We report the characterization of three Ralstonia TAL-like effectors, which mediate DNA binding and can be used as customizable architectures for DNA targeting. We determined DNA-binding specificities of novel repeat variable di-residues (RVDs) and devised a repeat assembly approach for engineering Ralstonia solanacearum TALE-like proteins (RTLs).
Key words: Ralstonia solanacearum, genome engineering, TAL effectors, TALE activators and repressors, TALE nucleases (TALENs), targeted genome modifications
Abstract
Transcription activator-like effectors (TALEs) from Xanthomonas sp. have been used as customizable DNA-binding modules for genome-engineering applications. Ralstonia solanacearum TALE-like proteins (RTLs) exhibit similar structural features to TALEs, including a central DNA-binding domain composed of 35 amino acid-long repeats. Here, we characterize the RTLs and show that they localize in the plant cell nucleus, mediate DNA binding, and might function as transcriptional activators. RTLs have a unique DNA-binding architecture and are enriched in repeat variable di-residues (RVDs), which determine repeat DNA-binding specificities. We determined the DNA-binding specificities for the RVD sequences ND, HN, NP, and NT. The RVD ND mediates highly specific interactions with C nucleotide, HN interacts specifically with A and G nucleotides, and NP binds to C, A, and G nucleotides. Moreover, we developed a highly efficient repeat assembly approach for engineering RTL effectors. Taken together, our data demonstrate that RTLs are unique DNA-targeting modules that are excellent alternatives to be tailored to bind to user-selected DNA sequences for targeted genomic and epigenomic modifications. These findings will facilitate research concerning RTL molecular biology and RTL roles in the pathogenicity of Ralstonia spp.
INTRODUCTION
Molecular tools that can be used to achieve precise, site-specific modifications of genomes are valuable for many basic research and biotechnological applications in agriculture and medicine. Developing genome-editing tools for targeted modifications of the genome requires the development of customizable DNA-binding modules that can be easily engineered to bind to any user-defined genomic sequence. These DNA-binding modules can be fused to functional domains to generate chimeric proteins for the desired type of genomic modification (Bogdanove and Voytas, 2011; Carroll, 2011). Chimeric transcriptional regulators, methylases, and nucleases have been generated and used to modulate gene expression or epigenetic status, or to induce double-strand breaks, in order to edit the genome in many useful ways (Nomura and Barbas, 2007; Christian et al., 2010; Morbitzer et al., 2010; Geissler et al., 2011; Li et al., 2011a; Mahfouz et al., 2011, 2012). Zinc finger DNA-binding domains, for example, have been used as customizable DNA-binding modules to obtain a variety of site-specific genomic modifications (Miller et al., 2007; Bozas et al., 2009; Shukla et al., 2009; Zhang and Voytas, 2011). The production of zinc fingers that bind to a user-defined DNA sequence, however, is labor-intensive and time-consuming and suffers from inadequate reproducibility and adaptability (Ramirez et al., 2008).
Recently, a group of transcription activator-like effector (TALE) proteins that are secreted by the phytopathogenic bacteria Xanthomonas spp. have been shown to bind to DNA via a central DNA-binding domain (DBD) of tandem repeats (Romer et al., 2007). All TALEs share the same structural features, including an N-terminal secretion signal and a C-terminal acidic activation domain (AAD), a bipartite nuclear localization signal (NLS), and the central domain of tandem repeats. The repeats of the DBD are 33–35 amino acids long and are nearly identical but are polymorphic at positions 12 and 13, which constitute the repeat variable di-residue (RVD). Each repeat binds to one DNA nucleotide in the target sequence. The DNA-binding specificity of the RVDs of these repeats has been deciphered by experimental and computational approaches (Boch et al., 2009; Moscou and Bogdanove, 2009). The RVDs HD, NI, NK, and NG mediate the binding to C, A, G, and T nucleotides, respectively (Boch et al., 2009; Moscou and Bogdanove, 2009; Morbitzer et al., 2010). Moreover, there is some binding degeneracy within the code; for example, the RVD NN binds to A or G nucleotides and the RVD NS binds to any nucleotide. The degeneracy of these RVDs could be advantageous for targeting multiple genomic DNA sequences using a single effector protein. Several TALE proteins have been used to create a scaffold for DNA binding by engineering the sequence and order of the DBD repeats (Morbitzer et al., 2010; Li et al., 2011a; Mahfouz et al., 2011). Because of the modular nature of the TALEs as DNA-binding modules and the relative ease of engineering their DBDs to bind to virtually any user-selected DNA sequence, a suite of precise and efficient molecular genome-modification tools applicable to eukaryotic systems has been assembled (Cermak et al., 2011; Li et al., 2011b; Mahfouz and Li, 2011; Li et al., 2012). For example, TAL effectors have been used to generate artificial transcriptional activators in a variety of eukaryotic systems (Morbitzer et al., 2010). Moreover, TAL effectors fused to other functional domains such as nucleases or repressor domains have been shown to produce double-strand breaks or to inhibit transcription (Li et al., 2011a; Mahfouz et al., 2011, 2012; Zhang et al., 2011a). A number of repeat assembly approaches have been reported that utilize restriction ligation cloning from either plasmid libraries or PCR-generated multi-repeat fragments. These methods have been used to reproducibly generate functional, custom TALEs with user-defined DNA targets with much less labor and time than required for zinc finger assembly approaches (Cermak et al., 2011; Li et al., 2011b; Morbitzer et al., 2011; Weber et al., 2011; Zhang et al., 2011a; Li et al., 2012; Reyon et al., 2012). Thus, TALEs may represent the DNA-binding module of choice for developing genome-editing tools across eukaryotic systems.
Several bacterial species contain TALE-like proteins or proteins with TALE-like structural features, particularly the repeat domain region (Bogdanove et al., 2010), and one of these is Ralstonia solanacearum. Ralstonia solanacearum is a soil-borne, gram-negative, pathogenic bacterium that colonizes the xylem of more than 200 species of plants, causing bacterial wilt disease (Genin and Denny, 2012). Ralstonia solanacearum type III effectors are required for its pathogenicity on Arabidopsis and tomato plants (Remigi et al., 2011; Zhang et al., 2011b). DNA sequences homologous to the Xanthomonas AvrBs3 TAL effectors were detected in strains of the R. solanacearum species complex (Salanoubat et al., 2002; Heuer et al., 2007). The repeats are composed of 35 amino acids and are similar to the Hax2 TAL effector of Xanthomonas campestris pv. armoraciae. Not all Ralstonia species contain TAL effectors, which suggests that these effectors were acquired through horizontal gene transfer (HGT) (Heuer et al., 2007). The DNA sequences of the repeats are prone to recombination, which might enhance the adaptive responses of the pathogen to different hosts through interactions with disease or resistance host regulators. It is largely unknown whether Ralstonia TAL-like effectors function as genuine transcriptional regulators and contribute to the organism’s pathogenicity. The identification of novel TALE architectures and RVDs from Ralstonia will facilitate studying the biology of TALEs interactions with DNA and their roles in the pathogenicity. Moreover, these studies will facilitate the production of highly efficient TALE architectures for genome-engineering applications.
