Abstract
Accurate genetic association studies are crucial for the detection and the validation of disease determinants. One of the main confounding factors that affect accuracy is population stratification, and great efforts have been extended for the past decade to detect and to adjust for it. We have now efficient solutions for population stratification adjustment for single-SNP (where SNP is single-nucleotide polymorphisms) inference in genome-wide association studies, but it is unclear whether these solutions can be effectively applied to rare variation studies and in particular gene-based (or set-based) association methods that jointly analyze multiple rare and common variants. We examine here, both theoretically and empirically, the performance of two commonly used approaches for population stratification adjustment—genomic control and principal component analysis—when used on gene-based association tests. We show that, different from single-SNP inference, genes with diverse composition of rare and common variants may suffer from population stratification to various extent. The inflation in gene-level statistics could be impacted by the number and the allele frequency spectrum of SNPs in the gene, and by the gene-based testing method used in the analysis. As a consequence, using a universal inflation factor as a genomic control should be avoided in gene-based inference with sequencing data. We also demonstrate that caution needs to be exercised when using principal component adjustment because the accuracy of the adjusted analyses depends on the underlying population substructure, on the way the principal components are constructed, and on the number of principal components used to recover the substructure.
Keywords: sequencing studies, gene-based association test, genomic control, principal component analysis, C-alpha test, burden test
Introduction
For the past years, thousands of genome-wide association (GWA) studies have been conducted on more than 200 human diseases and traits, and have achieved numerous successes in identifying susceptibility loci for many complex phenotypes [Hindorff et al., 2009]. Up to date, for most traits/diseases, the identified loci altogether can only explain part of the heritability. By design, the majority of the associated single-nucleotide polymorphisms (SNPs) are common, with minor allele frequencies (MAFs) ≥ 5%. That is, they are well represented in the population and thus are less likely to have very strong effects on disease risk [Feero et al., 2010; Manolio et al., 2009]. In order to further understand the genetic basis of human traits/diseases, new efforts have been put forward to identify novel associations with the next-generation sequencing data, in which the entire allele frequency spectrum is extensively examined [Gilad et al., 2009], placing great emphasis on rare alleles and their roles in disease risk.
State-of-the-art sequencing data provides an unparalleled opportunity to identify causal genetic risk variants, though the engagement to rare variants (MAF < 5%) brings new challenges and makes some issues shared with the GWA data more apparent [Teo, 2008]. On the one hand, it can be argued that highly penetrant risk alleles are more likely to have low fitness and have lower frequencies in the population, and as such rare variants would be of great interest for the detection of genetic factors associated with disease risk [King et al., 2010]. On the other hand, if an allele is very rarely observed in the collected samples, standard statistical approaches may not have sufficient power to detect its risk association, especially when considering the expanded number of variants and the stringent genome-wide significance threshold required in a typical sequencing study [Madsen and Browning, 2009; Morgenthaler and Thilly, 2007; Wu et al., 2011]. To improve power to detect risk associations, various gene- or set-based association tests have been proposed to jointly analyze multiple variants in a gene or a set, and have demonstrated great efficiency in real studies [Dering et al., 2011; King et al., 2010; Madsen and Browning, 2009; Morgenthaler and Thilly, 2007; Neale et al., 2011; Wu et al., 2011]. Besides the power concern, the potential confounding effect caused by population stratification is also nonneglectable and needs to be examined in the context of gene-based association analysis involving multiple rare and common variants [Mathieson and McVean, 2012;Tintle et al., 2011].
Population stratification refers to the systematic differences in allele frequencies between different subpopulations. In the presence of population stratification in a case-control GWA study, different compositions of subpopulations in cases and in controls could cause systematic differences in allele frequencies between cases and controls, and may confound the true associations to disease risk. As a result, unaccounted population stratification may increase the type I error rates in association analyses [Pritchard and Donnelly, 2001]. For single-SNP inference in GWA studies with common variants only, great efforts have been extended and efficient solutions have been implemented to detect and to adjust for population stratification [Devlin and Roeder, 1999; Price et al., 2006; Pritchard and Rosenberg, 1999]. However, most rare variants do not have a long-standing evolutionary history and may suffer from different levels of population stratification than common variants. Thus, it is unclear whether the solutions for GWA studies would continue to be appropriate in the analyses involving both rare and common variants, especially when jointly analyzing multiple variants in a gene. In this work, we will show that the effect of population stratification on gene-based tests depends on the method used for association testing, the number and the MAF spectrum of SNPs in a gene. Moreover, we will evaluate, both theoretically and empirically, the performance of two widely used approaches in GWA studies for adjusting stratification—genomic control [Devlin and Roeder, 1999] and principal component analysis (PCA) [Price et al., 2006]—in gene-based tests with sequencing data.
