Abstract
Deposition of Aβ in the brain is a pathological hallmark of Alzheimer's disease. There are two major isoforms of Aβ: the 42-residue Aβ42 and the 40-residue Aβ40. The only difference between Aβ42 and Aβ40 is that Aβ42 has two extra residues at the C-terminus. The amyloid plaques in Alzheimer's brains consist of mostly Aβ42 and some plaques contain only Aβ42, even though Aβ40 concentration is several-fold more than Aβ42. Using electron paramagnetic resonance, we studied the formation of amyloid fibrils using a mixture of Aβ42 and Aβ40 in vitro. We show that Aβ42 and Aβ40 form mixed fibrils in an interlaced manner, although Aβ40 is not as efficient as Aβ42 in terms of being incorporated into Aβ42 fibrils. Our results suggest that both Aβ42 and Aβ40 would be present in amyloid plaques if in vivo aggregation of Aβ were similar to the in vitro process. Therefore, there must be some mechanisms that lead to the preferential deposition of Aβ42 at the extracellular space. Identifying such mechanisms may open new avenues for therapeutic interventions to treat Alzheimer's disease.
Keywords: Senile plaques, protein aggregation, electron paramagnetic resonance, spin labeling
Introduction
Aggregation of amyloid β (Aβ) plays a key role in the pathogenesis of Alzheimer's disease (AD). Aβ is a proteolytic product of amyloid precursor protein by β- and γ-secretases. The imprecise cleavage of γ-secretase at C-terminus of Aβ sequence results in two major Aβ isoforms: Aβ42 (42 residues long) and Aβ40 (40 residues long). The only difference between Aβ42 and Aβ40 is the two additional C-terminal residues on Aβ42. The concentration of Aβ40 in cerebral spinal fluid has been found to be several-fold more than that of Aβ42. However, Aβ42 is the major component of amyloid plaques in AD brains (Iwatsubo et al. 1995, Iwatsubo et al. 1994, Gravina et al. 1995, Miller et al. 1993, Mak et al. 1994), while Aβ40 is detected only in a subset of plaques (Iwatsubo et al. 1994, Miller et al. 1993, Mak et al. 1994). These findings suggest that the Aβ42 deposition precedes Aβ40 deposition and the initial Aβ42 aggregation does not involve Aβ40.
The interplay between Aβ42 and Aβ40 has been generally considered to play a critical role in AD. Increased Aβ42/Aβ40 ratios appear to correlate with the early-onset familial AD cases caused by presenilin mutations (Kumar-Singh et al. 2006). Lowering Aβ42/Aβ40 ratios in transgenic mice decreases Aβ deposition (Kim et al. 2007). Higher neurotoxicity has been reported with samples of higher Aβ42/Aβ40 ratios (Kuperstein et al. 2010). Previous studies also suggest that Aβ42 and Aβ40 affect each other's aggregation rates and toxic activities (Snyder et al. 1994, Yoshiike et al. 2003, Yan & Wang 2007, Jan et al. 2008, Kuperstein et al. 2010, Pauwels et al. 2012). Furthermore, in vitro studies have shown that Aβ42 and Aβ40 form mixed aggregates (Frost et al. 2003).
Why is Aβ42 the major, and sometimes only, component in amyloid plaques, when Aβ40 is several-fold more abundant in the brain? One seemingly plausible explanation is that Aβ42 is more aggregation prone than Aβ40, and thus would be deposited before Aβ40. However, this explanation assumes that Aβ42 and Aβ40 preferentially aggregate with their own species even if both isoforms are present. To this end, we use electron paramagnetic resonance (EPR) spectroscopy to investigate the interactions between Aβ42 and Aβ40 in amyloid fibrils at molecular level. Our results suggest that Aβ42 and Aβ40 form interlaced amyloid fibrils in vitro, suggesting some unrecognized mechanisms may contribute to the preferential deposition of Aβ42 in AD brains.
