Abstract
Deposits of fibrils formed by disease-specific proteins are the molecular hallmark of such diverse human disorders as Alzheimer's disease, type II diabetes, or rheumatoid arthritis. Amyloid fibril formation by structurally and functionally unrelated proteins exhibits many generic characteristics, most prominently the cross β-sheet structure of their mature fibrils. At the same time, amyloid formation tends to proceed along one of two separate assembly pathways yielding either stiff monomeric filaments or globular oligomers and curvilinear protofibrils. Given the focus on oligomers as major toxic species, the very existence of an oligomer-free assembly pathway is significant. Little is known, though, about the structure of the various intermediates emerging along different pathways and whether the pathways converge towards a common or distinct fibril structures. Using infrared spectroscopy we probed the structural evolution of intermediates and late-stage fibrils formed during in vitro lysozyme amyloid assembly along an oligomeric and oligomer-free pathway. Infrared spectroscopy confirmed that both pathways produced amyloid-specific β-sheet peaks, but at pathway-specific wavenumbers. We further found that the amyloid-specific dye thioflavin T responded to all intermediates along either pathway. The relative amplitudes of thioflavin T fluorescence responses displayed pathway-specific differences and could be utilized for monitoring the structural evolution of intermediates. Pathway-specific structural features obtained from infrared spectroscopy and Thioflavin T responses were identical for fibrils grown at highly acidic or at physiological pH values and showed no discernible effects of protein hydrolysis. Our results suggest that late-stage fibrils formed along either pathway are amyloidogenic in nature, but have distinguishable structural fingerprints. These pathway-specific fingerprints emerge during the earliest aggregation events and persist throughout the entire cascade of aggregation intermediates formed along each pathway.
INTRODUCTION
Physical and biophysical research on the thermodynamic and kinetics of protein phase separation phenomena provides important insight into protein self-assembly from molecular all the way to system level length-scales, both in healthy and disease states. An intriguing recent example highlighting the interplay of protein phase separation and cell biology involved “esoteric” liquid-liquid protein demixing. Metastable liquid-liquid demixing of proteins was originally investigated experimentally and theoretically in the context of supramolecular self-assembly of proteins, protein crystallization, and hemoglobin sickle cell polymerization,1 with the latter two processes representing important protein phase separation phenomena in their own right. It has now been reported that liquid-liquid demixing of proteins in cells might be actively induced via regulated polymerization of ubiquitous multivalent signaling proteins and their interaction partners.2 Demixing thereby initiated spatially scaled-up cellular responses and promoted rapid assembly of highly concentrated signaling complexes. In fact, protein phase separation has been invoked as mechanism underlying the formation of various subcellular foci and puncta.3
The emphasis of this paper is on protein self-assembly during amyloid fibril formation. Amyloid fibril deposits have been linked to an ever-increasing variety of human pathologies ranging from Alzheimer's disease over type II diabetes to rheumatoid arthritis.4, 5 Amyloid fibril formation can also be biologically functional such as in extracellular adhesion of bacteria, transmission of inheritable traits in yeast,6 or tight packing of peptide hormones into electron-dense secretory granules.7 Unraveling the mechanisms driving amyloid fibril formation and determining the features that render this process either pathogenic or functional represents a significant scientific challenge. Amyloid fibrils are non-branching polymeric aggregates of proteins that can reach many microns in length and, typically, are stained by the dyes Congo Red or Thioflavin T. Besides these morphological and tinctorial features, amyloid fibrils share a characteristic cross-β sheet structure in which at least portions of the individual polypeptide backbones of the proteins are aligned perpendicular to the main fibril axis and are linked to each other via intermolecular hydrogen bonds.8 The structural and morphological similarities of amyloid fibrils are in stark contrast to the wide variety of functionally and structurally distinct proteins that can form amyloid fibrils.5, 9 This has lead to the supposition that amyloid fibril self-assembly is yet another example of a generic protein phase separation process.10 There is, however, an important difference between amyloid fibril formation and “standard” protein phase separation phenomena such as crystallization and liquid-liquid demixing. While the latter mostly preserve the native protein structure, the natively folded or intrinsically disordered protein needs to undergo significant restructuring in order to assume the cross-β sheet structure of the amyloid fibril state.11 How these conformational constraints on aggregation affect the propensity of a given protein to form amyloid fibrils, when and how such restructuring occurs, and whether restructuring precedes or is induced by the formation of specific aggregation intermediates remains an open question.
Related to the above questions surrounding structural evolution is another aspect of amyloid fibril growth: fibril formation tends to proceed along one of two distinct assembly pathways characterized by populations of different intermediates with distinct morphologies. One pathway proceeds via the nucleation and cross-assembly of long, straight filaments. A second pathway produces comparatively stable globular intermediates, called oligomers, which later on assemble into curvilinear polymers referred to as either protofibrils or worm-like fibrils. The oligomer-free and oligomeric pathways have been reported during in vitro fibril growth of various structurally and functionally distinct amyloid proteins.12, 13, 14, 15 In light of the critical role ascribed to oligomeric intermediates as mediator of cellular toxicity, the very existence of an oligomer-free assembly pathway is noteworthy.16 One contentious issue is whether oligomer formation is actually off-pathway or on-pathway towards amyloid fibril formation.17 The existence of two pathways also raises the question whether and to what extent the internal structures of their respective intermediates are distinct and whether structural evolution proceeds differently along the oligomer-free vs. oligomeric assembly pathway.
We set out to examine the structure and structural evolution of early-stage intermediates and late-stage amyloid fibrils formed during in vitro self-assembly of hen egg white lysozyme. We have previously shown that fibril-formation of hen egg white lysozyme can proceed along either an oligomer-free or an oligomeric assembly pathway.15 To probe the structural evolution of intermediates formed during in vitro fibril formation we chose a combination of attenuated total reflection Fourier-transform infrared spectroscopy (ATR-FTIR) and normalized Thioflavin T (ThT) fluorescence responses. FTIR spectra from various amyloid fibrils display characteristic β-sheet peaks near 1620 cm−1 wavenumber that are distinct from the range of peak wavenumbers for native β-sheet folds.18 Since the Amide-I region of the FTIR spectrum for native lysozyme is dominated by a prominent α-helix peak near 1655 cm−1, lysozyme represents a particularly favorable system for resolving structural features emerging within the amyloid-specific region around 1620 cm−1. It is well-known that the kinetics signatures obtained from the amyloid indicator dye thioflavin T along the oligomer-free vs. oligomeric assembly pathway are distinct. Here, we investigated whether ThT fluorescence responses contained information about the structure and structural evolution of amyloidogenic intermediates as well. Structural features derived from FTIR and ThT measurements were correlated against each other, and against the underlying composition of intermediates in these samples, obtained independently from static and dynamic light scattering and from atomic force microscopy.
MATERIALS AND METHODS
Protein and chemicals
Two times recrystallized, dialyzed, and lyophilized hen egg white lysozyme (HEWL) was purchased from Worthington Biochemicals (Lakewood NJ) and was used for all experiments. Ultrapure grade thioflavin T was obtained from Anaspec (Freemont, CA). All other chemicals were purchased from Fisher Scientific (Pittsburgh, PA) and were reagent grade or better. All solutions were prepared using 18 MΩ water from a reverse osmosis unit (Barnstead E-pure, Dubuque, IA).
Preparation of HEWL growth solutions and ThT solutions
HEWL was dissolved at twice its final concentration in either 25 mM KH2PO4 at pH 2 or 20 mM HEPES at pH 7 buffer and was briefly placed in a water bath at 45 °C to help dissolve preformed clusters.19 Samples were consecutively filtered through 220-nm and 50-nm pore size syringe filters. This concentrated HEWL stock was mixed 1:1 either with a NaCl /25 mM KH2PO4 pH 2 or with a NaCl /20 mM HEPES pH 7 stock solutions, with NaCl concentrations in this salt/buffer stock adjusted to twice their final concentrations. Actual lysozyme concentrations were determined from UV absorption measurements at 280 nm (a280 = 2.64 ml mg−1 cm−1). These solutions were placed in standard glass cuvettes (Starna Cell, Atascadero, CA) and were incubated at 50 °C (pH 2) or 65 °C (pH 7) for as little as 4 h and as long as 5 days using a dry bath or the themostated cuvette holder of a dynamic light scattering unit (Zetasizer S, Malvern Instruments, Worcestershire, UK). For accelerated growth of monomeric fibrils at pH 2, fibril seeds were grown at pH 2 in 100 mM NaCl for 2.5 days. A freshly prepared lysozyme solution was then mixed with 3% of the seeding solution and was incubated for 24 h under the same growth conditions. Accelerated growth of oligomeric samples within just hours, in turn, was achieved by raising NaCl concentrations to 325 mM.