Methods
Inflation Factor
Population stratification could cause spurious associations and inflate the test statistics and significance on a genome-wide scale. The level of inflation is commonly measured by an inflation factor λ that can be estimated as the ratio of the median (or the mean) of the observed test statistics to the median (or the mean) of the statistics with no inflation [Devlin and Roeder, 1999; Reich et al., 2001]. In gene- or set-based association tests, different genes or sets have different numbers of SNPs, and thus the gene- or set-level test statistics do not have generally the same null distributions, rendering the inflation factors measured on those statistics incomparable to each other. As such, we employ the following inflation factor measured on the log-transformed P -values for gene-or set-based tests:
| (1) |
When stratification is not present, the P -values are uniformly distributed and −2 log(P –values) are chi-squared, distributed with degrees of freedom 2, and is the median of chi-squared statistics with degrees of freedom 2. Using either inflation factor measure (on the test statistics or on the P -values), when population stratification is not present, the inflation factor would be one, λ = 1; otherwise, λ > 1. When both strategies are appropriate, there is a one-to-one map between the two factors.
Simulation of Case-Control Studies With Population Stratification
We used the ms simulator [Hudson, 2002] to simulate genome-wide genotype data with certain population stratification. The ms simulator uses a coalescent approach to generate genome-wide genotype data for different subpopulations given prespecified parameters. Those parameters include sample size for each subpopulation, total number of subpopulations, number of genes, mutation parameter, and migration parameters between pairs of subpopulations. We assume all subpopulations consist of the same number of individuals, and denote the number of individuals in each subpopulation as N0. The mutation parameter is defined as 4N0μ, where μ is the neutral mutation rate. The mutation parameter is set to be 2, which is equivalent to having two mutations per generation for any locus. The migration parameter is defined as 4N0m, where m is the fraction of new migrants each generation from one subpopulation to the other. A smaller m indicates less interpopulation migration and more severe population stratification.
We simulated, using ms, case-control studies from two different levels of population stratifications. In the first scenario, we simulated two subpopulations, A and B, with relatively distinct ancestries. Each has 500 individuals. The number of genes was set to be 3,000. We set the pairwise migration parameter to be 10 and the migration pattern is symmetric, i.e., the fraction of new migrants from one subpopulation to the other per generation is 0.5%. We simulated a case-control study with 500 cases and 500 controls, randomly sampled with different proportions from the two subpopulations: 58% of the cases are from subpopulation A and 42% are from subpopulation B; 42% of the controls are from subpopulation A and 58% are from subpopulation B.