Here, we functionally characterize three Ralstonia TAL-like proteins (RTLs) as transcriptional activators and we determine the DNA-binding specificities for the RVD sequences ND, HN, NP, and NT. The RVD ND interacts with C nucleotides, HN interacts with A and G nucleotides, and NP interacts with C, A, and G nucleotides. Characterization of novel DNA-binding architectures and RVD DNA-binding specificities will provide us with great flexibility in the generation of specific targeting modules. Moreover, we demonstrate that the RTL proteins can be tailored to bind to user-selected DNA target sequences and can thereby be used to generate invaluable genome-engineering tools with a variety of potential biotechnological applications in agriculture and genetic medicine. Furthermore, our data represent a major step towards further molecular characterization of the DNA-binding activities of RTLs and their biological roles in the pathogenicity of Ralstonia spp.
RESULTS
Identification of Ralstonia TALE-Like Proteins (RTLs)
The R. solanacearum species complex possesses a high degree of genetic diversity and is divided into four phylotypes that reflect geographical distribution. The full genome sequence of seven strains, including the reference strain GMI1000, has been published (Salanoubat et al., 2002; Remenant et al., 2010) and genomic sequences of additional strains are accessible online. Because TALE proteins exhibit distinct features, including an N-terminal secretion signal, a C-terminal transcriptional activation domain, and an NLS and central DNA-binding repeat domain, the available Ralstonia proteomes were searched for proteins with features similar to those of TALE proteins. Because the presence of the central DNA-binding repeat domain is the most distinctive feature of the TALEs, a k-means-based algorithm was used to identify tandem repeats in Ralstonia proteomic sequences available in databases (Jorda and Kajava, 2009). After tandem repeats were filtered, 16 potential TALE-like effectors were identified in the proteomes of various strains of Ralstonia spp. (Table 1). Three proteins (CAQ18687.1, NP_519936.1, and YP_003750492.1) that appeared to be derived from full-length clones and to contain TALE-like structural features, including the N-terminus, the central DNA-binding repeats, and the C-terminus domain, were selected for further analysis. The general structural features of the RTL proteins and the alignment of the repeats, including the consensus among the three RTL proteins, are shown in Figure 1. The length of the N-terminus of these three proteins varies significantly, with YP_003750492.1 being the shortest (only 134 amino acids) and NP_519936.1 the longest (354 amino acids) (Supplemental Figure 1). With few exceptions, all the identified RTLs possess 35 amino acid-long repeats. Because those exceptions are probably due to sequencing errors of the highly repetitive fragments, repeats that were not 35 amino acids long were not included in downstream analysis.
Table 1.
Potential TALE-like Effectors in Ralstonia.
| Ralstonia strain | Number of tandem repeat units | Number of AA in tandem repeat unit | Accession |
|---|---|---|---|
| GMI1000 | 16, 16 | 35 | CAD15517.12,4, NP_519936.13,4 |
| PSI07 | 13, 13 | 35 | CBJ35887.14, YP_003750492.14 |
| MolK2 | 11, 15 | 35 | CAQ18305.11,2, CAQ18687.1 |
| Blood disease bacterium R229 | 16 | 35 | CCA82456.1 |
| UW360 | 13 | 35 | ABO27067.11 |
| GD5061311 | 12 | 35 | ABO27073.11 |
| GX5072306 | 11 | 35 | ABO27070.11 |
| ICPM11121 | 10 | 35 | ABO27069.11 |
| ZJ1993Bn1 | 7 | 35 | ABO27071.11 |
| GX5072605 | 7 | 35 | ABO27074.11 |
| FPJ16 | 4 | 35 | ABO27068.11 |
| FJ4071619 | 3 | 35 | ABO27072.11 |
| SD5072402 | 1 | 35 | ABO27075.11 |
1 Partial sequence; 2 probable; 3 hypothetical; 4 sequences identical.
Figure 1.
Structural Representation of RTL Effectors.
(A) Structural features present in studied RTL effectors. A typical RTL effector contains a central DNA-binding domain consisting of 35 amino acid-long repeats (yellow boxes), putative –1 and 0 repeats upstream and +1 and +2 half-repeats downstream the DNA-binding domain (pink and green boxes), an acidic activation domain (AD, turquoise box), and a translocation signal (TS, blue box). The conserved RTL repeat sequence that was used for RTL repeats assembly is also shown. The repeat variable di-residues (RVD) are shown in red.
(B) Alignment of RTL1 (YP_003750492.1), RTL2 (NP_519936.1), and RTL3 (CAQ18687.1) consensus sequence of repeats. The consensus sequence of repeats among the three proteins is shown at the bottom of the panel below the alignment. The RVD is highlighted in the red box.
(C) The frequency of the naturally occurring RVDs in RTL effectors.
The 12th and 13th amino acids have been extracted from all identified RTLs containing canonical tandem repeats and their total number have been normalized per protein.
Repeats with a high similarity to the canonical central repeats were identified in the upstream and downstream regions flanking the modular repeat domain (Figure 1). These repeats, however, do not display the well-characterized RVDs, either because the 13th amino acid is missing or because the 13th position contains an amino acid that has not been previously shown to bind to DNA or to mediate protein–DNA interactions (Supplemental Figure 2).
To determine the conservation level of the tandem repeats in the DBD region among all the RTLs, we performed a multiple sequence alignment using Jalview (Waterhouse et al., 2009). The [LSTEQVVAIASXXGGKPALEAVKALLLELRAAPYE] consensus repeat sequence is highly conserved among all the reported RTLs. Putative functional domains also were identified, including an NLS composed of short stretches of positively charged amino acids surrounding the modular repeat domain. Positively charged amino acids constitute a significant fraction of the sequence flanking the tandem repeat domain (Supplemental Figure 2).