ThT stock solutions were prepared by dissolving 1 mM dye in distilled water and passing them through 220-nm syringe filters. Actual ThT concentrations were determined from absorption at 416 nm (α416 = 26 620 ml mg−1 cm−1). These solutions were further diluted to a final stock concentration of 212 μM.
Static and dynamic light scattering
Static and dynamic light scattering (SLS and DLS) measurements were performed using the back scattering geometry (θ = 173°) of a Zetasizer Nano S (Malvern Instruments, Worchestershire, UK) with a 4 mW He-Ne laser (λ = 633 nm) and a temperature-controlled sample compartment. During in situ measurements of fibril growth, autocorrelation functions of scattered light were collected typically every 10–30 min, using acquisition times of 180 s per measurement. Autocorrelation functions were converted into particle-size distributions (PSDs) using the “narrow modes” or “general purpose” algorithms provided with the Zetasizer Nano S. Changes in scattering intensity were monitored simultaneously using the count rates of the avalanche photodiode photon detector.
FTIR spectroscopy of lysozyme solutions
ATR-FTIR spectroscopy was performed on a Bruker Optik Vertex 70 (Ettlingen, Germany) spectrometer with a mid-infrared source and pyroelectric DLATGS (deuterated L-alanine doped triglycene sulphate) room temperature detector. Usually, 30–40 μL of protein solution was placed on the thermostated silicon crystal of a BioATRcell II (Harrick Scientific Products, Inc.; Pleasantville, New York) ATR accessory. FTIR spectra were acquired between 1000 cm−1 and 4000 cm−1 wavenumbers. All spectra were taken at 24 °C with an aperture setting of 8 mm and a scanner velocity of 10 kHz. Water vapor contamination of the spectra was minimized using the atmospheric compensation algorithm within the OPUS software analysis package (version 6.5, Bruker Optik). Background spectra of the buffer solution without protein were recorded over 200–400 scans at 2 cm−1 resolution and subtracted from the sample spectra. Typically, three such runs were averaged prior to data analysis.
The Amide I and Amide II bands (1500–1700 cm−1) were subjected to a combined peak and deconvolution analysis. Peak positions in the Amide I band of the spectra were identified using the Fourier Self-deconvolution (FSD) (Bandwidth 6 cm−1; Enhancement 2.4) and second derivative (13 smoothing points) algorithms within the OPUS software analysis package (version 6.5, Bruker Optik). After a horizontal baseline correction, Gaussian curves were fitted from 1550 cm−1 to 1700 cm−1. We accounted for the overlap of the Amide I and Amide II band by fitting the entire Amide I band together with the high-frequency portion of the Amide II band. Peak positions were fixed to the values identified by FSD and second derivative algorithms while the intensity and widths of the Gaussian curves were optimized using the Levenberg-Marquardt Algorithm. To avoid unreasonable broadening of Gaussian peaks observed for some spectra, peak widths were restricted to values near those obtained with native lysozyme. For comparing spectra from different fibril samples (Fig. 3a), baseline corrected spectra were normalized to the integral over their Amide I bands. Difference spectra were obtained by subtracting the normalized reference spectrum of monomeric lysozyme from the spectra of the aggregated samples (Fig. 3b). These spectra were renormalized by dividing each by the values of their respective β-sheet peaks. For measurements of the temporal evolution of the FTIR difference spectra (Fig. 3c) sample aliquots were removed at different times (4 h, 1 day, 4 days, 7 days, 11 days) and the initial monomer FTIR spectrum at 25 °C was used for subtraction.
Figure 3.
Difference spectra for lysozyme solution undergoing oligomeric fibril growth. (a) Renormalized amide I peak for monomeric lysozyme at pH 2 (solid black curve) and for late-stage fibrils grown along either the oligomer-free (solid red curve) or oligomeric pathway (solid blue curve). (b) Corresponding FTIR difference spectra of the Amide I region for oligomer-free fibrils (red curves) and for oligomeric protofibrils (blue curves) at pH = 2.0. The two difference spectra for oligomer-free fibril growth are for samples either incubated for multiple days (solid red curve) or derived from rapid growth (<12 h) via seeding with pre-formed fibrils (short-dashed red curve). Similarly, oligomeric difference spectra are shown for growth with 175 mM NaCl (solid blue curve), during rapid growth (<3 h) in 325 mM NaCl (short-dashed blue curve), or for growth at pH 7/ 65 °C (blue dotted curve) (c) Temporal evolution of the Amide I difference spectrum for lysozyme undergoing oligomeric fibril growth.
Thioflavin T fluorescence spectroscopy
ThT fluorescence measurements were performed using a FluoroMax-4 spectrofluorometer (Horiba Jobin Yvon, Edison, NJ). ThT fluorescence was excited at 445 nm and emission spectra were measured between 465 nm and 565 nm. For each ThT fluorescence measurement, small sample aliquots were removed from the HEWL growth solution and were mixed with the ThT stock solution. In order to minimize potential changes to the particle size distribution arising from dilution during ThT measurements, only 3.3 μl of the 212 μM ThT stock solution was added directly to 66.7 μl of the undiluted lysozyme aliquots, resulting in a final ThT concentration of 10 μM. ThT/HEWL mixtures were transferred into small volume quartz cuvettes and allowed to equilibrate for 5 min before recording ThT fluorescence emission. For each data point, at least two separate ThT/HEWL solutions were prepared and their fluorescence analyzed. Off-line ThT fluorescence measurements were taken at regular intervals (typ. 2-6 times/day) while light scattering was monitored in situ as described above. Correlated DLS/SLS and ThT measurements ceased when light scattering indicated the onset of sample gelation. All ThT measurements were taken at room temperature. Using DLS we confirmed that the addition of ThT, the minimal dilution of the samples for ThT measurements, or the lower temperature used during fluorescence measurements did not noticeably alter the size distribution of HEWL aggregates.
Atomic force microscopy
Amyloid fibrils were imaged in air with a MFP-3D atomic-force microscope (Asylum Research, Santa Barbara, CA) using NSC36/NoAl (Mikromasch, San Jose, CA) or PFP-FMR-50 (Nanosensor, Neuchatel, Switzerland) silicon tips with nominal tip radii of 10 nm and 7 nm, respectively. The cantilever was driven at 60–70 kHz in alternating current mode and at a scan rate of 0.5 Hz, acquiring images at 512 × 512 pixel resolution. Raw image data were corrected for image bow and slope. For imaging, 75 μl of fibril solution was diluted 20 to 100-fold into the same salt/buffer combination used during growth, deposited onto freshly cleaved mica for 3–5 min, rinsed with deionized water, and dried with dry nitrogen. Amplitude and phase images as well as height images were collected for the same sample area. False-color height images were subsequently superimposed over either amplitude or phase images.
Gel electrophoresis
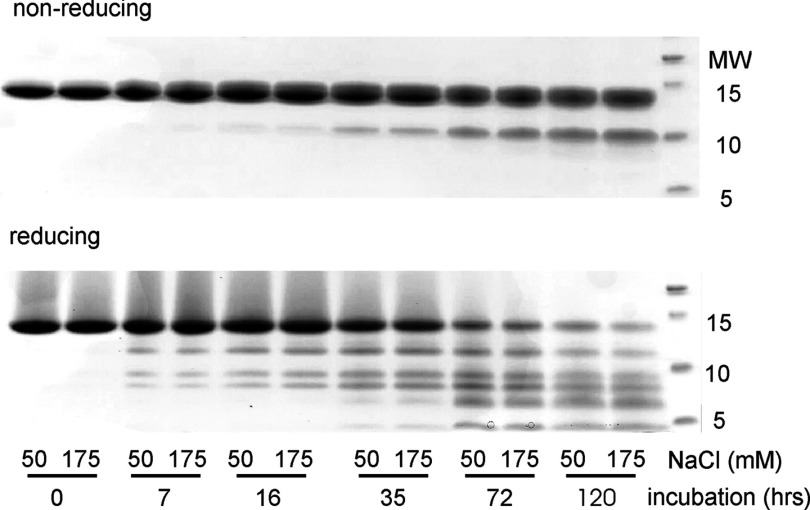
HEWL samples were analyzed using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS PAGE) using 10–20% gradient Tris-tricine gels (Criterion, Bio-Rad) and a sodium dodecyl sulfate (SDS) running buffer without glycine. An aliquot of 20 μg was mixed with Laemmli sample buffer (Bio-Rad) with or without reducing agent (β-mercapto-ethanol). Samples were boiled for 5 min, loaded onto precast gels, and stained with BioSafe Coomassie (Bio-Rad) per manufacturer's instructions. For assessing the extent and time course of lysozyme hydrolysis we incubated 1 mg/ml lysozyme solutions at pH 2.0 and 50 °C for 0 to 120 h under either oligomer-free filamentous (50 mM NaCl) or oligomeric (175 mM) fibril growth conditions. The distribution of lysozyme fragments in solution and the extent of overall hydrolysis were determined using both non-reducing and reducing SDS gel electrophoresis.