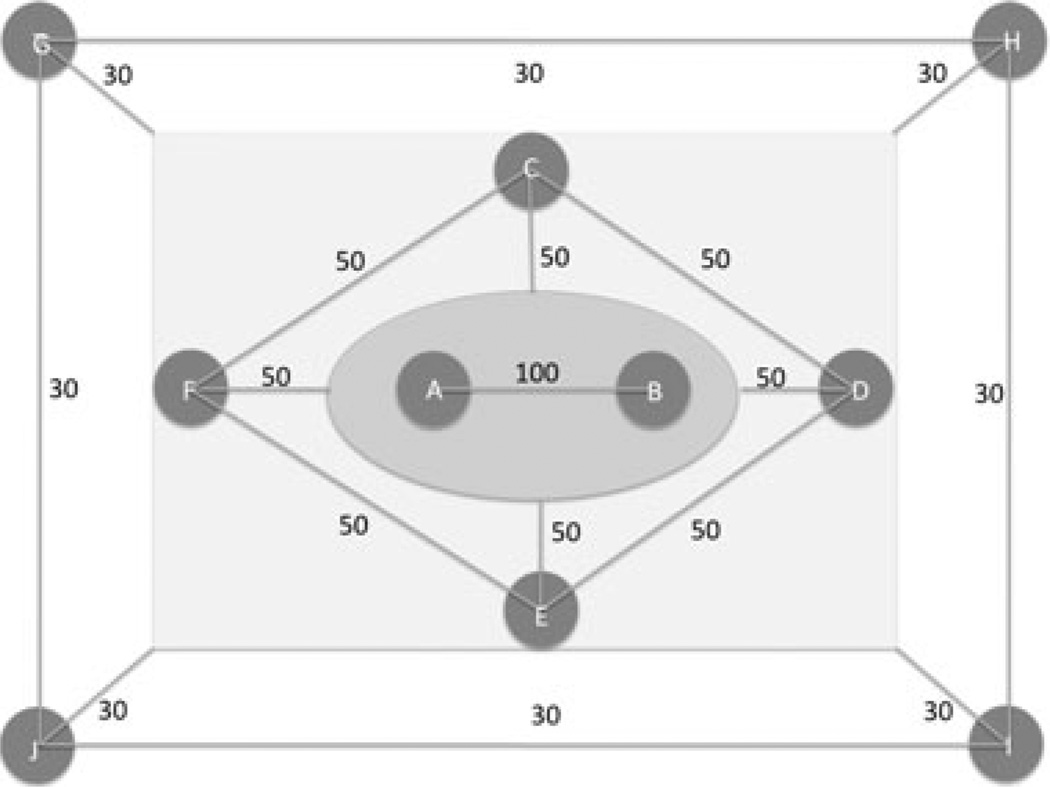
The second simulated scenario is slightly more complicated, consisting of 10 subpopulations with 500 individuals each. The number of genes was set to be 500. Figure 1 shows the symmetric interpopulation migration patterns among the 10 subpopulations. The migration parameters range from 30 to 100, corresponding to the fractions of new migrants per generation ranging from 1.5% to 5%. Subpopulations with larger migration parameters are more related in ancestry, e.g., subpopulations A and B are more related than others. We simulated a case-control study with 500 cases and 500 controls. For the cases, 300 were sampled from subpopulation A and 200 were randomly sampled from the other nine subpopulations. The 500 controls were all randomly sampled from the remaining individuals in the 10 subpopulations. This simulation resembles a study of a disease with higher prevalence in a certain subpopulation, and as such most of the cases are from there, although the rest of the cases and all controls are randomly sampled from the general population.
Figure 1.
A simulation of 10 subpopulations with complex migration patterns. A to J represent 10 subpopulations, each with 500 individuals. The numbers connecting any two subpopulations represent the migration parameters between them. When a subpopulation is connected with a group of subpopulations, it has the same migration parameter to each of the subpopulation in the group. We randomly select 300 individuals from subpopulation A and 200 individuals from the other nine subpopulations to be the cases; then randomly select 500 individuals from the remaining subjects in the 10 subpopulations to be the controls.
The simulation parameters were chosen so that, without adjustment for population stratification, the two simulated case-control studies (one from the two subpopulation and the other from the 10 subpopulation scenario) suffer from similar levels of inflation in single-SNP inference.
In order to study the effect of gene size on the inflation factor, we simulated genes of various sizes, i.e., different number of SNPs, based on the simulated genotype data described above. We simulated genes of a certain size k by randomly sampling k SNPs from the simulated genome-wide genotype data. For each gene size under investigation (k from 2 to 50), we generated 50,000 genes. We calculated the burden and the C-alpha gene-based statistics for these simulated genes. For each gene size, the inflation factor is calculated based on the median of the P -values given by equation (1).
