Abstract
Tumor-infiltrating regulatory T cells (Tregs) promote immune evasion and are associated with poor disease outcome in patients affected by various malignancies. We have recently demonstrated that several, inherited single nucleotide polymorphisms affecting Treg-related genes influence the survival of ovarian cancer patients, providing novel insights into possible mechanisms of immune escape.
Keywords: CD134, CD80, GARP, GITR, LRRC32, OX40, RGS1, TNFRSF18, TNFRSF4, inherited variants, ovarian cancer, regulatory T cells
Despite considerable progresses in our understanding of the factors that influence ovarian cancer progression, the survival rate of patients affected by this deadly cancer has not yet appreciably improved. Multiple factors influence patient survival in this setting, including the histological subtype of the tumor, stage at diagnosis and genetic predisposition.1 It has become clear that different histological subtypes of ovarian cancer represent distinct diseases, bearing specific genetic lesions, responding differently to chemotherapy and exhibiting dissimilar survival rates. Many studies have focused on serous ovarian cancers or have grouped all other subtypes together, hence failing to take into account the heterogeneous nature of the disease and obscuring subtype-specific relevant results.1
Elevated levels of tumor-infiltrating T cells also influence the survival of ovarian cancer patients. In particular, tumor infiltration by cytotoxic CD8+ T cells has been associated with improved survival, whereas increased numbers of regulatory T cells (Tregs) have been associated with dismal prognosis.2,3 Tumors halt immune responses via several pathways including the induction and recruitment of Tregs. In turn, Tregs block immune effectors through several mechanisms, including the secretion of soluble mediators such as interleukin (IL)-10 and transforming growth factor β (TGFβ), the induction of T-cell anergy, the depletion of IL-2 (which is needed for the survival, proliferation and differentiation of naïve T cells) and the conversion of naïve T cells into induced Tregs (iTregs).4
Based on evidence suggesting that tumor-infiltrating Tregs influence the survival of ovarian cancer patients, we reasoned that common, inherited single nucleotide polymorphisms (SNPs) affecting genes involved in the regulation of Treg functions may also be associated with disease outcome in this setting. To assess this issue, we genotyped > 1,500 SNPs relevant to 54 genes related to Tregs using germline DNA isolated from the blood of women affected by primary epithelial ovarian, peritoneal or fallopian tube cancer, representing all the major histological subtypes of ovarian neoplasms. Over 3,000 individuals were genotyped at SNPs which either (1) tagged underlying variations in or (2) were known to correlate with the expression of genes relevant for Treg induction, functions or trafficking. Additionally, we correlated tumor mRNA expression data with genotype in a subset of cases, in order to determine the association between survival-linked SNPs and gene expression at the tumor level.5
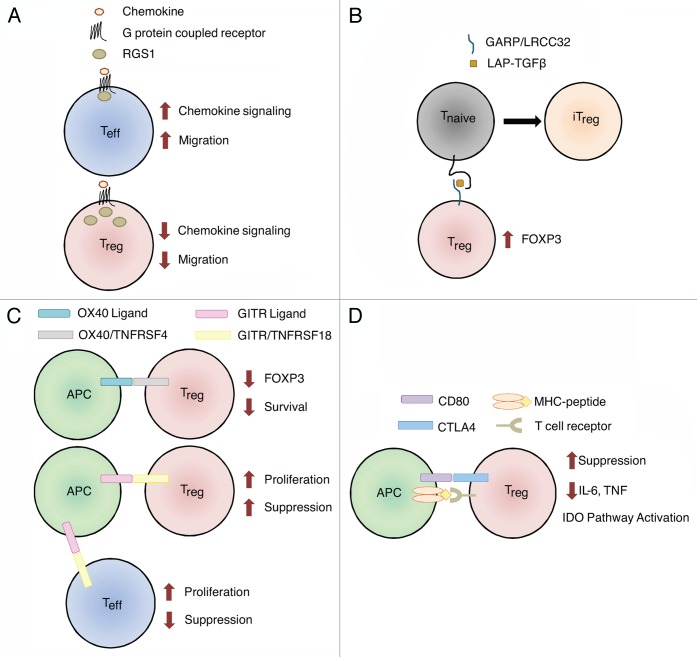
We identified six independent SNPs (R2 < 0.3) in four chromosomal regions associated with ovarian cancer patient survival. The strongest association was between the RGS1 gene and the survival of clear cell carcinoma patients (p = 2.7 × 10−5). The minor allele of an intronic SNP, rs10921202, was indeed associated with a 2.93-fold increased risk of death. Variants in RGS1 may influence the trafficking of Tregs and other immune cells by limiting the duration of G-protein signaling (Fig. 1A).6 Lymphocyte migration is indeed mediated by chemotactic factors that operate via G-protein-coupled receptors, and mutations in these systems may perturb the influx/efflux of immune cells into/from the tumor microenvironment.