RESULTS
HEWL is a small enzyme of 14.3 kD molecular weight with well-defined biochemical and physical characteristics. Point mutations of lysozyme underlie non-neuropathic systemic forms of human amyloidosis, with dangerous disruptions to kidney and liver function.20 The amino acids mutations implicated in human lysozyme amyloidosis occur in regions that are highly homologous to hen egg-white lysozyme.21 Amyloid fibrils of lysozyme readily form in vitro in acidic solutions and at elevated temperatures.22 For most experiments described below amyloid fibril growth was initiated by incubating 1.4 mM of lysozyme at pH 2.0 and 50 °C, with incubation periods ranging from 3–4 h up to several days. To control for potential effects of harsh growth conditions on structural features, we also investigated fibril structures generated using accelerated seeding protocols or grown at physiological pH values.
Morphological features of intermediates for oligomer-free vs. oligomeric fibril assembly
A detailed description of the two fibril assembly pathways of lysozyme and their intermediates was presented in Ref. 15. A brief synopsis is provided here to aid the subsequent discussion of structural features. Lysozyme incubated at pH 2 reproducibly follows either an oligomer-free filamentous or oligomeric fibril assembly pathways as a function of salt concentration.15 Fibril formation at low salt concentrations (≤150 mM) proceeds in the absence of any detectable oligomeric intermediates. In this pathway, a lag period lasting days is followed by a prominent nucleation event which results in the formation of monomeric filaments of two distinct lengths. Three of these monomeric filaments can later cross-assemble into thicker fibrils. An atomic force microscopy image taken during the late-stages of this fibril growth pathway (Fig. 1a) shows the presence of both short and long monomeric filaments among the taller cross-assembled mature fibrils.
Figure 1.
AFM images of monomeric filaments, oligomers, and protofibrils. Superposition of AFM phase and height images of various intermediates and late stage aggregates of lysozyme amyloid growth. (a) Mixture of monomeric filaments (orange) and taller, wider mature fibrils (green) grown in 50 mM NaCl at pH = 2 and T = 50 °C. Both fibril populations are very straight and mechanically stiff. (b) Early-stage oligomeric intermediates (orange/green) and (c) late stage protofibrils (green) obtained during incubation at 175 mM NaCl. Protofibrils are much more curvilinear despite being noticeably thicker than the stiff monomeric filaments shown in (a). A detailed analysis of all intermediates and their morphological features were presented in Ref. 15. Scale bars represent 1 μm in (a) and (c) and 250 nm in (b). False color height scales are identical in all images.
As ionic strength exceeds 150 mM, assembly abruptly switches over to oligomeric fibril growth. In this latter pathway, oligomers composed of about eight monomers form immediately and at a steady rate (Fig. 1b), which eventually results in nucleation of oligomeric protofibrils. As shown in Fig. 1c, the protofibrils obtained under oligomeric growth conditions develop highly curvilinear geometries, particularly during the later stages of growth. We have previously shown that the cross-sectional area of the initial low-salt filaments agree with that of lysozyme monomers while protofibrils nucleating under oligomeric growth conditions have cross-sectional areas matching those of their oligomeric precursors.15 As protofibrils increase in length they occasionally cross-assemble pairwise into mature fibrils. Aside from the above differences both pathways involve a prominent nucleation event (monomers to monomeric filaments or oligomers to oligomeric protofibrils) and develop cross-linked fibril populations via cross-assembly of either three monomeric filaments or two oligomeric protofibrils.
Infrared spectroscopy (ATR-FTIR) of intermediates in the oligomer-free vs. oligomeric fibril assembly pathways
Figure 2 displays the Amide I peak region of the FTIR spectra for monomeric lysozyme (Fig. 2a) together with the corresponding spectra for late stage fibrils grown in the presence of either 50 mM NaCl (Fig. 2b, oligomer-free assembly) or 175 mM NaCl (Fig. 2c, oligomeric assembly). To reduce contributions from lysozyme monomers to fibril spectra, fibril samples were subjected to ultracentrifugation and the resulting pellets were resuspended prior to measurement. When inspecting the raw spectra even prior to any quantitative analysis several general trends emerge. Most noticeable, both oligomer-free and oligomeric fibril assembly resulted in the formation of a prominent β-sheet peak around 1620 cm−1. This indicates that both pathways produce β-sheet structures considered diagnostic of amyloid fibril formation.18 In addition, the native α-helix content of the fibril spectra did not disappear. Dynamic light scattering measurements performed on the re-suspended fibril pellets confirmed that the residual monomeric content in these pellets was drastically reduced. Hence, the persistent α-helical peak in the Amide-I band is not due to contributions from monomers but implies that lysozyme fibrils preserved secondary structure elements from their native state. This observation agrees with NMR structure determinations for mature lysozyme fibrils obtained under comparable growth conditions.23 Finally, the region of the Amide-I peak between 1700 and 1660 cm−1 was typically more elevated for late-stage protofibrils than for the oligomer-free filaments and fibrils.
Figure 2.
Infrared spectra of late stage fibrils in either pathway show characteristic β-sheet peaks. Amide I peak for (a) native lysozyme in buffer at pH 2.0, and for mature fibrils analyzed after ultracentrifugation and incubation under (b) oligomer-free or (c) oligomeric growth conditions. Black lines represent the raw spectra while gray lines indicate the gaussian decomposition of the spectra into α-helix, β-turn, and β-sheet peaks as well as the resulting overall fit to the raw spectra. The bar graphs to the right of each spectrum represent the relative amplitudes of the α-helix and β-sheet peaks derived from spectral decomposition.
We decomposed the raw spectra (solid lines) into individual gaussian peaks (gray lines) by simultaneously fitting the Amide-I and Amide-II peaks using second-derivatives and direct fits (see Materials and Methods section for details). The relative peak area for the α-helix peak (1655 cm−1) and amyloid β-sheet peak (∼1620 cm−1) shown next to each spectrum recapitulates the main feature already seen in the raw spectra: fibril formation in either pathway induces a significant decrease in the α-helix content and concurrent rise of the amyloid-specific β-sheet region. We also observed that the β-sheet peak for fibrils grown in the oligomer-free pathway occurred at a slightly lower wavenumber when compared to the corresponding peak for the protofibril pathway (see Figs. 2b, 2c).
To confirm this pathway-specific difference in the β-sheet position we generated difference spectra of the fibril FTIR spectra and their corresponding monomer spectra, collected under the same solution conditions. Figure 3a shows a superposition of the area-normalized FTIR spectra for monomers, late-stage “oligomer-free” fibrils, and ‘oligomeric” fibrils while Fig. 3b displays the corresponding difference spectra matched to the maxima of their amyloid β-sheet peak (for details of the normalization and subtraction procedure see Materials and Methods section). These difference spectra directly yield the peak position for the emerging amyloid β-sheet peak without the detour over spectral decomposition (and its potential artifacts). As seen in Fig. 3b, the amyloid β-sheet peaks differ by 5 cm−1, with the peak for oligomer-free assembly developing at (1623 ± 1) cm−1, while oligomeric assembly induced a peak at (1618 ± 1) cm−1. The additional difference spectra in Fig. 3b were obtained from samples subjected to accelerated fibril growth or from fibrils grown at pH 7. For all these different growth conditions the difference spectra yielded subtle but reproducible structural differences in the FTIR spectra of oligomer-free filaments and oligomeric protofibrils which, in turn, correlated with the pronounced differences in their respective fibril morphologies.