Materials and Methods
Preparation of Aβ42 peptides and spin labeling
The DNA constructs of GroES-ubiquitin-Aβ (Shahnawaz et al. 2007) and the deubiquitylating enzyme Usp2cc (Baker et al. 2005) were kindly provided by Dr. Il-Seon Park at Chosun University (South Korea) and Dr. Rohan T. Baker at Australian National University (Australia). The L17C mutation was introduced into Aβ42 sequence using QuikChange kit (Agilent) and confirmed with DNA sequencing.
Expression of GroES-ubiquitin-Aβ and Usp2cc proteins in E. coli and their purification were performed as previously described (Ngo & Guo 2011, Agopian & Guo 2012). Full-length Aβ42 was cleaved from the fusion protein with Usp2cc in a buffer containing 19 mM phosphate, 3 M urea, 2 mM TCEP, pH 10.0. Usp2cc was added to the fusion protein at a Usp2cc:Aβ molar ratio of 1:100. The digestion reaction was allowed to proceed at 37°C for 15 min. The reaction mixture was then immediately filtered with 0.2-μm filter (Whatman) and loaded on a 5-mL HisTrap column (GE Healthcare) equilibrated with PSU buffer (50 mM phosphate, 0.3 M NaCl, 8 M urea, pH 10.0). Aβ peptide was eluted with 25 mM imidazole. Purified Aβ was checked with SDS-PAGE, and no non-cleaved proteins were detected. Wild-type (WT) Aβ peptides were buffer exchanged to 30 mM ammonium acetate, pH 10.0, and then lyophilized.
For spin labeling of Aβ42 L17C mutant, dithiothreitol was added to purified protein fraction to a final concentration of 10 mM and was allowed to incubate at room temperature for 20 min. Then the Aβ42 sample was buffer exchanged to labeling buffer (20 mM MOPS, 7 M guanidine hydrochloride, 50 mM NaCl, pH 6.8) using a 5-mL HiTrap desalting column (GE Healthcare). The spin labeling reagent MTSSL (1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl methanethiosulfonate, Enzo Life Sciences) was added at 10-times molar excess and then incubated at room temperature for 1 h. The spin label is named R1. The spin-labeled Aβ42 was further buffer exchanged to 30 mM ammonium acetate, pH 10.0. Spin-labeled Aβ42 was lyophilized and stored at -80°C. MALDI-TOF mass spectrometry was performed to ensure that the mass of Aβ42 is correct, and the extent of labeling is >95%.
Fibril growth
To mix spin-labeled Aβ with WT Aβ, lyophilized Aβ42 L17R1 and WT Aβ were dissolved separately in 30 mM ammonium acetate, pH 10.0 and then mixed at molar ratios of 1:1 and 1:3 as described in the text. Then the mixture is lyophilized. For fibril formation, the mixture was suspended in 100% 1,1,1,3,3,3 hexafluoro-2-propanol (HFIP) at 1 mM and bath sonicated for 5 min. Then the sample was incubated at room temperature for 30 min. HFIP was removed by evaporation overnight in the fume hood and then under vacuum for 1 h. Finally the Aβ sample was dissolved in PG buffer (20 mM CAPS, 7 M guanidine hydrochloride, pH 11) to 1 mM and then diluted 20× to HBS buffer (50 mM HEPES, 140 mM NaCl, pH 7.4) to a final concentration of 50 μM. Then the Aβ solution was placed on a digital vertex mixer with a speed of 600 rpm at room temperature. Fibrils were collected by centrifugation at 14,000 g for 20 min after thioflavin T binding has plateaued (∼5–7 days). Soluble proteins were removed by washing the pellet with HBS buffer.
Transmission electron microscopy
The Aβ fibril sample (5 μl) was placed on glow-discharged copper grids covered with 400 mesh formvar/carbon film (Ted Pella). The sample was negatively stained with 2% uranyl acetate. Samples were examined using a JEOL JEM-1200EX transmission electron microscope at 80 kV.