Design, Synthesis, and Construction of RTL Proteins
To characterize the RTL effectors and analyze their molecular function, we designed three artificial RTLs based on the amino acid sequences of the YP_003750492.1, NP_519936.1, and CAQ18687.1 clones. Codon-optimized versions of the clones, herein referred to as RTL1 (YP_003750492.1), RTL2 (NP_519936.1), and RTL3 (CAQ18687.1), were generated (Supplemental Figure 3). Each RTL protein is composed of three major parts: the N-terminus, the C-terminus, and the central DNA-binding repeats. The backbone of each RTL effector was custom-synthesized as two DNA fragments that correspond to the N- and C-termini of the protein. The first fragment corresponds to the sequence of the N-terminus of the clone and was engineered to contain ApaI, NdeI, and SpeI (for RTL1) or SalI, NdeI, and SpeI (for RTL2 and RTL3) restriction-enzyme recognition sequences at the 3’ end. The second fragment sequence corresponds to the C-terminus of the clone and was flanked by NdeI and SpeI restriction-enzyme recognition sequences. For generation of the backbone clone (full-length minus the central repeat domain), the second fragment was digested with NdeI and SpeI and ligated into the first fragment, which had been digested with NdeI and SpeI; this produced the backbone clones RTL1∆R, RTL2∆R, and RTL3∆R.
Two assembly approaches were used to generate RTL central repeat fragments. First, a protocol for an ordered assembly of RTL di-repeats was devised. Six fragments of di-repeats (210 bp) were designed based on the sequence that is conserved among the RTL effector repeats (LSTEQVVAIASXXGGKPALEAVKALLLELRAAPYE). All the di-repeat DNA fragments were synthesized in the pUC19 minus MCS vector with the proper flanking sequences. Fragment 1 was digested with ApaI or SalI and BsmAI, fragments 2–5 were digested with BsmAI, and fragment 6 was digested with BsmAI and NdeI. The restricted fragments were gel-purified and ligated into an RTL∆R backbone clone digested with ApaI or SalI and NdeI. A library containing fragments 1–6 with each of the desired RVD combinations was generated to allow the RVDs to be assembled in the desired sequence and order. The details of the ordered assembly of the di-repeats are provided in Figure 2B and in the Supplementary Data.
Figure 2.
Design and Construction of RTL Effectors and the Ordered Assembly of Their Repeats.
(A) Each RTL backbone clone (RTL minus-repeats, RTL∆R) was custom-synthesized as an N-terminus fragment (NTF) and C-terminus fragment (CTF). The CTF was restricted with NdeI/SpeI and subcloned into the NTF clone to generate the RTL∆R clone. RTL∆R clones are flanked with L1 and L2 gateway recombination sites to allow cloning into desired destination vectors.
(B) Schematic representation of the ordered assembly of RTLs repeats showing the six restriction fragments and the overhang sequence of each fragment produced by a BsmAI restriction digestion.
For the second assembly approach, a PCR-based procedure was developed to clone RTL pre-assembled repeats into the Xanthomonas backbone, as described below. The central repeat fragments were PCR-amplified using forward and reverse primers that contain ApaI or SalI and NdeI restriction-enzyme recognition sequences. The fragments were then digested and ligated into the RTL∆R backbone clones to generate the full-length RTL DNA clones.
Determining whether RTL repeats (RRs) would recognize and bind to DNA in the Xanthomonas TAL backbone was essential. To study the DNA-binding specificities of the RRs in the Xanthomonas dHax3 backbone, we designed three types of constructs. The first construct contains part of the N-terminus of the dHax3 backbone and the first repeat, the second construct type represents a single repeat, and the third construct contains the last half repeat and the C-terminus of the dHax3 backbone. The repeats with identical or mixed RVD types in the dHax3 backbone can be assembled by sequential restriction digestion, concatamerization of the second single-repeat type, and ligation of these three types of fragments to generate the desired repeat number and RVD order (Supplemental Figure 1). Clones of dHax3 containing repeats of variable length (ranging from 2 to 15 repeats) were selected by restriction and Sanger sequence analysis. These repeat concatemer clones were generated for all desired RVDs of the RRs. Using the two repeat assembly approaches described above, we generated clones with the desired repeat number and RVD order in both the Xanthomonas and Ralstonia TAL backbones. It should be noted that all the backbone clones are flanked by gateway L1 and L2 sequences to allow LR gateway cloning in desired destination vectors.
RTLs Are Nuclear Proteins
As indicated in Figure 1, the general structural features of the RTL effectors are very similar to those of the Xanthomonas TALEs. In addition to a central DNA-binding region consisting of a variable number of nearly identical 35 amino acid repeats, RTL effector structural features also include multiple putative NLSs and an activation domain (AD) in the C-terminus. All of these features were previously shown to be essential for the functionality of TALEs, indicating that, if the RTLs are truly transcriptional activators, these features should be essential for the activity of the RTLs as well (Boch and Bonas, 2010). Because the functionality of the RTL NLS motif has not been experimentally verified, we determined whether RTL effectors possess a functional NLS and thereby localize to the cell nucleus. The RTL1, RTL2, and RTL3 clones fused to GFP were transiently expressed in Nicotiana benthamiana tobacco leaves. Two days post agroinfiltration, leaf discs were collected and the samples were analyzed by confocal laser scanning microscopy. The three RTL:GFP proteins localized to the nucleus, indicating the functionality of their NLS sequences (Figure 3). The nuclear localization of the RTLs suggests that these proteins may be transcriptional activators that modulate host gene expression for their own benefit.
Figure 3.
RTL Effector Proteins Localize in the Plant Cell Nucleus.
RTL1, RTL2, and RTL3 were fused in frame to green fluorescent protein, respectively, to generate RTL:GFP and were transiently expressed in tobacco leaves via Agrobacterium-mediated delivery. Two days post agroinfiltration, confocal laser scanning microscopy was used to analyze samples. 4′,6′-Diamidino-2-phenylindole (DAPI) staining indicates nuclei. Scale bars = 30 μm. RTL:GFP fusion proteins localize in the plant cell nucleus.
RRs Mediate DNA Binding in the Xanthomonas TALE Backbone
To determine whether the repeats of the RTL DBDs mediate DNA binding in the Xanthomonas TAL backbone, we engineered the RRs to include Xanthomonas RVD sequences with known DNA-binding specificities. Eleven identical RRs were cloned with the RVD sequence HD in the Xanthomonas dHax3 backbone to generate the dHax3.RR.HD12 clone. To determine whether the RRs containing the RVD HD mediate DNA binding and recognize the C nucleotide, we cloned all possible effector-binding elements (EBEs) (polyA, polyT, polyG, or polyC) into the Bs3 minimal promoter, which drives uidA expression (Supplementary Data). The clones of the dHax3.RR.HD12 effector were co-agroinfiltrated with each of the promoter clones of the EBEs (polyA, polyG, polyC, or polyT:Bs3:uidA) in Nicotiana benthamiana leaves. Two days after agroinfiltration, the leaf discs were collected for qualitative and quantitative analysis of the transcriptional activation of the GUS reporter. As indicated in Figure 4, dHax3.RR.HD12 bound to the polyC EBE and strongly activated the expression of the uidA reporter in vivo. These data demonstrate that the Ralstonia repeats mediate DNA binding and that the Xanthomonas RVD DNA-binding specificity code can be applied to the Ralstonia repeats. The ability of RRs to mediate DNA binding strongly suggests that RTL effector proteins may function as transcriptional activators that bind to their genomic targets to reprogram host gene expression to the benefit of the phytopathogen.