Structural evolution along the oligomeric fibril assembly pathway
As mentioned in the Introduction, it remains unclear at what stage of the assembly process the characteristic amyloid β-sheet structure emerges. We, therefore, decided to follow structural evolution along the oligomer assembly pathway in which oligomeric intermediates begin to form without lag phase while protofibrils only nucleate after several hours of incubation. DLS provided a direct read-out of the lag period and at what moment protofibrils nucleated. Analyzing FTIR spectra collected at different time points during fibril assembly was challenging since the monomeric contributions to the FTIR spectra kept changing with the sample-specific ratio of aggregates to monomers. Using FTIR difference spectra reliable removed this monomeric background and did not require any assumptions that were intrinsic to decomposition analysis. Figure 3c displays the resulting difference Amide-I spectra as a function of incubation period for lysozyme samples undergoing oligomeric fibril growth. While the number of time points was limited, the overall trend is obvious: The main structural features observed during late stages of fibril growth, i.e., the loss of α-helix content and the emergence of the amyloid-specific β-sheet peak around 1620 cm−1, emerged even during the lag phase dominated by oligomer formation. Only the amplitude of these two features of the difference spectra increased as incubation progressed and oligomers became superseded by protofibrils. The spectra also suggested that the region around 1720–1680 cm−1 undergoes structural evolution during the aggregation process. We did not systematically explore this feature due to the intrinsic noise limitations in the data.
Pathway-specific structural features are independent of growth conditions or lysozyme hydrolysis
Under the acidic solution conditions commonly used for in vitro fibril growth, lysozyme can be subject to concurrent hydrolysis. For fibril growth at pH 1.6 and 65 °C, lysozyme hydrolysis was shown to accelerate fibril formation.24 Exposure of mature fibril to acid pH during extended incubations caused portions of the native structure to be trimmed off the fibrils.23 We, therefore, investigated whether the existence of the oligomer-free vs. oligomeric pathway, or the specific features of their FTIR spectra shown here, were affected by lysozyme hydrolysis. To test for the extent of hydrolysis, we incubated lysozyme under the acid growth conditions used in our experiments (pH 2, T = 50 °C) and at two salt concentrations inducing either oligomer-free filamentous (50 mM NaCl) or oligomeric (175 mM) fibril growth. Lysozyme concentrations were kept below 1 mg/ml in order to suppress aggregate formation. Aliquots from both solutions were collected at a series of time points and the amount of lysozyme hydrolysis was assessed with SDS-PAGE gel electrophoresis under both non-reducing and reducing conditions. As indicated in Fig. 4 lysozyme hydrolysis was minimal during the initial 24 h of incubation, but became significant after 3–5 days. The rate of hydrolysis was independent of salt concentrations and, therefore, the assembly pathway. Hence, lysozyme hydrolysis does not correlate with the sharp, salt-mediated transition from oligomer-free to oligomeric fibril assembly.
Figure 4.
Rate of lysozyme hydrolysis during oligomer-free vs. oligomeric fibril growth. Lysozyme monomers (1 mg/ml) were incubated for a total of 120 h under either oligomer-free (50 mM NaCl) or oligomeric (175 mM NaCl) fibril growth conditions (pH 2, 507 °C). Aliquots were removed at various time points and analyzed with SDS PAGE electrophoresis under either (top) non-reducing or (bottom) reducing conditions (1 μl β-mercaptoethanol).
We further explored whether the incorporation of hydrolyzed or nicked material accumulating during extended incubation might affect features of the FTIR spectra. Since lysozyme hydrolysis was negligible during the initial 24 h of incubation (Fig. 4), we grew fibrils within this shortened time-frame and analyzed the corresponding IR spectra. For oligomeric fibrils, increasing salt concentration to 325 mM generated fibril material for FTIR analysis within 4 h. For the oligomer-free fibril pathway, we circumvented the long lag phase by seeding solutions with 3% of preformed fibrils and allowed growth for 24 h. In either case, the amount of aggregated material was significant and had noticeably depleted the monomer pools, while the amount of hydrolyzed material formed over this time frame was negligible. The corresponding FTIR difference spectra for these rapidly formed fibrils are superimposed with the original difference spectra in Fig. 3b. None of the overall similarities or pathway-specific differences of their FTIR spectra changed when fibrils were grown sufficiently fast to render hydrolysis irrelevant.
Fibril growth at physiological pH has same structural fingerprint
Last but not the least, we determined whether lysozyme fibrils grown near physiological pH values would yield structural features comparable to those at pH 2. At pH 7 only the oligomeric assembly pathway was accessible as determined from the characteristic temporal evolution of the particle size distribution and AFM images. The corresponding FTIR difference spectra for fibrils grown for 2.5 days at pH 7 and 20 mM NaCl are superimposed on the pH 2 data in Fig. 3b as well. Again, the structural features match almost perfectly with those observed for oligomeric fibrils growth at acidic pH. We, therefore, conclude that the similarities and pathway-specific structural differences reported by FTIR are robust features of specific fibril assembly pathways and do not depend on their growth conditions. At least for the growth conditions and incubation periods used for our experiment, lysozyme hydrolysis did not affect the pathway-specific structural features in the FTIR spectra.
Characteristic thioflavin T responses during oligomer-free vs. oligomeric fibril assembly
ThT fluorescence has become a universally accepted indicator dye for mature amyloid fibrils due to its pronounced increase in fluorescence emission upon binding to various amyloid fibrils.25 ThT binding is considered to occur at surface grooves of mature amyloid fibrils. Hence, ThT binding monitors a structural motif that is related to, but is distinct from, the cross-β sheet structure considered the hallmark of amyloid fibrils. It has been established that the time course of ThT fluorescence responses is different for assembly along the oligomer-free vs. oligomer assembly pathway.26 The goals of the subsequent Thioflavin T experiments were two-fold. Our main objective was to determine whether ThT showed differences in its intrinsic sensitivity to different intermediates formed either within a given pathway or across the two different assembly pathways. Such aggregate-specific differences in ThT amplitude responses, in turn, would suggest underlying differences in aggregate structure and, thereby, speak to the structural evolution from the native to the fibril state along either one of the two assembly pathways.