EPR spectroscopy
EPR measurements were performed at X-band frequency on a Bruker EMX spectrometer equipped with the ER 4102ST cavity. A modulation frequency of 100 kHz was used. Measurements were performed at 20 mW microwave power at room temperature. Modulation amplitude was optimized to individual spectrum (typically ∼4 G). Approximately 20 μL of fibril sample was loaded into glass capillaries (VitroCom) sealed at one end. EPR spectra in each figure panel were normalized to the same number of spins.
Spectral simulations
Spectral simulations were performed using the program MultiComponent of Dr. Christian Altenbach, which provides a LabVIEW (National Instruments) interface of the program NLSL developed by Freed and co-workers (Schneider & Freed 1989, Budil et al. 1996). A microscopic order macroscopic disorder model was used as previously described (Budil et al. 1996). A least-squares fit of the user-defined spectral parameters was performed using the Levenberg-Marquardt algorithm. For all fits, the values for the magnetic tensors A and g were fixed as Axx = 6.2, Ayy = 5.9, Azz = 37.0, and gxx = 2.0078, gyy = 2.0058, gzz = 2.0022, which were determined previously for spin label R1 (Columbus et al. 2001). An anisotropic model for the motion of the spin label was assumed and was found to give better fits than isotropic models. For anisotropic simulations, diffusion tilt angles were fixed to (α,β,γ) = (0,36°,0) for z-axis anisotropy as previously reported (Columbus et al. 2001). The diffusion tilt angels are the Euler angles relating the axes of the diffusion tensor and the magnetic tensor. The number of fitted parameters was kept at a minimum, which in this work includes the rotational diffusion constant (R), an order parameter (S), and Heisenberg exchange frequency (ω). We found that satisfactory fits were obtained with only these three parameters. Rotational correlation time (τ) was calculated using τ = 1/(6R). For two-components fitting of the 1:1 mixture of spin-labeled Aβ42 and WT Aβ, the parameters (R, S, and ω) for the single-line component were fixed at the fitted parameters for the fully labeled Aβ42 L17R1, and the parameters for the three-line component was allowed to vary.
The fitted values for ω, τ, and S are as follows. For Aβ42 L17R1, ω = 160.4 ± 1.4 MHz, τ = 5.7 ± 0.1 ns, S = 0.55 ± 0.01. For 1:1 spin dilution with Aβ42 WT three-line component, ω = 70.9 ± 3.5 MHz, τ = 2.5 ± 0.4 ns, S = 0.77 ± 0.03. For 1:3 spin dilution with Aβ42 WT, ω = 68.2 ± 0.6 MHz, τ = 3.3 ± 0.2 ns, S = 0.72 ± 0.01. For 1:1 spin dilution with Aβ40 WT three-line component, ω = 70.9 ± 4.9 MHz, τ = 5. ± 0.4 ns, S = 0.55 ± 0.05. For 1:3 spin dilution with Aβ40 WT, ω = 64.8 ± 0.8 MHz, τ = 5.0 ± 0.1 ns, S = 0.46 ± 0.02. The 1:1 dilution was repeated once with the following fitted parameters. For 1:1 spin dilution with Aβ42 WT, ω = 72.5 ± 3.1 MHz, τ = 2.2 ± 0.3 ns, S = 0.77 ± 0.02. For 1:1 spin dilution with Aβ40 WT, ω = 71.4 ± 5.4 MHz, τ = 5.4 ± 0.3 ns, S = 0.487 ± 0.05.
Statistical analysis
Data are presented as mean ± SD. Statistical significance was defined at p < 0.05, using the Student's t-test.