Figure 1. Genes with variants associated with ovarian cancer survival. Normal functions of genes involved in the biology of regulatory T cells (Tregs) are shown. (A) Tregs express more RGS1 than effector T cells (Teffs), resulting in reduced chemotactic migration. SNPs in RGS1 may increase the responsiveness of Tregs to chemotactic signals, allowing them to accumulated within tumors and to establish an immunosuppressive microenvironment. (B) Garpin (GARP) is a surface-bound receptor for latency-associated peptide (LAP)-bound transforming growth factor β (TGFβ). TGFβ-bound GARP can promote the acquisition of a regulatory phenotype by naïve T cells (Tnaïve) cells, hence converting them in induced Tregs (iTregs) and upregulate the transcription factor FOXP3 in Tregs. SNPs in LRRC32 (encoding GARP) may increase its expression levels, stability or activation status, resulting in the propagation of an immunosuppressive phenotype in the microenvironment of mucinous tumors. (C) The ligation of OX40 on the surface of Tregs results in decreased FOXP3 expression and limits their survival. SNPs in TNFRSF4 (encoding OX40) may abrogate this process, resulting in increased Treg survival and functions in mucinous tumors. GITR ligation may affect Tregs directly or indirectly. The ligation of GITR expressed on Tregs increases their proliferation and immunosuppressive functions. The activation of GITR expressed on effector T cells also increases their proliferation, which in turn can prevent the expansion of Tregs. SNPs in TNFRSF18 (encoding GITR) may increase GITR levels or activity, amplifying the immunosuppressive phenotype of mucinous tumors. Alternatively, SNPs in TNFRSF18 may reduce downstream Teff functions, resulting in decreased tumor clearance. (D) CD80 expressed on antigen-presenting cells (APCs) can bind CTLA4 on the surface of Tregs, triggering multiple mechanisms of immunosuppression, including a decreased expression of pro-inflammatory cytokines and the activation of the indoleamine 2,3-dioxygenase (IDO) pathway. SNPs affecting CD80 may increase the affinity of CD80 for CTLA4 in endometrioid tumors, while SNPs in MAD1L1 (which are linked to CD80 expression), may influence the CD80 levels in ovarian cancer patients at large.
Three SNPs were associated with poor survival in patients affected by mucinous ovarian cancers. The first two were located in the 3′ UTR and in an intron of LRRC32 (rs3781699 and rs7944357) and resulted in a more than 2-fold increased risk of death. rs3781699 and rs7944357 were modestly correlated with each other (R2 = 0.26). LRRC32 encodes garpin (GARP), the receptor for latency-associated peptide (LAP)-bound TGFβ on activated Tregs. GARP is part of a positive feedback loop involving FOXP3 that contributes to the maintenance of an immunosuppressive microenvironment. Furthermore, LAP-bound TGFβ on the surface of Tregs can stimulate the acquisition of immunosuppressive functions by naïve T cells via infectious tolerance (Fig. 1B).7
The third SNP associated with mucinous cancer patient survival was an intergenic SNP (rs3753348) located between TNFRSF4 (encoding OX40) and TNFRSF18 (encoding GITR), which was associated with a 3.41-fold increased risk of death. This SNP tagged variations in both genes. OX40 signaling results in decreased expression of FOXP3 by Tregs and increases the amount of IL-2 that is required for their survival. Thus, OX40 can modulate both Treg survival and immunosuppressive functions.8 The role of GITR in the biology of Tregs remains a matter of debate, with reports supporting positive as well as negative regulatory functions.9 Functional variants or OX40 or GITR, however, might significantly influence both the survival and the immunosuppressive functions of Tregs (Fig. 1C).