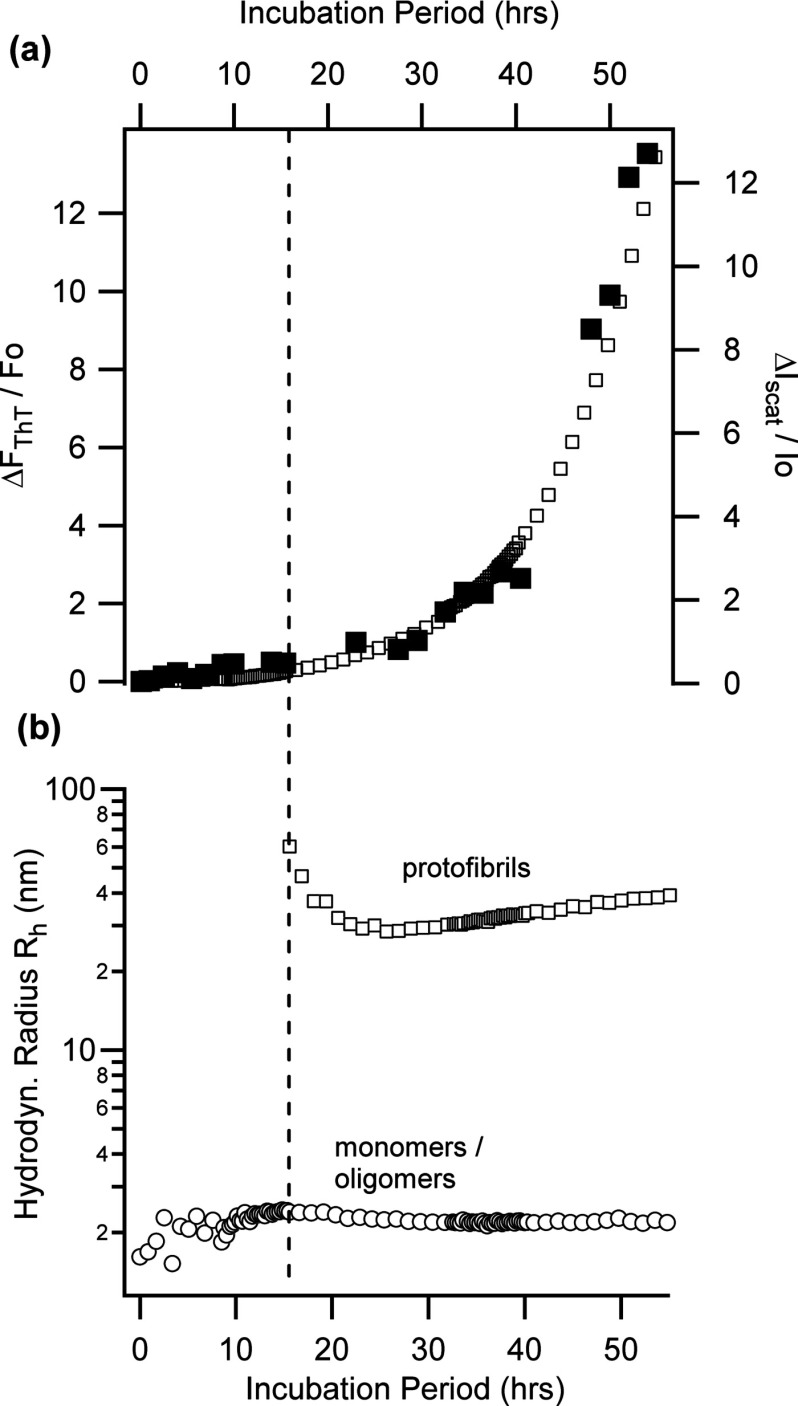
At first we determined which of the various intermediates other than mature fibrils evoked a ThT fluorescence response. Towards that end, we incubated lysozyme samples under either oligomer-free filamentous or oligomeric growth conditions and monitored aggregation-induced changes in total scattering intensity and in particle size distributions using static (SLS) and dynamic (DLS) light scattering. ThT fluorescence responses were assessed off-line from the same samples by withdrawing small aliquots from the incubating solutions at regular intervals. Figure 5a displays the corresponding changes in ThT fluorescence intensity (solid squares) obtained from off-line measurements typical for a lysozyme sample undergoing monomeric filament growth (T = 50 °C, 50 mM NaCl, pH 2.0). Here, ΔF/F0 is the relative increase in fluorescence over the resting fluorescence of the same lysozyme/ThT solution prior to incubation. The rapid rise in the ThT fluorescence ratio ΔF/F0 commences in unison with the nucleation of monomeric filament in these samples, as detected by DLS (Fig. 5b). Superimposed with the ThT fluorescence ratios are the corresponding fractional changes in the light scattering intensity (ΔIscat/I0) from the sample. In this pathway the fractional change in scattering intensity over the entire incubation period is quite modest (≤2.5-fold). While the fractional amplitude changes of these two signals are very different in amplitude, the superposition of the two signals (see Fig. 5a) indicates that their respective time courses are directly correlated.
Figure 5.
ThT fluorescence, light scattering intensity, and particle size distributions during oligomer-free fibril growth. (a) Fractional changes in ThT fluorescence intensity (■) and light scattering intensity (□) measured for lysozyme undergoing oligomer-free filament growth (50 mM NaCl, pH 2, 50 °C). The sudden increase in both signals coincides with the nucleation of monomeric filaments shown below. (b) Temporal evolution of the particle size distribution (PSD) for the same sample. Each point represents the dominant hydrodynamic radius for separable peaks in the PSD obtained from DLS measurements. During an extended lag period a single peak near the monomeric radius of lysozyme (Rh = 1.9 nm) persists until two additional peaks emerge with maximal radii near 400 nm and 40 nm, respectively. This nucleation event coincides with the appearance of monomeric filament populations in AFM images with lengths distributions consistent with the two DLS peaks.
Figure 6 displays the same set of measurements for lysozyme undergoing oligomeric fibril assembly (175 mM NaCl, pH 2.0 at 50 °C). The superposition of the fractional changes in ThT fluorescence amplitudes and light scattering intensities is shown in Fig. 6a. The ThT fluorescence signal in the oligomeric assembly path undergoes a continuous but accelerating increase which after 55 h of incubation reaches ∼12 times the background fluorescence. As before, the time courses for the fractional increases in scattered light intensity and ThT fluorescence are identical. The temporal evolution of the particle size distribution (Fig. 6b) shows a latency period of several hours prior to the nucleation of a single protofibril peak. Atomic force microscopy during the latency period in this regime reveals the formation of oligomers assembled from about eight lysozyme monomers (note that the relative population of oligomers is low enough and their hydrodynamic radius sufficiently close to those of monomer that DLS cannot separate oligomers from the monomeric background).15 In contrast to monomeric filament growth, though, neither the ThT fluorescence nor the light scattering traces display obvious features indicating the nucleation of the polymeric protofibrils. In addition, the fractional increase in ThT fluorescence and scattering intensity under oligomeric growth conditions are of comparable magnitude while they were widely different during oligomer-free filament growth. As observed with other amyloidogenic proteins, the time course of ThT fluorescence signals was dramatically different along the oligomer-free filamentous vs. oligomeric assembly pathway. Furthermore, ThT fluorescence increased either linearly (oligomer-free filament growth) or superlinearly (oligomeric growth) with time, and this time course matched the corresponding fractional increases in light scattering intensity. The lack of any signs of signal saturation for either pathway indicated that signal distortions due to saturation of dye binding or inner filter effects (i.e., attenuation of the excitation intensity via light scattering from protein aggregation or due to increases in ThT absorption) were negligible.27 Hence, the differences in both the time course and the relative amplitude of the ThT signals were diagnostic of the specific fibril assembly pathway they occurred in. Most importantly, though, ThT fluorescence changes matched perfectly with changes in scattering intensity, indicating that the former faithfully responded to all intermediates formed along either the oligomer-free or oligomeric assembly pathway.
Figure 6.
ThT fluorescence, light scattering intensity, and particle size distributions during oligomeric fibril growth. (a) Fractional changes in ThT fluorescence intensity (■) and light scattering intensity (□) measured from lysozyme undergoing oligomeric fibril growth (175 mM NaCl, pH 2, 50 °C). Both ThT fluorescence and light scattering intensity show a continuous, accelerating increase with no discernible signature of the prominent nucleation event detected by DLS. (b) Dominant hydrodynamic radii for separable peaks in particle size distributions obtained during oligomeric fibril assembly (175 mM NaCl). The subtle upward drift in the monomer peak results from the formation of small populations of oligomers readily seen in AFM. The new peak emerging around 15 h indicates the nucleation of protofibrils formed from oligomers.
Structure-specific ThT responses during oligomer-free vs. oligomeric fibril growth
Beyond the well-established pathway-specific differences in ThT kinetics, the above data imply that the fractional increases of ThT fluorescence amplitudes themselves display pathway-specific enhancements. While oligomer-free filamentous growth typically caused a nearly 100-fold augmentation in ThT fluorescence (Fig. 5a) oligomeric fibril growth yielded a comparatively muted 12-fold increase in ThT response (Fig. 6a). To extract aggregate-specific changes in ThT responses from the overall increases in ThT emission the latter need to be corrected for the total amount of amyloidogenic material contributing to the ThT signal. Towards that goal, we took advantage of the observed one-to-one temporal correlation between the fractional increases in light scattering intensity and ThT fluorescence. This correlation suggests that the scattering intensity and the ThT response are both direct measures of the number of aggregates present in the sample. This seems surprising, at first, since scattering intensity is heavily weighted towards larger aggregates and, to a first approximation, measures the weight-average of aggregates.28
The particle size distributions derived from simultaneous DLS measurements suggest how the SLS signal might be reporting changes to the number- and not to the weight-average of the aggregates. Following nucleation, changes in the particle size distribution measured with DLS were dominated by increases in the peak area while the average hydrodynamic radii changed surprisingly little. This was true for both oligomer-free and oligomeric fibril growth (see Fig. 7). This behavior suggests that changes in the particle size distribution were dominated by nucleation of new aggregates while increases in particle size itself were limited by concurrent fibril fragmentation. Such fragmentation was apparent in our monomeric fibril pathway (see Fig. 5b), has been experimentally observed in other systems, and is an integral part of nucleated polymerization models of amyloid fibril growth.29 As a result, scattering intensity reports the predominant increases in the number-averages of aggregates within an essentially stationary aggregate peak. Light scattering intensity increases also correlated well with the total amount of aggregates obtained via ultracentrifugation, further buttressing our conclusion. Therefore, rationing ThT fluorescence with the light scattering intensity normalizes ThT amplitudes by the total number of aggregates contributing to the ThT signal at any given moment in time. The resulting “ratio of ratios” tracks the intrinsic increase in ThT fluorescence arising from aggregate-specific changes in ThT responses. These aggregate-specific increases could arise via a variety of different mechanisms, such as changes in the aggregate-specific number of available ThT binding sites, differences in the structure or identity of the binding site, the binding affinity or quantum yield of ThT upon binding. Pursuing the complex mechanisms underlying the observed changes in ThT photochemistry is outside the scope of this work. Here, we only utilized the aggregate-specific ThT responses for identifying structural differences among amyloid intermediates.