Evaluation of Genomic Control and PCA With Gene-Based Tests
Many gene-based association tests have been proposed in the literature. To illustrate the diverse effects of population stratification on gene-based association tests, we discuss two representative methods, the burden test [Madsen and Browning, 2009] and the C-alpha test [Neale et al., 2011]. The burden test forms an association statistic for a gene by collapsing (with weights) the rare allele counts of individual variants in the gene and testing the association of the collapsed rare allele counts with a binary or a quantitative trait. The burden test or other collapsing type of procedures [Li and Leal, 2008; Morgenthaler and Thilly, 2007; Wang and Elston, 2007] are most effective when all rare variants in a gene are associated with disease risk in the same direction, e.g., all mutations are deleterious. Whereas, the C-alpha test is based on the sum of individual variant statistics that compare the observed and the expected variance of rare allele counts in cases for each variant [Neale et al., 2011]. That is, the C-alpha test is a test for overdispersion of rare alleles in cases and its power is not affected by the direction of association effects for variants in a gene. For both the burden and the C-alpha tests, we calculated all P -values based on 50,000 permutations.
There are two widely used approaches to detect and adjust for population stratification in single-SNP inference based on GWA studies, genomic control [Devlin and Roeder, 1999] and PCA [Price et al., 2006]. For single-SNP inference, genomic control assumes that the effect of population stratification is roughly constant for SNPs in the genome and it adjusts the association statistics by a common inflation factor. Though controversial [Marchini et al., 2004], this overarching inflation estimate and the adjustment is commonly used in many GWA studies. To evaluate the performance of genomic control in gene-based tests, we examined the constant inflation assumption by studying the inflation factors in genes with various sizes and different MAF spectrum.
The PCA adjustment does not assume constant inflation for tests performed in the genome. It uses the top few principal components (PCs; often less than 10) constructed from genome-wide data to capture the unmeasured substructure in the study, and includes those PCs as covariates in regression-based tests. To measure the inflation of the burden test with PCA adjustment, we adapted the burden test to a regression setting. We used the collapsed burden score as the predictor in a logistic regression, where wi is the weight on the ith variant (all weights were set as 1 in our simulations) and Gij is the rare allele count of the ith variant for individual j, and we included the top q PCs (q = 5, 10) as covariates. We measured the inflation factor of the burden test by the inflation in the P -values of the predictors in the regression, defined by (1). To adjust for population stratification for the C-alpha test with PCs as covariates, we used the biasedUrn sampling method [Epstein et al., 2012], which treats the PCs as modeled population substructure and permutes the data set in a way to preserve the modeled substructure under the null for association testing. That is, if the PCs completely captures the subpopulation structure, the biasedUrn method could adjust for population stratification in nonregression-based approaches, such as the C-alpha test.