In addition, CD80 was of particular interest. First, MAD1L1 SNP rs7804190 was genotyped because other studies have shown a correlation between this SNP and CD80 expression. rs7804190 was associated with a 1.14-fold increased risk in death for patients affected by all ovarian cancer subtypes. Second, an intronic SNP (rs13071247) affecting CD80 itself was associated with a 1.73-fold increased risk of death among endometrioid ovarian cancer patients. CD80 can exert multiple biological functions, depending on both the receptor it binds to and the cell type expressing such receptor (Fig. 1D). Tregs constitutively express CTLA4, one of the CD80 receptors. When activated, CTLA4 mediates immunosuppressive functions by promoting the translocation of the FOXO3 transcription factor to the nucleus, in turn repressing the expression of IL-6 and tumor necrosis factor α (TNFα), and by activating the indoleamine 2,3-dioxygenase (IDO) pathway, hence depleting extracellular tryptophan and stimulating anergy in naïve T cells.10
Understanding the mechanisms that underpin these associations remains a challenge. Only one of these SNPs, the intronic CD80 SNP, was associated with mRNA expression in ovarian tumors. In particular, rare alleles at rs13071247, which were associated with poor patient survival, resulted in an increased expression of the CD80-coding mRNA, perhaps boosting CTLA4 signaling and promoting immunosuppression. Causality however cannot be assumed and whether the assessed SNPs or nearby, correlated SNPs are responsible for the reduction in patient survival must be examined by precise mechanistic studies. Further investigation is also needed to clearly delineate which effects these SNPs have on the induction, function, and trafficking of Tregs.
In summary, our study highlights the importance of investigating ovarian cancer subtypes as distinct diseases. Indeed, we identified only one SNP that was associated with survival in patients affected by all the histological subtypes of the disease, while the association with patient survival for other five SNPs was limited to a single disease subtype. Thus, the ability of the immune system to respond to ovarian tumors may differ with histological subtype. Determining which genes are important for the immune response to each ovarian cancer subtype may be required for the development of precisely tailored immunotherapies.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/oncoimmunology/article/24535
References
- 1.Vaughan S, Coward JI, Bast RC, Jr., Berchuck A, Berek JS, Brenton JD, et al. Rethinking ovarian cancer: recommendations for improving outcomes. Nat Rev Cancer. 2011;11:719–25. doi: 10.1038/nrc3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sato E, Olson SH, Ahn J, Bundy B, Nishikawa H, Qian F, et al. Intraepithelial CD8+ tumor-infiltrating lymphocytes and a high CD8+/regulatory T cell ratio are associated with favorable prognosis in ovarian cancer. Proc Natl Acad Sci U S A. 2005;102:18538–43. doi: 10.1073/pnas.0509182102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Curiel TJ, Coukos G, Zou L, Alvarez X, Cheng P, Mottram P, et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat Med. 2004;10:942–9. doi: 10.1038/nm1093. [DOI] [PubMed] [Google Scholar]
- 4.Janikashvili N, Bonnotte B, Katsanis E, Larmonier N. The dendritic cell-regulatory T lymphocyte crosstalk contributes to tumor-induced tolerance. Clin Dev Immunol. 2011;2011:430394. doi: 10.1155/2011/430394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goode EL, DeRycke M, Kalli KR, Oberg AL, Cunningham JM, Maurer MJ, et al. Inherited variants in regulatory T cell genes and outcome of ovarian cancer. PLoS One. 2013;8:e53903. doi: 10.1371/journal.pone.0053903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hollinger S, Hepler JR. Cellular regulation of RGS proteins: modulators and integrators of G protein signaling. Pharmacol Rev. 2002;54:527–59. doi: 10.1124/pr.54.3.527. [DOI] [PubMed] [Google Scholar]
- 7.Andersson J, Tran DQ, Pesu M, Davidson TS, Ramsey H, O’Shea JJ, et al. CD4+ FoxP3+ regulatory T cells confer infectious tolerance in a TGF-β-dependent manner. J Exp Med. 2008;205:1975–81. doi: 10.1084/jem.20080308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.So T, Lee S-W, Croft M. Immune regulation and control of regulatory T cells by OX40 and 4-1BB. Cytokine Growth Factor Rev. 2008;19:253–62. doi: 10.1016/j.cytogfr.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walker LSK. Regulatory T cells overturned: the effectors fight back. Immunology. 2009;126:466–74. doi: 10.1111/j.1365-2567.2009.03053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen L. Co-inhibitory molecules of the B7-CD28 family in the control of T-cell immunity. Nat Rev Immunol. 2004;4:336–47. doi: 10.1038/nri1349. [DOI] [PubMed] [Google Scholar]