Figure 7.
Evolution of aggregate peaks during oligomer-free vs. oligomeric fibril growth. Particle size distributions (PSDs) with DLS for either (a) oligomer-free filamentous growth at 50mM NaCl or (b) oligomeric fibril assembly at 175 mM NaCl. The gray bars represent the PSD obtained shortly after (a) nucleation of the two populations of rigid filaments (short and long filaments) or (b) nucleation of protofibrils. The empty bars, in turn, represent the corresponding PSDs near the end of the measurement period for either pathway.
Figure 8a displays the ratio of the ΔF/F0 amplitudes divided by the corresponding ΔIscat/I0 amplitudes for late-stage fibrils grown at different salt concentrations. All oligomer-free filamentous samples (50, 75, 125 mM NaCl) showed fractional fluorescence increases that exceeded the corresponding fractional light scattering increases by factors of 20–80. The ∼100-fold increase in ThT fluorescence in the oligomer-free filamentous pathway is considered typical for binding of ThT to mature amyloid fibrils. In contrast, the fractional ThT and light scattering increases for oligomeric growth conditions (175 or 200 mM NaCl) were of similar magnitudes, yielding ratios near 1. Finally, amorphous precipitation (400 mM NaCl) yielded ΔF/F0 over ΔIscat/I0 ratios hovering around 0.1. This steep and abrupt decrease in pathway-specific, intrinsic ThT responses with increasing salt concentration was opposite to the gradual net increase in ThT fluorescence upon non-specific binding to lysozyme monomers under the same solution conditions (Fig. 8a, inset). Therefore, when calibrating ThT responses against aggregate production, the ThT enhancements fell within two clearly distinct “bands” for samples undergoing filamentous vs. oligomeric fibril growth. Hence, ThT binding recognized distinct structural motifs for fibrils formed along the oligomer-free vs. oligomeric assembly pathway.
Figure 8.
Correlation of ThT fluorescence and light scattering in different assembly pathways. (a) Ratio of fractional ThT over fractional light scattering increments for late-stage fibrils vs. the NaCl concentrations used during their growth. ThT/light scattering ratios for fibrils after oligomer-free (<150 mM NaCl) vs. those after oligomeric fibril growth (>150 mM NaCl) differ by approximately two orders of magnitude (note logarithmic scale). The onset of amorphous precipitation near 400 mM NaCl causes another sudden drop in this ratio by a factor of ten. (Inset) Salt-dependent changes in ThT fluorescence emission (■) in presence of lysozyme, (◯) for ThT without lysozyme and (□) for lysozyme auto-fluorescence, all measured at 485 nm. (b) Log-log plot of the fractional changes in ThT fluorescence vs. their corresponding fractional light scattering intensity as they evolve during the entire aggregation process. Results are shown for multiple lysozyme samples incubated under conditions of either monomeric filament growth (solid symbols) or oligomeric fibril growth (open symbols). The dashed lines represent linear fits through data at a fixed salt concentration. ThT ratios display strong and persistent linear correlations to their corresponding light scattering ratios throughout the entire growth process. (c) ThT and light scattering ratios measured for fibril growth of lysozyme at physiological pH (pH 7, 20 mM NaCl, T = 65 °C).
The above analysis only considered the relative ThT responses at the endpoints of the fibril growth reaction. However, similar to the structural evolution we explored in Fig. 3c using FTIR, we also investigated changes in relative ThT responses occurring throughout the assembly process. We, therefore, plotted the fractional changes in ThT emission against the corresponding fractional light scattering changes (Fig. 8b). Using Figs. 5a, 6a, these ratios can be related back to specific time points within the fibril assembly process. In Fig. 8b, the fractional change in light scattering intensity serves as a “reaction coordinate” indicating how far the aggregation has progressed – independent of the aggregation rates for different samples. This plot thereby quantifies the ThT response in proportion to the amount of aggregated material formed over the entire incubation period. The different symbols indicate results for different samples grown along either the oligomer-free (closed symbols) or the oligomeric fibril pathway (open symbols). While we used a logarithmic display to accommodate the wide range of ThT and Iscat rations, the solid lines represent simple linear fits (slope = 1), through each individual dataset. This suggests that the fractional ThT response, when normalized by the total amount of aggregated material, remained nearly constant throughout the entire aggregation process within each pathway. This is noteworthy since the specific mix of intermediates did change significantly over the course of fibril growth. In the oligomer-free pathway the initial population of short and long monomeric filaments was supplanted by cross-assembled mature fibrils. In the oligomeric pathway, monomers and oligomers were the only species present prior to protofibril formation while protofibrils dominated both ThT and light scattering responses in the late stages. If the structure of intermediate aggregates within a pathway would change noticeably as a result of the aggregation process, the aggregate-specific responses of ThT should have changed, as well. For examples, monomeric fibrils emerging in the protofibril pathway should have caused the ThT response in Fig. 8 to “hop” from the lower protofibril band onto the oligomer-free filament band above it. The persistent linear relation suggests, instead, that within a given pathway the distinct structural motifs recognized by ThT become established during the earliest aggregation step, i.e., during filament nucleation or oligomer formation, respectively. These structural motifs then persisted or underwent only subtle restructuring throughout the entire hierarchy of intermediates emerging later in the assembly process. This observation is consistent with the steady evolution of the ATR-FTIR difference spectra in Fig. 3c.
Independent of pathway-specific ThT responses on growth conditions or protein hydrolysis
Just as for the structural differences in infrared spectroscopy we confirmed that pathway-specific differences in ThT responses were independent of the growth conditions, of protein hydrolysis, and persisted near physiological growth conditions of pH 7. The specific changes in particle size distribution and AFM images of fibrils formed at pH 7 indicated that fibril formation followed the oligomeric assembly pathway. Measurements of ThT and SLS ratios at pH 2 for varying salt concentrations and, therefore, growth rates are shown in Fig. 5b, and for fibril growth at pH 7 are shown in Fig. 8c. Both the time course and the relative amplitudes of the ThT to SLS signals were determined by the assembly pathway and not the specific solution conditions used to generate the fibrils. Hence, the pathway specific structural motifs recognized by ThT displayed the same behavior as the amyloid-specific β-sheet peaks in the FTIR spectra.
DISCUSSION AND CONCLUSION
The existence of two fibril assembly pathway generating intermediates with distinct morphologies comparable to the oligomer-free vs. oligomeric pathways discussed here has been described for a variety of different amyloid and prion systems.13, 14, 30 One of the persistent questions, though, has been whether the oligomeric pathway is “on pathway” towards fibril formation or represents a separate, potentially competing pathway not leading to mature fibril formation.13, 31 The dominant feature of the FTIR spectra for late-stage lysozyme fibrils formed along either of these two pathways was the prominent β-sheet peak right around 1620 cm−1. This peak wavenumber is considered “diagnostic” of amyloidogenic β-sheet formation.18 In addition, both early and late-stage aggregates in either pathway generated significant responses from the amyloid indicator-dye thioflavin T. These observations suggest that both assembly pathways, while distinct and despite the clear morphological differences of their intermediates, are “amyloidogenic” in nature.
