J, Lebedova V, Prochazka B. Characteristics of Streptococcus pneumoniae, Haemophilus influenzae, Moraxella catarrhalis and Staphylococcus aureus isolated from the nasopharynx of healthy children attending day-care centres in the Czech Republic. Epidemiol Infect. 2006;134:1179–1187. doi: 10.1017/S0950268806006157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.