Figure 4.
dHax3.RR.HD12 DNA-Binding Activity and Efficiency.
Sequence of effector dHax3.RR.HD12 and T-preceded EBE boxes, polyA, G, T, and C, in minimal Bs3 promoter (mBs3) that drives uidA reporter gene expression. The constitutive 35S-driven dHax3.RR.HD12 clone was co-delivered, separately, with mBs3 containing polyA, G, T, or C EBE clone via A. tumefaciens into N. benthamiana leaves. dHax3.RR.HD12 alone and dHax3.RR.HD12 with mBs3 served as negative controls and dHax3 with mBs3.dHax3.EBE served as positive control. Leaf discs were collected 2 d post infiltration; half were stained with X-Gluc and half were used for quantitative measurement of GUS activities. The results show that the dHax3.RR.HD12 effector strongly activated the expression of mBs3polyC-driven uidA reporter. These experiments were repeated at least three times, with similar results. RR, Ralstonia repeat; EBE, DNA-binding element.
RTLs May Function as Transcriptional Activators
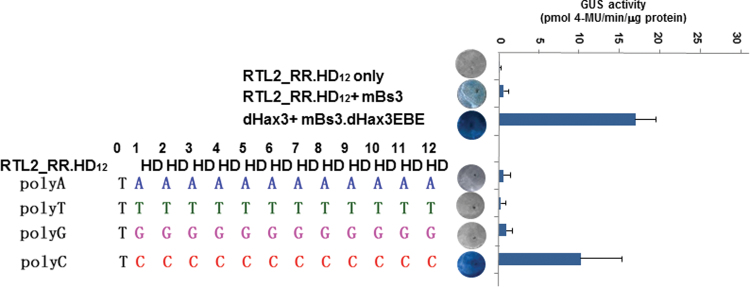
Because the RRs mediated DNA binding in the Xanthomonas dHax3 backbone, we determined whether RTL effectors can function as genuine transcriptional activators. An RTL protein (RTL2_RR.HD12) was engineered that contained the previously reported Xanthomonas HD RVD in the RTL backbone. The RTL effectors were separately co-infiltrated into Nicotiana benthamiana leaves with mBs3 clones that contained polyA, polyT, polyG, or polyC EBEs. Two days post infiltration, leaf discs were collected and qualitative and quantitative GUS assays were performed. As shown in Figure 5, the RTL2_RR.HD12 was able to bind to the promoter target sequence and activate the expression of the uidA gene. These data provide strong evidence that the RTLs may function as transcriptional activators and used by the Ralstonia phytopathogen to reprogram host transcription.
Figure 5.
RTLs May Function as Transcriptional Activators.
The effector clone of RTL2_RR.HD12 and clones of EBE boxes, T-preceded polyA, G, T, and C, in mBs3 promoter-driven uidA reporter were co-argoinfiltrated into N. benthamiana leaves, respectively. GUS activities were analyzed qualitatively and quantitatively 2 d post infiltration. RTL2_RR.HD12 alone and RTL2_RR.HD12 with mBs3 served as negative controls and dHax3 with mBs3.dHax3.EBE as positive control. RTL2_RR.HD12 strongly activated the expression of mBs3polyC-driven uidA reporter gene.
The RVD ND Interacts Specifically with Cytosine Nucleotides
RTLs possess unique RVDs, and the DNA-binding specificities of the equivalent RVDs from TAL effectors of the Xanthomonas phytopathogen have not been determined. Moreover, the frequency of certain repeats is much higher in the RTLs than in the TAL effectors of Xanthomonas (Figure 1C). To determine the DNA-binding specificity of the ND RVD, we generated a DBD that possesses 12 RRs containing the RVD ND in the dHax3 backbone (dHax3.RR.ND12). Two days after co-agroinfiltration of the effector with each of the polyC-, G-, T-, or A- EBE promoter clones, the leaf discs were analyzed by GUS assays as described above. The dHax3.RR.ND12 selectively and strongly activated the polyC EBE-containing promoter (Figure 6A), indicating that the RVD ND specifically recognizes and selectively binds to C nucleotides. To test whether the binding specificity of the other RVDs can be determined using the same approach, we co-expressed effectors containing poly(RVDs) (HN, NP, and NT) in the dHax3 backbone along with promoter clones that contain DNA-binding boxes of polyC, G, A, and T nucleotides. dHax3.RR.HN12 showed a weak binding and activation to A and G nucleotides (Figure 6C). Interestingly, dHax3.RR.HN12 RVD repeats more strongly bind to and activate a target EBE-containing polyAG nucleotides stronger than to a target EBE with only polyA or polyG nucleotides (Figure 6C). Moreover, dHax3.RR.NP12 RVD showed moderate interaction and activation of C, A, and G (Figure 6D), and dHax3.RR.NT12 RVD showed weak binding and activation of T, A, G and C (Figure 6E).
Figure 6.
Determining the DNA-Binding Specificities of Ralstonia RVDs.
The RVDs sequences of effectors, dHax3.RR.ND12, dHax3.RR.HN12, dHax3.RR.NP12, and dHax3.RR.NT12, and DNA sequences of EBE boxes in mBs3 or mBs4 promoter that drive uidA reporter expression are shown. Clones of the RTL effectors and EBE-containing mBs3 were co-argoinfiltrated into N. benthamiana leaves, respectively, as shown. dHax3.RR.ND12 strongly and specifically activated the expression of polyC, preceded by T, -mBs3-driven uidA reporter gene (A, B), but not polyCdT-Bs3 (B). dHax3.RR.HN12 showed a weak activation to A and G (C); dHax3.RR.NP12 showed moderate interaction and activation of C, A, and G (D); and dHax3.RR.NT12 showed weak binding and activation of T, A, G, and C (E).
To function, Xanthomonas TAL effectors require the presence of the T nucleotide preceding the DNA-binding box in the promoter region (Boch et al., 2009). We therefore determined whether the T nucleotide that corresponds to the cryptic repeat 0 (T0) is important for DNA binding and hence for transcriptional activation of RTL effectors. Several EBE DNA-binding boxes were generated, with or without (T0 is replaced by G) the T nucleotide, in Bs4 minimal promoters fused to the uidA reporter and were cloned into binary vectors for plant expression. These EBE promoter constructs were co-delivered into tobacco leaves with their respective RTLs via agroinfiltration (Figure 6B). RTL effectors with EBEs lacking the T0 nucleotides showed minimal transcriptional activation background as negative controls (Figure 6B). Thus, the T nucleotide that corresponds to repeat 0 is essential for RTL binding and transcriptional activation.