The performance of PCA adjustment can heavily depend on how the PCs are constructed. To illustrate this point, we obtained two sets of PCs: (1) PCs constructed from the standardized genotype matrix [Price et al., 2006] of common variants with MAF ≥ 5%, and referred as “common PCs” hereafter; and (2) PCs constructed from the standardized genotype matrix of rare variants with MAF < 5%, and referred as “rare PCs.” In constructing common and rare PCs, we used roughly the same number of variants in each setting. We will show with simulations that the performance of PCA adjustment in gene-based tests with sequencing data may depend on several factors including the nature of the population substructure, the gene-based association-testing method that is used, the number of PCs used in the adjustment, and the way the PCs are constructed.
Results
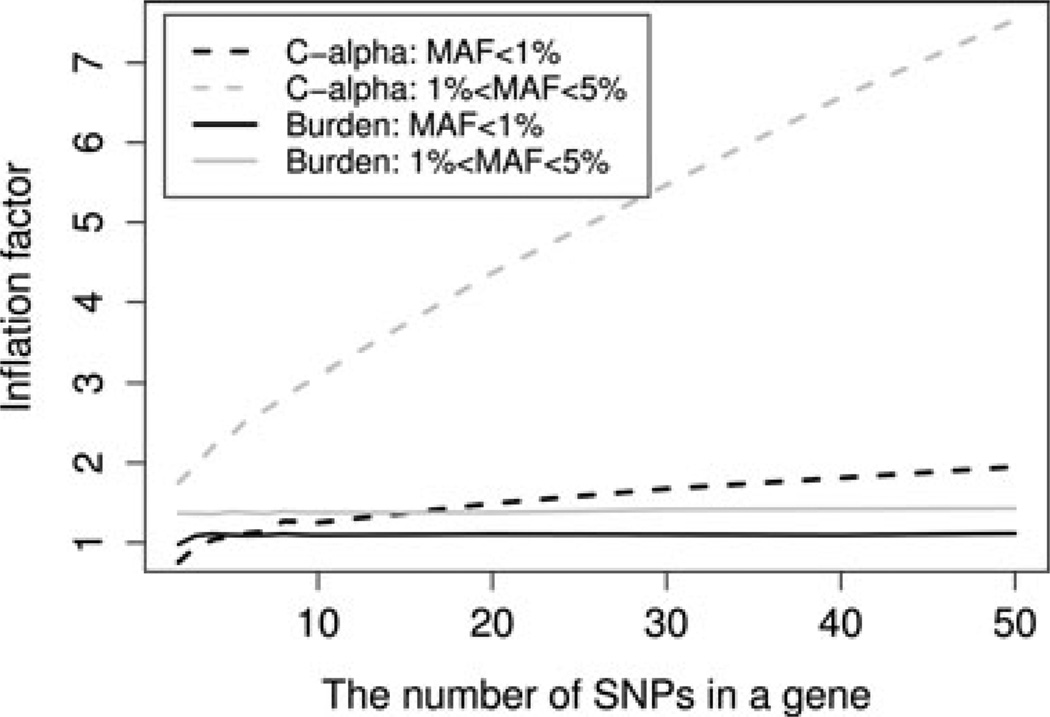
As shown in Figure 2, the inflation factor of the burden test depends heavily on the MAFs of the variants, and is less dependent on the number of variants in a gene. In this simulation, the inflation is larger for the more common SNPs (1% < MAF < 5%). In Appendix, we showed mathematically that as the number of variants in a gene, k, is expanding, the inflation factor for the burden statistic converges to a distribution that always takes values greater than 1 under population stratification, and the distribution is dependent on the MAFs of variants in cases and in controls. The inflation factor of the burden statistic is a measure of the effect of population stratification on statistics based on collapsed variants. Though less dependent on gene size, the inflation could differ substantially across genes with different proportions of rare and very rare variants.
Figure 2.
The inflation factors for different gene-based methods vs. the number of variants in a gene. The inflation factors for the C-alpha statistics (indicated by the dash lines) increase with the number of variants in a gene under population stratification, regardless of the MAF spectrum. The inflation factors for the burden statistics (indicated by the solid lines) quickly converge, but the limit depends on the MAF spectrum in cases and controls.
In contrast, the inflation factor of the C-alpha test grows rapidly as the number of variants in a gene increases, indicating that when using the C-alpha test, larger genes are more prone to spurious association findings caused by stratification. We have also shown mathematically that, as the number of variants in a gene increases, the inflation factor of the C-alpha statistic will amplify on the order of k (see Appendix). Intuitively, the C-alpha statistic is based on the summation of variant statistics measuring the overdispersion of each variant. Under stratification, as each variant suffers from population stratification, the C-alpha statistics for larger genes would accumulate more evidences of overdispersion/stratification, showing a higher level of inflation.
In summary, for both the burden and the C-alpha test, the inflation caused by population stratification is unlikely to remain constant across genes that vary in size and MAF spectrum. Therefore, the constant inflation assumption for genomic control does not hold in gene-based association tests. As such, directly applying genomic control in gene-based tests could be problematic because a universal inflation adjustment may underestimate the true inflation for some genes although overestimating for some others, leading to overall biased inference on a genome-wide scale.