At the same time, both FTIR spectra and thioflavin T responses point to pathway-specific structural differences. The FTIR difference spectra for oligomer-free filaments vs. those from oligomeric protofibrils consistently displayed peaks at (1623 ± 1) cm−1 vs. (1618 ± 1) cm−1, respectively. This shift was independent of growth conditions and highly reproducible. Specifically, fibrils grown for less than 24 h either via seeding (oligomer-free pathway) or via increased salt concentration (oligomeric pathway) showed the same spectral signature as those incubated for long periods. In contrast, the amount of hydrolyzed lysozyme accumulated during these different time periods increases from barely noticeable to very prominent (see Fig. 4). Furthermore, the oligomeric fibrils obtained at physiological pH values showed the same FTIR peak position as those grown at highly acidic pH values. Reducing SDS-PAGE gels at pH 7 showed no signs of lysozyme hydrolysis. We, therefore, conclude that, under our growth conditions, hydrolysis does not affect the structural features reported by either FTIR or ThT. The shift of the amyloid β-sheet peak for oligomer-free vs. oligomeric lysozyme fibrils to higher wavenumbers seen here also agrees with spectral shifts in the IR spectra reported for long straight vs. worm-like fibrils of β2-microglobulin.12
In comparison to the more subtle spectral differences seen in FTIR, the two pathways were readily distinguishable by their intrinsic ThT response amplitudes. The dramatic pathway-specific difference in intrinsic ThT responses imply that ThT recognized distinct structural motives of intermediates and mature fibrils formed along either the oligomer-free vs. the oligomeric assembly pathway. Similar to the FTIR signatures, these structure-specific ThT responses were unaffected by changes in the rate of fibril formation, the amount of lysozyme hydrolysis, or solution pH. Therefore, fibril formation leaves structural “fingerprints” that are specific to the pathway along which the fibrils have grown. These fingerprints can be interrogated either with FTIR spectroscopy or ThT fluorescence. This opens the possibility that either FTIR or ThT measurements can resolve which pathway(s) fibril formation follows in vivo. In fact, FTIR spectra of late-stage β2-microglobulin fibrils isolated from patients have been shown to match closely with those of long-straight but not worm-like fibrils grown in vitro.12
While NMR, for example, provides much more structural resolution, FTIR spectroscopy and the intrinsic ThT responses used in this study permit following the temporal evolution of intermediate structures along a given amyloid fibril pathway. As shown in Fig. 8b, the intrinsic ThT responses remained essentially unchanged during the entire growth process along either assembly pathway, despite significant changes to the relative mixture of intermediates present in solution. FTIR spectroscopy indicated a similarly uniform evolution of the pathway-specific β-sheet peak (Fig. 3c). For the oligomer-free pathway, these intermediates include short and long monomeric filaments and mature fibers which cross-assemble from three monomeric filaments. For the oligomeric assembly pathway, the range of intermediates includes pre-nucleation oligomers to post-nucleation protofibrils at all stages. Our results suggest that the dominant pathway-specific structural features recognized by ThT and FTIR get established by the earliest intermediates emerging in either pathway and remain preserved through subsequent generations of amyloidogenic intermediates. This implies that the main “structural transition” from a near-native to an amyloidogenic state occurred within a single step. This step takes place either right before or as a result of the very first aggregation event, i.e., nucleation of monomeric filaments or formation of compact oligomeric intermediates. The above findings are consistent with reports that partially denatured proteins can develop extended β-sheet conformations even prior to the onset of aggregation.32 The first atomic resolution structure for oligomers grown from a segment of αB crystalline is also consistent with the above observations.33 These oligomers displayed a structure resembling β-sheet barrels and yielded X-ray diffraction patterns compatible with the cross-β sheet structures of mature amyloid fibrils. Therefore, normalized ThT responses and FTIR spectroscopy offer an appealing way to follow the structural evolution of amyloid fibril formation and represent a unique opportunity to determine which assembly pathway amyloid formation follows in vivo.
ACKNOWLEDGMENTS
We acknowledge Dr. Garrett Matthews for generously making his AFM system available to us and Ben Muschol for assisting with processing of the AFM images. This work was supported, in part, by a grant from The Neuroscience Collaborative (NSC) at USF and the National Institutes of Health (NIH) Grant No. R15-GM097723 (M.M.). Additional support was provided by the National Science Foundation (NSF) REU program Grant No DMR-1004873 (C.P.) and by a departmental start-up grant (S.W.).
References
- Berland C. R., Thurston G. M., Kondo M., Broide M. L., Pande J., Ogun O., and Benedek G. B., Proc. Natl. Acad. Sci. U.S.A. 89, 1214 (1992); 10.1073/pnas.89.4.1214 [DOI] [PMC free article] [PubMed] [Google Scholar]; Muschol M. and Rosenberger F., J. Chem. Phys. 107 (6), 1953 (1997); 10.1063/1.474547 [DOI] [Google Scholar]; Galkin O., Chen K., Nagel R. L., Hirsch R. E., and Vekilov P. G., Proc. Natl. Acad. Sci. U.S.A. 99(13), 8479 (2002); 10.1073/pnas.122055299 [DOI] [PMC free article] [PubMed] [Google Scholar]; tenWolde P. R. and Frenkel D., Science 277(5334), 1975 (1997); 10.1126/science.277.5334.1975 [DOI] [PubMed] [Google Scholar]; Rosenbaum D., Zamora P. C., and Zukoski C. F., Phys. Rev. Lett. 76(1), 150 (1996); 10.1103/PhysRevLett.76.150 [DOI] [PubMed] [Google Scholar]; Sciortino F., Prasad K. U., Urry D. W., and Palma M. U., Chem. Phys. Lett. 153(6), 557 (1988); 10.1016/0009-2614(88)85260-6 [DOI] [Google Scholar]; Sciortino F., Urry D. W., Palma M. U., and Prasad K. U., Biopolymers 29(10–11), 1401 (1990); 10.1002/bip.360291007 [DOI] [PubMed] [Google Scholar]; Vaiana S. M., Palma-Vittorelli M. B., and Palma M. U., Proteins: Struct., Funct., Bioinf. 51(1), 147 (2003). 10.1002/prot.10306 [DOI] [PubMed] [Google Scholar]
- Li P., Banjade S., Cheng H.-C., Kim S., Chen B., Guo L., Llaguno M., Hollingsworth J. V., King D. S., Banani S. F., Russo P. S., Jiang Q.-X., Nixon B. T., and Rosen M. K., Nature (London) 483(7389), 336 (2012). 10.1038/nature10879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell J. D., Zhao A., Ellington A. D., and Marcotte E. M., Annu. Rev. Cell Dev. Biol. 28(1), 89 (2012). 10.1146/annurev-cellbio-101011-155841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly J. W., Curr. Opin. Struct. Biol. 6(1), 11 (1996). 10.1016/S0959-440X(96)80089-3 [DOI] [PubMed] [Google Scholar]
- Chiti F. and Dobson C. M., Annu. Rev. Biochem. 75(1), 333 (2006). 10.1146/annurev.biochem.75.101304.123901 [DOI] [PubMed] [Google Scholar]