Determining the DNA-Binding Specificities of the RTL RVDs
The binding of the poly(RVD) appears to depend on the nature of the RVD. Thus, the use of RTLs containing poly(RVDs) may not be the best experimental approach to determine the DNA-binding specificities of RVDs. Consequently, RTL effectors were designed and assembled that contain mixed RVDs of known and unknown binding specificities. Following the same procedure that we used to determine the binding specificity of the RVD ND, we assembled an RTL2 effector with the RVD [HN-HN-ND-ND-HN-HN-ND-ND-HN-HN-ND-ND]. The possible EBEs for this assembly were fused to the Bs3 minimal promoter that was used to drive uidA expression. The RTL2_HN-ND12 effector was agroinfiltrated with each of the possible Bs3 promoters (Figure 7A). The RTL2_HN-ND12 effector induced GUS expression only in combination with the Bs3 promoter containing the [TAACCAACCAACC] or [TGGCCGGCCGGCC] EBE. The HN-type RVD therefore mediates specific binding for A and G nucleotides. To determine the binding specificity of the NP-type RVD, we generated a dHax3_NP-ND-NT-NG11 effector containing the RVD sequence [NP-ND-ND-ND-NP-NP-NP-NT-NP-NP-NG] and fused the possible EBEs for this assembly to the Bs3 minimal promoter. Our findings demonstrate that the NP-type RVD mediates DNA binding to C, A, and G nucleotides, when: ND binds to C, NG binds to T, and NT binds to T nucleotides (Figure 7B). To further confirm the binding specificities, we assembled RTL2_RR.HN-NP12 (HN-HN-NP-NP-HN-HN-NP-NP-HN-HN-NP-NP), cloned possible EBEs in the mBs3 minimal promoter (Figure 7C), and separately co-expressed the effector with each possible EBE respectively via agroinfiltration in N. benthamiana tobacco leaves. The GUS assays showed that HN binds to A and G, NP binds more strongly to C than to G and A (Figure 7C).
Figure 7.
Determine the DNA-Binding Specificities of RTL RVDs.
RTL2_RR.HN-ND12 (A), dHax3_NP-ND-NT-NG11 (B), and RTL2_RR.HN-NP12 (C) were constructed with sequences as shown. Possible target EBE boxes were cloned in mBs3 promoter with uidA reporter downstream. Clones of 35S promoter-driven RTLs were co-agroinfiltrated with possible EBEs, respectively. Leaf discs from 2 d post infiltration were collected for qualitative and quantitative analyses. RTL2_RR.HN-ND12 activated promoters containing EBEs that HN corresponds to A or G strongly (A); dHax3_NP-ND-NT-NG11 activated promoters containing EBE that NP corresponds to C stronger, but to A and G weaker, when ND binds to C and NT binds to T, but, when NT was changed to A, G, or C, dHax3_NP-ND-NT-NG11 did not activate uidA reporter expression (B); RTL2_RR.HN-NP12 activated promoters containing EBEs that HN corresponds to A or G, NP corresponds to A, G, and C, strongly (C).
DISCUSSION
Developing customizable DNA-binding modules that can be easily engineered to bind to any user-selected DNA sequence is of paramount importance in the development of tools and reagents for genome-engineering applications. TAL effectors from Xanthomonas phytopathogenic bacteria have been recently used as programmable DNA-binding scaffolds for genome-editing applications (Bogdanove et al., 2010; Bogdanove and Voytas, 2011; Mahfouz and Li, 2011). Although the TAL DNA-binding code is surprisingly simple, many questions remain about the biology of TAL effectors and their molecular function. Identifying novel TAL-like DNA-binding architectures from other phytopathogens would advance our understanding of the function of TAL effectors in disease biology and facilitate their use in customizable platforms for DNA targeting. Here, we report the identification and characterization of R. solanacearum TAL-like effectors (RTLs) and demonstrate the DNA-binding specificities of several of their naturally occurring RVDs, including ND, HN, NP, and NT.
Based on their overall structural similarity to Xanthomonas TAL effectors, three TAL-like Ralstonia proteins (RTL1, RTL2, and RTL3) were selected for molecular characterization and in vivo testing of their DNA-binding specificities. To determine whether RTLs function as transcriptional factors, we first assessed the ability of their DNA repeats to mediate DNA binding. The data demonstrate that the RRs in the Xanthomonas TALE backbone do mediate DNA binding as inferred by the transcriptional activation of their targets (Figure 4). These data prompted us to study whether the RTLs are genuine DNA-binding proteins that mimic eukaryotic transcriptional factors. Cellular localization studies indicate that the RTLs are localized to the nucleus (Figure 3), which provides evidence that those RTLs may possess functional NLSs. Because our data showed that the RRs in the Xanthomonas backbone mediate DNA binding and possess functional NLSs, we next determined whether they function as genuine transcription factors and activate the expression of target genes. RTLs with poly(HD)-containing repeats induced the transcriptional activation of promoters that contained the respective DNA-binding sequences (Figure 5). These data indicate that RTLs may function as transcriptional activators. The finding that RTLs may function as transcriptional activators prompted us to determine the DNA-binding specificities of RTL RVDs. The ND, HN, NP, and NT RVDs were selected for analysis of their DNA-binding specificities. Our data clearly demonstrate that the ND RVD binds to the C nucleotide with high specificity. Surprisingly, RTL1, RTL2, and RTL3 containing poly(NP), poly(HN), and poly(NT) did not noticeably induce the expression of the polyA/T/G/C:mBs3::uidA. These data indicate that the RTL binding may depend on the composition of the RVDs. It should be noted that RVDs possess different binding affinities (Deng et al., 2012a; Mak et al., 2012). Because the RTLs containing poly(ND) bind specifically and strongly to the C nucleotide, the inability of RTLs containing other poly(RVDs) to bind to DNA might be the result of weak binding of those RVDs. In other words, RTLs composed of strong RVDs may bind strongly to DNA, and RTLs composed of weak RVDs may bind weakly to DNA, which could affect the overall function of the RTL. Because the HD and ND RVD types bind to DNA with high affinity, their poly(RVDs) were able to bind to DNA and activate the expression of the GUS reporter with high efficiency.
To exclude the possibility that the inability to bind DNA is due to the nature of the RTL scaffold and that the binding might be scaffold-dependent, we generated poly(RVDs) in the Xanthomonas dHax3 backbone and tested their binding activities. As observed with the RTL scaffold, only dHax3.poly(ND) showed strong binding to the C nucleotide, whereas the dHax3.poly(NT), (HN), and (NP) did not show strong binding with any nucleotide (Figure 6). These data clearly indicate that, when the repeats contain a single RVD type, the binding of the TAL or RTL effectors will be weak if the binding of that particular RVD is naturally weak.