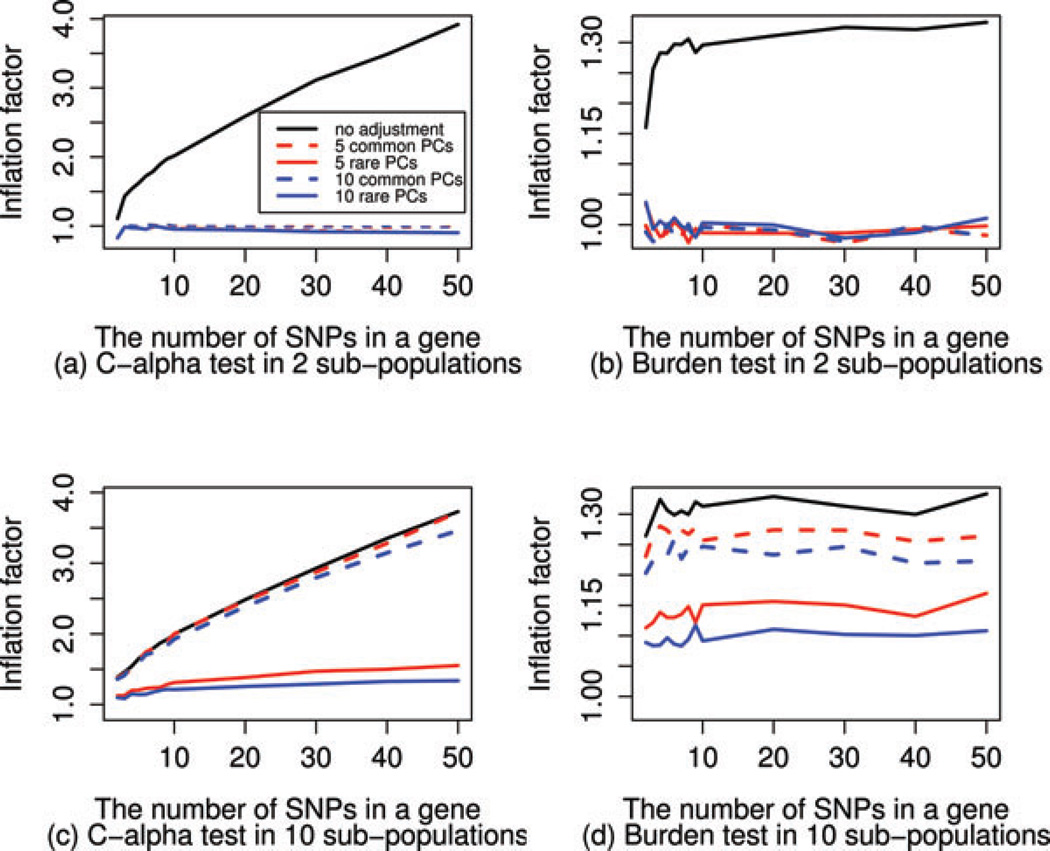
Figure 3 shows the performance of PCA adjustment for the burden test and the C-alpha test in the two simulation settings. In the simulation with two subpopulations, either common or rare PCs could completely capture the substructure in the samples, and adjusting for the top few PCs would remove the inflation in the gene-level statistics, even for the C-alpha test where inflation may accumulate over variants. However, in the more complicated simulation with 10 subpopulations shown in Figure 3(c) and (d), based on either common or rare PCs, the top few (up to 10) PCs may not completely recover the subtle substructure in the samples, and as such can not entirely remove the inflation in individual variant statistics, leaving different levels of inflation in the gene-based statistics. From our simulations, we observed different performance of PCA adjustment based on common and rare PCs, demonstrating that PCA adjustment can be affected by the way PCs are constructed, a conclusion consistent with Mathieson and McVean [2012].
Figure 3.
The performance of PCA adjustment for the burden and C-alpha tests in two simulations with different population structures. For the simple substructure in the two subpopulations simulation, PCA adjustment is adequate to remove stratification, as shown in (a) and (b). For the more complicated simulation with 10 subpopulations, adjusting for rare PCs outperforms the adjustment by common PCs, but does not completely remove the effect of population stratification. The results from PCA adjustment based on PCs constructed from all variants are between those adjusting for rare PCs and those adjusting for common PCs.
Discussion
We examined the effect of population stratification in sequencing data on two classes of gene-based methods, represented by the burden and the C-alpha tests, respectively. The constant inflation assumption underlying genomic control is apparently violated in sequencing studies with gene-based tests. Different genes in the genome may display marbled stratification effects that would depend on the method used in gene-based testing, and may also depend on the number of variants and/or the allele frequency composition of the set of variants in a gene. Genes in the genome vary substantially on their sizes, and even among the genes with the same number of variants, some may have more rare or more very rare variants. Therefore, a universal inflation factor is not adequate to adjust for stratification in this context, making a direct application of the genomic control approach less appropriate.