- Halfmann R., Jarosz D. F., Jones S. K., Chang A., Lancaster A. K., and Lindquist S., Nature (London) 482(7385), 363 (2012). 10.1038/nature10875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler D. M., Koulov A. V., Alory-Jost C., Marks M. S., Balch W. E., and Kelly J. W., PLoS Biol. 4(1), e6/1 (2005); 10.1371/journal.pbio.0040006 [DOI] [PMC free article] [PubMed] [Google Scholar]; Otzen D., Prion 4(4), 256 (2010); 10.4161/pri.4.4.13676 [DOI] [PMC free article] [PubMed] [Google Scholar]; Maji S. K., Perrin M. H., Sawaya M. R., Jessberger S., Vadodaria K., Rissman R. A., Singru P. S., Nilsson K. P. R., Simon R., Schubert D., Eisenberg D., Rivier J., Sawchenko P., Vale W., and Riek R., Science 325(5938), 328 (2009). 10.1126/science.1173155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn T. R., Makin O. S., Morris K. L., Marshall K. E., Tian P., Sikorski P., and Serpell L. C., J. Mol. Biol. 395(4), 717 (2010); 10.1016/j.jmb.2009.09.039 [DOI] [PubMed] [Google Scholar]; Sunde M. and Blake C. C. F., Adv. Protein Chem. 50, 123 (1997); 10.1016/S0065-3233(08)60320-4 [DOI] [PubMed] [Google Scholar]; Fändrich M., Cell. Mol. Life Sci. 64(16), 2066 (2007). 10.1007/s00018-007-7110-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo E. H., Lansbury P. T., and Kelly J. W., Proc. Natl. Acad. Sci. U.S.A. 96, 9989 (1999). 10.1073/pnas.96.18.9989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson C. M., Trends Biochem. Sci. 24, 329 (1999). 10.1016/S0968-0004(99)01445-0 [DOI] [PubMed] [Google Scholar]
- Uversky V. N., Fernandez A., and Fink A. L., in Protein Misfolding, Aggregation and Conformational Diseases. Part A: Protein Aggregation and Conformational Diseases, edited by Vladimir N. U. and Fink A. L. (Springer, New York, 2006); [Google Scholar]; Uversky V. N. and Fink A. L., Biochim. Biophys. Acta 1698, 131 (2004). 10.1016/j.bbapap.2003.12.008 [DOI] [PubMed] [Google Scholar]
- Jahn T. R., Tennent G. A., and Radford S. E., J. Biol. Chem. 283(25), 17279 (2008). 10.1074/jbc.M710351200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosal W. S., Morten I. J., Hewitt E. W., Smith D. A., Thompson N. H., and Radford S. E., J. Mol. Biol. 351, 850 (2005). 10.1016/j.jmb.2005.06.040 [DOI] [PubMed] [Google Scholar]
- Goldsbury C., Frey P., Olivieri V., Aebi U., and Müller S. A., J. Mol. Biol. 352(2), 282 (2005); 10.1016/j.jmb.2005.07.029 [DOI] [PubMed] [Google Scholar]; Hess S., Lindquist S., and Scheibel T., EMBO Rep. 8, 1196 (2007). 10.1038/sj.embor.7401096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill S. E., Miti T., Richmond T., and Muschol M., PLoS ONE 6(4), e18171/1 (2011). 10.1371/journal.pone.0018171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira M. N. N., Forny-Germano L., Saraiva L. M., Sebollela A., Martinez A. M. B., Houzel J.-C., De Felice F. G., and Ferreira S. T., J. Neurochem. 103(2), 736 (2007); 10.1111/j.1471-4159.2007.04809.x [DOI] [PubMed] [Google Scholar]; Kayed R., Head E., Sarsoza F., Saing T., Cotman C., Necula M., Margol L., Wu J., Breydo L., Thompson J., Rasool S., Gurlo T., Butler P., and Glabe C., Mol. Neurodegeneration 2(1), 18 (2007); 10.1186/1750-1326-2-18 [DOI] [PMC free article] [PubMed] [Google Scholar]; Cleary J. P., Walsh D. M., Hofmeister J. J., Shankar G. M., Kuskowski M. A., Selkoe D. J., and Ashe K. H., Nat. Neurosci. 8, 79 (2005); 10.1038/nn1372 [DOI] [PubMed] [Google Scholar]; Kayed R., Sokolov Y., Edmonds B., McIntire T. M., Milton S. C., Hall J. E., and Glabe C. G., J. Biol. Chem. 279(45), 46363 (2004); 10.1074/jbc.C400260200 [DOI] [PubMed] [Google Scholar]; Kayed R., Head E., Thompson J. L., McIntire T. M., Milton S. C., Cotman C. W., and Glabe C. G., Science 300, 486 (2003); 10.1126/science.1079469 [DOI] [PubMed] [Google Scholar]; Walsh D. M., Klyubin I., Fadeeva J. V., Cullen W. K., Anwyl R., Wolfe M. S., Rowan M. J., and Selkoe D. J., Nature (London) 416(6880), 535 (2002). 10.1038/416535a [DOI] [PubMed] [Google Scholar]
- Kodali R. and Wetzel R., Curr. Opin. Struct. Biol. 17(1), 48 (2007). 10.1016/j.sbi.2007.01.007 [DOI] [PubMed] [Google Scholar]
- Zandomeneghi G., Krebs M. R. H., McCammon M. G., and Fändrich M., Protein Sci. 13(12), 3314 (2004). 10.1110/ps.041024904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmar A. S., Gottschall P. E., and Muschol M., Biophys. Chem. 129, 224 (2007). 10.1016/j.bpc.2007.06.002 [DOI] [PubMed] [Google Scholar]
- Booth D. R., Sunde M., Bellotti V., Robinson C. V., Hutchinson W. L., Fraser P. E., Hawkins P. N., Dobson C. M., Radford S. E., Blake C. F. F., and Pepys M. B., Nature (London) 385, 787 (1997); 10.1038/385787a0 [DOI] [PubMed] [Google Scholar]; Gillmore J. D., Booth D. R., Madhoo S., Pepys M. B., and Hawkins P. N., Nephrol. Dial Transplant 14(11), 2639 (1999); 10.1093/ndt/14.11.2639 [DOI] [PubMed] [Google Scholar]; Pepys M. B., Hawkins P. N., Booth D. R., Vigushin D. M., Tennent G. A., Soutar A. K., Totty N., Nguyen O., Blake C. C. F., Terry C. J., Feest T. G., Zalin A. M., and Hsuan J. J., Nature (London) 362(6420), 553 (1993). 10.1038/362553a0 [DOI] [PubMed] [Google Scholar]
- Trexler A. J. and Nilsson M. R., Curr. Protein Pept. Sci. 8, 537 (2007). 10.2174/138920307783018659 [DOI] [PubMed] [Google Scholar]
- Arnaudov L. N. and de Vries R., Biophys. J. 88(1), 515 (2005); 10.1529/biophysj.104.048819 [DOI] [PMC free article] [PubMed] [Google Scholar]; Hill S. E., Robinson J., Matthews G., and Muschol M., Biophys. J. 96, 3781 (2009); 10.1016/j.bpj.2009.01.044 [DOI] [PMC free article] [PubMed] [Google Scholar]; Krebs M. R. H., Wilkins D. K., Chung E. W., Pitkeathly M. C., Chamberlain A. K., Zurdo J., Robinson C. V., and Dobson C. M., J. Mol. Biol. 300(3), 541 (2000). 10.1006/jmbi.2000.3862 [DOI] [PubMed] [Google Scholar]
- Frare E., Polverino de Laureto P., Zurdo J., Dobson C. M., and Fontana A., J. Mol. Biol. 340(5), 1153 (2004). 10.1016/j.jmb.2004.05.056 [DOI] [PubMed] [Google Scholar]
- Mishra R., Sörgjerd K., Nyström S., Nordigården A., Yu Y.-C., and Hammarström P., J. Mol. Biol. 366, 1029 (2007). 10.1016/j.jmb.2006.11.084 [DOI] [PubMed] [Google Scholar]
- Biancalana M. and Koide S., Biochim. Biophys. Acta 1804(7), 1405 (2010); 10.1016/j.bbapap.2010.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]; Vassar P. S. and Culling C. F., Arch. Pathol. 68, 232 (1959). [PubMed] [Google Scholar]
- Smith D. P., Jones S., Serpell L. C., Sunde M., and Radford S. E., J. Mol. Biol. 330(5), 943 (2003). 10.1016/S0022-2836(03)00687-9 [DOI] [PubMed] [Google Scholar]
- Sulatskaya A. I., Kuznetsova I. M., and Turoverov K. K., J. Phys. Chem. B 115(39), 11519 (2011). 10.1021/jp207118x [DOI] [PubMed] [Google Scholar]
- Cummins H. Z., in Photon Correlation and Light Beating Spectroscopy, edited by Cummins H. Z. and Pike E. R. (Plenum Press, New York, 1973). [Google Scholar]
- Tanaka M., Collins S. R., Toyama B. H., and Weissman J. S., Nature (London) 442(7102), 585 (2006); 10.1038/nature04922 [DOI] [PubMed] [Google Scholar]; Masel J., Jansen V. A. A., and Nowak M. A., Biophys. Chem. 77, 139 (1999); 10.1016/S0301-4622(99)00016-2 [DOI] [PubMed] [Google Scholar]; Knowles T. P. J., Waudby C. A., Devlin G. L., Cohen S. I. A., Aguzzi A., Vendruscolo M., Terentjev E. M., Welland M. E., and Dobson C. M., Science 326, 1533 (2009). 10.1126/science.1178250 [DOI] [PubMed] [Google Scholar]
- Jahn T. R. and Radford S. E., Arch. Biochem. Biophys. 469, 100 (2008). 10.1016/j.abb.2007.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Necula M., Kayed R., Milton S., and Glabe C. G., J. Biol. Chem. 282(14), 10311 (2007). 10.1074/jbc.M608207200 [DOI] [PubMed] [Google Scholar]
- Yang W. Y., Larios E., and Gruebele M., J. Am. Chem. Soc. 125(52), 16220 (2003). 10.1021/ja0360081 [DOI] [PubMed] [Google Scholar]
- Laganowsky A., Liu C., Sawaya M. R., Whitelegge J. P., Park J., Zhao M., Pensalfini A., Soriaga A. B., Landau M., Teng P. K., Cascio D., Glabe C., and Eisenberg D., Science 335(6073), 1228 (2012). 10.1126/science.1213151 [DOI] [PMC free article] [PubMed] [Google Scholar]