When mixed repeats containing strong and weak RVDs were generated, the RTL effector strongly induced GUS expression, indicating strong DNA binding. These data suggest that the presence of strong RVDs could stabilize RTLs and allow the weak RVDs to bind specifically (Figure 7). Thus, the binding strength of an engineered RTL could be increased by including strong RVDs spaced throughout the repeats. Similar conclusions regarding DNA-binding specificities and efficiencies were drawn recently from work on Xanthomonas TAL effectors and repeats (Streubel et al., 2012). Further studies, however, are needed to determine the positional effects of the RVDs on the DNA-binding strength and on transcriptional functions of the RTLs or TALEs.
Recently, the structural basis of TAL binding to DNA has been reported. These studies indicated that the 12th residue of the RVD makes the conformation more rigid, whereas the 13th residue mediates base-specific contacts (Deng et al., 2012a; Mak et al., 2012). Our data indicate that Ralstonia TAL effectors may assume a similar structure, because RVDs that end with D (e.g. ND) specifically recognize the C nucleotide, whereas the RVDs that end with N (e.g. HN) recognize the A and G nucleotides. At least two important questions remain to be answered with structural and molecular genetic approaches. First, what are the functions of the two repeats (repeat 0 and repeat –1) that precede the canonical repeats containing the DBD? These two repeats are quite similar to the canonical repeats but lack the RVD residues. Also, at the end of the C-terminus of the repeat domains, there are two half-repeats, +1 and +2, that lack known RVDs. It will be quite important to determine the molecular functions of these repeats and whether they contribute to the RTL binding to DNA. Second, what are the molecular mechanisms of the conformational plasticity of the TALEs and RTLs (such plasticity might be important for their binding specificity to epigenetically modified DNA in its cellular chromatic environment (Deng et al., 2012b))? Answers to these questions would guide the design of RTLs and TALEs to ensure their conformational plasticity and thereby their specificity and molecular activity in vivo.
The Xanthomonas TAL effectors function mainly in virulence, but some induce plant resistance and hence are termed avirulence (Avr) genes. It is likely that the Avr genes, including the RTLs, were transferred into the Ralstonia genome via a stochastic HGT process. Some of these genes might have been selected for pathogenic functions. Whether RTLs are involved in the pathogenicity of Ralstonia spp. or whether they target host disease or resistance genes is not known. It should be feasible to determine whether RTLs are functioning as avirulence genes that activate molecular pathways of disease resistance. Thus, molecular characterization of the RTL effector proteins and their DNA-binding specificities would greatly enhance our understanding of the biology of disease and disease resistance among the many hosts of Ralstonia spp.
In conclusion, we have identified and characterized three RTL effectors, which may function as transcription factors, and identified and experimentally verified the DNA-binding specificities of novel RVDs. Therefore, we are presenting three novel structures that can be used as targeting modules and four novel RVDs that can be used to engineer RTLs to bind to any user-selected DNA sequence. These findings should facilitate the development of several immediate biotechnological applications, including the use of RTLs as customizable DNA-binding scaffolds to generate a variety of chimeric proteins for targeted genomic and epigenomic modifications. RTLs could also be tailored to generate sequence-specific transcriptional activators and could be fused to other functional domains, such as transcriptional repressors, nucleases, methylases, demethylases, recombinases, or chromatin modifiers. In addition to supporting the development of new technologies, our findings may enhance the basic understanding of the molecular interactions between pathogenicity-related genes and RTLs.
METHODS
Generation and Cloning of the RTL Repeat Concatemers in the dHax3 Backbone
To assemble RRs in the dHax3 backbone, a collection of plasmids (pUCminusMCS) that contains three types of fragments was generated. The first fragment type contains the first repeat plus an overlapping part of the N-terminus of the dHax3 backbone to facilitate cloning using the PpuMI restriction-enzyme. The second fragment type contains a single-repeat clone that can be cleaved with and can concatamerize in the ligation reaction, thereby generating different fragment lengths. The third repeat type contains the last repeat plus the C-terminus of the dHax3 backbone to facilitate cloning using the SacI enzyme. All RVD possibilities were generated in these fragments to facilitate the assembly of the desired number and order of the Ralstonia RVDs in the dHax3 backbone. Sequence information for all of these fragments is provided in the Supplementary Data. To assemble the repeats, three 20-μl digestion reactions containing 1000–1500 ng of DNA were used. The first, second, and third fragments were released from the plasmid by digestion with PpuMI and BsmAI, BsmAI, and SacI and BsmAI, respectively. All digested fragments were purified from the agarose gel. To allow concatemer formation of the second fragment, each type of RVD was ligated to the last fragment in a 1:3 ratio. After 4 h of incubation with T7 ligase, the same RVD and ligation buffer was added, and the ligation reaction was repeated three times. After the completion of the ligation reaction, 1.2–1.5-kb products were separated and eluted from the E-gel Ex 2% Agarose (Life Technologies, Grand Island, NY, USA), ligated to the first fragment (which had been digested with PpuMI–BsmAI), and finally ligated to the backbone (pENTR221.dHax3 digested with PpuMI–SacI) with T7 ligase. After 2 h of ligation, the product was used to transform ONE-shot Top 10 cells (Life Technologies), which were spread on Kan+ LB plates. The repeat fragment size was confirmed either by colony PCR or by PpuMI–HindIII restriction digestion. Sanger sequencing confirmed the RVD number and order. NEB (New England Biolab) restriction enzymes were used for all restriction digestions.
Design and Construction of the Backbones of RTL Proteins
To facilitate the synthesis and restriction ligation assembly of overlapping fragments, the codons of the Ralstonia clones (YP_003750492.1, NP_519936.1, and CAQ18687.1) were optimized. The sequences of the optimized clones are provided in the Supplementary Data. Each of the RTL clones was divided into fragments containing the N-terminus, the central repeats, and the C-terminus, with overlapping ends containing unique restriction enzymes. For example, to assemble the YP clone, the N-terminus fragment (NTF) was synthesized and cloned into pUC19minusMCS flanked by the L1 gateway cloning sequence at the 5’ end, and by a sequence containing ApaI–NdeI–SpeI restriction recognition sequences at the 3’ end. The C-terminus fragment (CTF) was designed and synthesized to be flanked by an NdeI site at the 5’ end and by the L2 gateway cloning sequence followed by a SpeI site at the 3’ end. The pUCminusMCS.L1.NTF.ApaI-NdeI-SpeI clone was used as a backbone to subclone the CTF using NdeI and SpeI to generate the pUCminusMCS.L1.NTF.ApaI/SalI-NdeI-CTF.L2.SpeI backbone clone. This RTL- minus-repeats clone (RTL∆R) was used as a scaffold to assemble the repeats in a random or defined order. The other RTL clones were assembled using the same approach, and the details of their assembly are described in the Supplementary Data.