The performance of PCA adjustment depends heavily on how well the ancestry information is captured by linear principal components. In our simulations, the same number of rare PCs often outperforms the same number of common PCs. This is consistent with previous work [Mathieson and McVean, 2012] and with our intuition that rare variants may be recently derived in the evolutionary history and may capture more ancestry information. However, rare PCs constructed from linear PCA can be easily influenced by extreme values, and the standardized sequencing data may contain many extreme values generated from rare allele counts after standardization, and so they may not always perform better than common PCs. Moreover, even though adjusting for 10 PCs constructed from rare or common variants could alleviate the effects of population stratification, the inflation may not be completely removed if the data has a slightly complicated substructure. When using even more PCs to capture that structure, power for detecting real associations could be hurt. Besides, including too many PCs as covariates may also violate the restriction on the number of covariates allowed in a logistic regression [Peduzzi et al., 1996].
Therefore, the inflation in the PCA-adjusted analysis, similarly to the unadjusted analysis, may depend on the gene-based testing methods, the number of variants and/or their MAFs in the genes, and additionally, it may depend on the complication of substructure in the data, on the way the PCs are constructed and on how many PCs are adjusted. To reach a more definitive conclusion on how to efficiently construct PCs from rare and common variants, or more generally on how to adjust for population stratification in gene-based tests with sequencing data, additional work is needed, and is beyond the scope of this work. In order to avoid spurious findings in gene-based inference with sequencing data, the aforementioned issues should be kept in mind, and novel methods should be considered for analysis in gene-based association discovery studies.
Acknowledgments
This research was supported in part by NIH grants HL087665, MH090937, and HG005773.
Appendix
The Burden Test and Its Inflation Factor Under Stratification
We derive the inflation factor for the burden statistic based on analysis using t-tests. The burden score for individual j is
where wi is the weight on the ith variant, Gij is the rare allele count of the ith variant for individual j (we use Gij, A and Gij, U for cases and controls, respectively), and k is the total number of variants in the gene. The weight does not alter our conclusion, and we use equal weights: wi = 1, i = 1, … , k.
Suppose, we have N cases and N controls. Let PiA denote the MAF of the ith SNP in the cases and PiU denote the MAF in the controls. We use the following test statistic:
For a given set of SNPs, by the central limit theorem under the assumption of no linkage disequilibrium (LD),
Note that, under the null hypothesis of no association, and when there is no population stratification, we have that PiA = PiU for all i, and as such, the burden statistic S has zero mean. It is easy to see that the inflation factor corresponding to this test statistic is given by,
| (A1) |
where .
In the following, we derive the limit distribution for this inflation factor, as the number of SNPs in a gene (or a set) increases (k → ∞). We assume that SNPs are sampled independently, and we use E (PA), e.g., to denote the mean MAF in cases for the set of SNPs where we sample from. From (A1),
By the central limit theorem, as k → ∞, the numerator converges in distribution to a scaled distribution, and by the strong law of large numbers, the denominator converges almost surely. Using Slutsky’s theorem,
Note that, population stratification leads to Var(PA − PU) > 0, and to a limit distribution shifted away from 1.
The C-alpha Test and Its Inflation Factor Under Stratification
Let p0 be the proportion of cases among all samples and ni be the total rare allele counts of variant i, ni = ∑j (Gij, A + Gij, U). The individual variant test statistic contrasts the observed and the expected variances of rare allele counts for variant i in cases. The C-alpha statistic is given by .
Let Vi denote the variance of individual statistic, Vi = Var(Ti) > 0. When there is no association and no population stratification, . Let , with μi > 0 under stratification. Assuming all variants are independent,
Similar to (A1), the inflation factor can be measured by
Assuming there is μ0 > 0, such that μi > μ0 for all variants (in fact we need this condition only for a nonvanishing proportion of the SNPs), then
We assume E (Vi) < ∞, which is natural for a fixed number of subjects. By the law of large numbers, as k → ∞,
The derivations above assume that all SNPs are independent. In the presence of LD, the conclusions do not change as long as the “effective” number of SNPs is O(k), i.e., of the same order as the total number of SNPs. The “effective” number of SNPs corresponds to the number of independent single-SNP tests, and the assumption is valid as long as the size of the LD blocks does not increase with the total number of SNPs.
Footnotes
The authors declare no conflict of interest.