Subcloning of the RRs into RTL Backbone Clones
The RRs in the dHax3 backbone were amplified by primers that contain the ApaI/SalI and NdeI enzyme sites at the 5’ and 3’ ends, respectively, and were cloned into a TOPO TA vector. The RTL repeat fragments were digested with ApaI or SalI and NdeI and subcloned into the RTL∆R backbone. Sanger sequencing was used to confirm the identity of the full-length clones with the in silico assembled clone. The RTL clones were subcloned into the pK2GW7 and pK7WGF2 plant overexpression vectors or the pET32a–GW bacterial expression vector using the LR gateway cloning reaction.
Ordered Assembly Cloning of the RRs in the RTL Backbone Clones
To achieve the ordered assembly of the RRs in the RTL backbone clones, a library of di-repeat clones was assembled that contains the Ralstonia RVDs in all possible combinations. The di-repeat clones, flanked by BsmAI sites, produce distinct overhangs after digestion to ensure the ordered assembly of 12 repeats. The sequences of the di-repeats in the library are provided in the Supplementary Data. Several RTL clones were generated with repeats in the desired order, and full-length clones were confirmed by Sanger sequencing. Generating an extended library of the di-repeats will make it possible to generate any RTL with any desired RVD sequence to target any DNA sequence of interest.
Cloning of Minimal Bs3 and Bs4 Promoters
Minimal Bs3 (mBs3) and Bs4 promoters containing different EBEs, which were designed to test the transcriptional activation of different effectors, were PCR-amplified using specific primers listed in the Supplementary Data) and cloned into a D/TOPO vector. After the clones were confirmed by Sanger sequencing, they were transferred by LR reaction to the gateway destination vector pKGWFS7 to drive the expression of uidA reporter genes.
RTL Cellular Localization Studies
To confirm whether RTLs function as transcription activators, pK7WGF2/RTL clones that express GFP–RTL fusion proteins in planta were generated. The clones were transformed into Agrobacterium tumefaciens GV3101 and used for transient expression in tobacco leaves. Two days post agroinfiltration, infiltrated discs were collected for confocal laser scanning microscopy analysis. The discs were prepared on slides, embedded with 50% glycerol, and covered with coverslips. The specimens were examined and imaged with an LSM 710 laser scanning confocal microscope (Zeiss, Germany) with ZEN 2009 software. Stained DAPI was excited by a 405-nm laser, while the detection range was set at 410–483 nm. The expressed GFP was activated by a 488-nm laser with an emission range of 493–598 nm. Images were recorded with a 2000 × 2000 pixel frame size and 8 bits of depth.
Measurement of RTL Transcriptional Activity Using GUS Reporter Assays
To test the transcriptional activity of the RTLs in planta, pK2GW7/RTLs were generated. The clones were transformed into A. tumefaciens GV3101 and used for transient expression in tobacco leaves. A. tumefaciens harboring mBS3/Bs4::uidA and its RTL activator were co-infiltrated or separately infiltrated into tobacco leaves. Infiltrated discs were collected at 48 h post inoculation. The qualitative GUS activity assay was performed by immersing leaf discs in GUS staining solution (10 mM sodium phosphate, pH 7, 10 mM EDTA, 0.1% Triton X-100, 0.1% 5-bromo-4-chloro-3-indolyl-β-D-glucuronide, 1 mM potassium ferricyanide, 1 mM potassium ferrocyanide) at 37°C for 24 h. The discs were cleared in ethanol. For the quantitative GUS analysis, three leaf discs were collected, and the tissue was homogenized, diluted, and incubated with 4-methyl-umbelliferyl-β-D-glucuronide (MUG) as previously described (Kay et al., 2007).
SUPPLEMENTARY DATA
Supplementary Data are available at Molecular Plant Online.
FUNDING
This research is funded from the Center for Desert Agriculture baseline funding. The work in J.-K.Z.’s lab is supported by the National Institute of Health grant R01GM070795.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Bioscience Core Facility at KAUST for technical assistance. We also thank members of the genome-engineering group at KAUST for their helpful discussions and technical assistance throughout the preparation of the manuscript. No conflict of interest declared.
Footnotes
The original version of this paper was removed because of author-generated errors in Figures 6 and 7. These figures have been corrected in the present version.
REFERENCES
- Boch J., Bonas U. (2010). Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu. Rev. Phytopathol. 48, 419–436 [DOI] [PubMed] [Google Scholar]
- Boch J., Scholze H., Schornack S., Landgraf A., Hahn S., Kay S., Lahaye T., Nickstadt A., Bonas U. (2009). Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 326, 1509–1512 [DOI] [PubMed] [Google Scholar]
- Bogdanove A.J., Voytas D.F. (2011). TAL effectors: customizable proteins for DNA targeting. Science. 333, 1843–1846 [DOI] [PubMed] [Google Scholar]
- Bogdanove A.J., Schornack S., Lahaye T. (2010). TAL effectors: finding plant genes for disease and defense. Curr. Opin. Plant Biol. 13, 394–401 [DOI] [PubMed] [Google Scholar]
- Bozas A., Beumer K.J., Trautman J.K., Carroll D. (2009). Genetic analysis of zinc-finger nuclease-induced gene targeting in Drosophila . Genetics. 182, 641–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll D. (2011). Genome engineering with zinc-finger nucleases. Genetics. 188, 773–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak T., Doyle E.L., Christian M., Wang L., Zhang Y., Schmidt C., Baller J.A., Somia N.V., Bogdanove A.J., Voytas D.F. (2011). Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 39, e82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian M., Cermak T., Doyle E.L., Schmidt C., Zhang F., Hummel A., Bogdanove A.J., Voytas D.F. (2010). Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 186, 757–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng D., Yan C., Pan X., Mahfouz M., Wang J., Zhu J.K., Shi Y., Yan N.(2012a). Structural basis for sequence-specific recognition of DNA by TAL effectors. Science. 335, 720–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng D., Yin P., Yan C., Pan X., Gong X., Qi S., Xie T., Mahfouz M., Zhu J.K., Yan N., et al. (2012b). Recognition of methylated DNA by TAL effectors. Cell Res. 22, 1502–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler R., Scholze H., Hahn S., Streubel J., Bonas U., Behrens S.E., Boch J. (2011). Transcriptional activators of human genes with programmable DNA-specificity. PLoS One. 6, e19509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genin S., Denny T.P. (2012). Pathogenomics of the Ralstonia solanacearum species complex. Annu. Rev. Phytopathol. 50, 67–89 [DOI] [PubMed] [Google Scholar]
- Heuer H., Yin Y.N., Xue Q.Y., Smalla K., Guo J.H. (2007). Repeat domain diversity of avrBs3-like genes in Ralstonia solanacearum strains and association with host preferences in the field. Appl. Environ. Microbiol. 73, 4379–4384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorda J., Kajava A.V. (2009). T-REKS: identification of Tandem REpeats in sequences with a K-meanS based algorithm. Bioinformatics. 25, 2632–2638 [DOI] [PubMed] [Google Scholar]
- Kay S., Hahn S., Marois E., Hause G., Bonas U. (2007). A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Science. 318, 648–651 [DOI] [PubMed] [Google Scholar]
- Li L., Piatek M.J., Atef A., Piatek A., Wibowo A., Fang X., Sabir J.S., Zhu J.K., Mahfouz M.M. (2012). Rapid and highly efficient construction of TALE-based transcriptional regulators and nucleases for genome modification. Plant Mol. Biol. 78, 407–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Huang S., Jiang W.Z., Wright D., Spalding M.H., Weeks D.P., Yang B. (2011a). TAL nucleases (TALNs): hybrid proteins composed of TAL effectors and FokI DNA-cleavage domain. Nucleic Acids Res. 39, 359–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Huang S., Zhao X., Wright D.A., Carpenter S., Spalding M.H., Weeks D.P., Yang B. (2011b). Modularly assembled designer TAL effector nucleases for targeted gene knockout and gene replacement in eukaryotes. Nucleic Acids Res. 39, 6315–6325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahfouz M.M., Li L. (2011). TALE nucleases and next generation GM crops. GM Crop. 2, 99–103 [DOI] [PubMed] [Google Scholar]
- Mahfouz M.M., Li L., Piatek M., Fang X., Mansour H., Bangarusamy D.K., Zhu J.K. (2012). Targeted transcriptional repression using a chimeric TALE-SRDX repressor protein. Plant Mol. Biol. 78, 311–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahfouz M.M., Li L., Shamimuzzaman M., Wibowo A., Fang X., Zhu J.K. (2011). De novo-engineered transcription activator-like effector (TALE) hybrid nuclease with novel DNA binding specificity creates double-strand breaks. Proc. Natl Acad. Sci. U S A. 108, 2623–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak A.N., Bradley P., Cernadas R.A., Bogdanove A.J., Stoddard B.L. (2012). The crystal structure of TAL effector PthXo1 bound to its DNA target. Science. 335, 716–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.C., Holmes M.C., Wang J., Guschin D.Y., Lee Y.L., Rupniewski I., Beausejour C.M., Waite A.J., Wang N.S., Kim K.A., et al. (2007). An improved zinc-finger nuclease architecture for highly specific genome editing. Nat. Biotechnol. 25, 778–785 [DOI] [PubMed] [Google Scholar]
- Morbitzer R., Elsaesser J., Hausner J., Lahaye T. (2011). Assembly of custom TALE-type DNA binding domains by modular cloning. Nucleic Acids Res. 39, 5790–5799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morbitzer R., Romer P., Boch J., Lahaye T. (2010). Regulation of selected genome loci using de novo-engineered transcription activator-like effector (TALE)-type transcription factors. Proc. Natl Acad. Sci. U S A. 107, 21617–21622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscou M.J., Bogdanove A.J. (2009). A simple cipher governs DNA recognition by TAL effectors. Science. 326, 1501 [DOI] [PubMed] [Google Scholar]
- Nomura W., Barbas C.F., III (2007). In vivo site-specific DNA methylation with a designed sequence-enabled DNA methylase. J. Am. Chem. Soc. 129, 8676–8677 [DOI] [PubMed] [Google Scholar]
- Ramirez C.L., Foley J.E., Wright D.A., Muller-Lerch F., Rahman S.H., Cornu T.I., Winfrey R.J., Sander J.D., Fu F., Townsend J.A., et al. (2008). Unexpected failure rates for modular assembly of engineered zinc fingers. Nat. Methods. 5, 374–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remenant B., Coupat-Goutaland B., Guidot A., Cellier G., Wicker E., Allen C., Fegan M., Pruvost O., Elbaz M., Calteau A., et al. (2010). Genomes of three tomato pathogens within the Ralstonia solanacearum species complex reveal significant evolutionary divergence. BMC Genomics. 11, 379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remigi P., Anisimova M., Guidot A., Genin S., Peeters N. (2011). Functional diversification of the GALA type III effector family contributes to Ralstonia solanacearum adaptation on different plant hosts. New Phytol. 192, 976–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyon D., Tsai S.Q., Khayter C., Foden J.A., Sander J.D., Joung J.K. (2012). FLASH assembly of TALENs for high-throughput genome editing. Nat. Biotechnol. 30, 460–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romer P., Hahn S., Jordan T., Strauss T., Bonas U., Lahaye T. (2007). Plant pathogen recognition mediated by promoter activation of the pepper Bs3 resistance gene. Science. 318, 645–648 [DOI] [PubMed] [Google Scholar]
- Salanoubat M., Genin S., Artiguenave F., Gouzy J., Mangenot S., Arlat M., Billault A., Brottier P., Camus J.C., Cattolico L., et al. (2002). Genome sequence of the plant pathogen Ralstonia solanacearum . Nature. 415, 497–502 [DOI] [PubMed] [Google Scholar]
- Shukla V.K., Doyon Y., Miller J.C., DeKelver R.C., Moehle E.A., Worden S.E., Mitchell J.C., Arnold N.L., Gopalan S., Meng X., et al. (2009). Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature. 459, 437–441 [DOI] [PubMed] [Google Scholar]
- Streubel J., Blucher C., Landgraf A., Boch J. (2012). TAL effector RVD specificities and efficiencies. Nat. Biotechnol. 30, 593–595 [DOI] [PubMed] [Google Scholar]
- Waterhouse A.M., Procter J.B., Martin D.M., Clamp M., Barton G.J. (2009). Jalview Version 2: a multiple sequence alignment editor and analysis workbench. Bioinformatics. 25, 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber E., Gruetzner R., Werner S., Engler C., Marillonnet S. (2011). Assembly of designer TAL effectors by Golden Gate cloning. PLoS One. 6, e19722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Voytas D.F. (2011). Targeted mutagenesis in Arabidopsis using zinc-finger nucleases. Methods Mol. Biol. 701, 167–177 [DOI] [PubMed] [Google Scholar]
- Zhang F., Cong L., Lodato S., Kosuri S., Church G.M., Arlotta P.(2011a). Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat. Biotechnol. 29, 149–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Kiba A., Hikichi Y., Ohnishi K.(2011b). prhKLM genes of Ralstonia solanacearum encode novel activators of hrp regulon and are required for pathogenesis in tomato. FEMS Microbiol. Lett. 317, 75–82 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.