References
- Dering C, Hemmelmann C, Pugh E, Ziegler A. Statistical analysis of rare sequence variants: an overview of collapsing methods. Genet Epidemiol. 2011;35:S12–S17. doi: 10.1002/gepi.20643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- Epstein MP, Duncan R, Jiang Y, Conneely KN, Allen AS, Satten GA. A permutation procedure to correct for confounders in case-control studies, including tests of rare variation. Am J Hum Genet. 2012;91:215–223. doi: 10.1016/j.ajhg.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feero W, Guttmacher A, Manolio T. Genomewide association studies and assessment of the risk of disease. N Engl J Med. 2010;363:166–176. doi: 10.1056/NEJMra0905980. [DOI] [PubMed] [Google Scholar]
- Gilad Y, Pritchard J, Thornton K. Characterizing natural variation using next-generation sequencing technologies. Trends Genet. 2009;25:463–471. doi: 10.1016/j.tig.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics. 2002;18:337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- King C, Rathouz P, Nicolae D. An evolutionary framework for association testing in resequencing studies. PLoS Genet. 2010;6:e1001202. doi: 10.1371/journal.pgen.1001202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Leal S. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am J Hum Genet. 2008;83:311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen B, Browning S. A group wise association test for rare mutations using a weighted sum statistic. PLoS Genet. 2009;5:e1000384. doi: 10.1371/journal.pgen.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio T, Collins F, Cox N, Goldstein D, Hindorff L, Hunter D, McCarthy M, Ramos E, Cardon L, Chakravarti A, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J, Cardon LR, Phillips MS, Donnelly P. The effects of human population structure on large genetic association studies. Nat Genet. 2004;36:512–517. doi: 10.1038/ng1337. [DOI] [PubMed] [Google Scholar]
- Mathieson I, McVean G. Differential confounding of rare and common variants in spatially structured populations. Nat Genet. 2012;44:243–246. doi: 10.1038/ng.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenthaler S, Thilly W. A strategy to discover genes that carry multi-allelic or mono-allelic risk for common diseases: a cohort allelic sums test (CAST) Mut Res. 2007;615:28–56. doi: 10.1016/j.mrfmmm.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Neale B, Rivas M, Voight B, Altshuler D, Devlin B, Orho-Melander M, Kathiresan S, Purcell S, Roeder K, Daly M. Testing for an unusual distribution of rare variants. PLoS Genet. 2011;7:e1001322. doi: 10.1371/journal.pgen.1001322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peduzzi PN, Concato J, Kemper E, Holford T, Feinstein A. A simulation study of the number of events per variable in logistic regression analysis. J Clin Epidemiol. 1996;49(12):1373–1379. doi: 10.1016/s0895-4356(96)00236-3. [DOI] [PubMed] [Google Scholar]
- Price A, Patterson N, Plenge R, Weinblatt M, Shadick N, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Pritchard J, Donnelly P. Case-control studies of association in structured or admixed populations. Theor Popul Biol. 2001;60:227–237. doi: 10.1006/tpbi.2001.1543. [DOI] [PubMed] [Google Scholar]
- Pritchard J, Rosenberg N. Use of unlinked genetic markers to detect population stratification in association studies. Am J Hum Genet. 1999;65:220–228. doi: 10.1086/302449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich DE, Goldstein DB. Detecting association in a case-control study while correcting for population stratification. Genet Epidemiol. 2001;20:4–16. doi: 10.1002/1098-2272(200101)20:1<4::AID-GEPI2>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Teo Y. Common statistical issues in genome-wide association studies: a review on power, data quality control, genotype calling and population structure. Cur Opin Lipidol. 2008;19:133–143. doi: 10.1097/MOL.0b013e3282f5dd77. [DOI] [PubMed] [Google Scholar]
- Tintle N, Aschard H, Hu I, Nock N, Wang H, Pugh E. Inflated type I error rates when using aggregation methods to analyze rare variants in the 1000 genomes project exon sequencing data in unrelated individuals: summary results from group 7 at Genetic Analysis Workshop 17. Genet Epidemiol. 2011;35 doi: 10.1002/gepi.20650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Elston R. Improved power by use of a weighted score test for linkage disequilibrium mapping. Am J Hum Genet. 2007;80:353–360. doi: 10.1086/511312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]