Results and Discussion
Rationale for using EPR to study interactions between Aβ42 and Aβ40 in amyloid fibrils
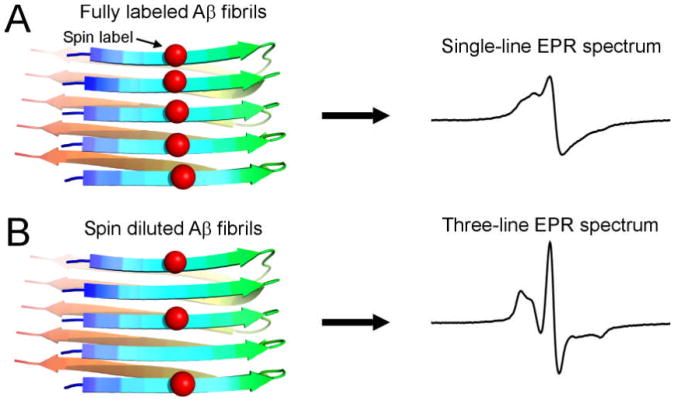
Both Aβ42 and Aβ40 have been shown to form fibrils with parallel in-register β-sheet structures (Tycko 2006, Agopian & Guo 2012), in which the side chains from the same residue positions stack in ladder-like arrangement. When a spin label is introduced into the Aβ sequence, the stacking of spin labels lead to strong spin exchange interactions. Normally, a nitroxide spin label gives rise to an EPR spectrum with three spectral lines, but the strong exchange interaction causes the three lines to collapse into a single spectral line (Figure 1A). The single-line spectrum serves as a signature for the strong exchange interaction (Margittai & Langen 2008). The spin exchange interaction is related to inter-spin distance by an exponential function and quickly diminishes when inter-spin distances are beyond ∼7 Å. Therefore, if the fibrils are formed by a mixture of spin-labeled and unlabeled Aβ, the EPR spectrum will lose the single-line feature (Figure 1B). The mixing experiment of spin-labeled and unlabeled samples is referred to as “spin dilution”.
Figure 1. Effect of spin dilution on the EPR lineshape of spin-labeled fibrils.
Red balls represent spin labels. (A) In a parallel in-register β-sheet structure of amyloid fibrils, the spin label side chains pack closely against each other and lead to strong spin exchange interactions between spin labels. As a result, the EPR spectrum shows a characteristic single-line feature. (B) When the fibrils are formed by spin-labeled Aβ and unlabeled Aβ (i.e. spin dilution), the inter-spin label spacing is more than ∼7 Å, and thus spin exchange interactions are weak, leading to the normal three-line spectrum.
Spin dilution provides a way to study the interactions between Aβ42 and Aβ40 at molecular level. When spin-labeled Aβ42 is mixed with Aβ42 WT, the EPR spectrum will change from a single-line to a three-line feature. When spin-labeled Aβ42 is mixed with Aβ40 WT, the EPR lineshape depends on whether Aβ40 WT is incorporated into spin-labeled Aβ42 fibrils. If Aβ40 WT forms fibrils only with Aβ40 WT, then no spin dilution effect will be seen and the EPR spectrum will be single-line. On the other hand, if Aβ40 WT forms interlaced fibrils with Aβ42, then we will observe similar effect of spin dilution as with Aβ42 WT. With spin dilution, we can study the interactions of Aβ42 and Aβ40 at high spatial resolution. A simple co-aggregation without interlacing of Aβ42 and Aβ40 in the fibril would not generate any spin dilution effect.
Similar morphologies for spin-labeled Aβ42 fibrils in the presence and absence of spin dilution
We introduced a spin label named R1 at position 17 of Aβ42 sequence. The chemical structure of R1 is shown in Figure 2A. Previous studies show that Aβ40 L17R1 gives rise to a single-line spectrum, suggesting strong spin exchange interactions at this position (Agopian & Guo 2012). Residue Leu-17 is located at the beginning of the central hydrophobic cluster, and previous studies suggest that Leu-17 is well ordered in Aβ42 fibrils (Lührs et al. 2005, Olofsson et al. 2006). To study the effect of spin dilution with Aβ42 and Aβ40 WT proteins, we prepared five fibril samples under agitated condition: (1) Aβ42 L17R1; (2) Aβ42 L17R1 with Aβ42 WT at 1:1 molar ratio; (3) Aβ42 L17R1 with Aβ42 WT at 1:3 molar ratio; (4) Aβ42 L17R1 with Aβ40 WT at 1:1 molar ratio; (5) Aβ42 L17R1 with Aβ40 WT at 1:3 molar ratio.
Figure 2. Characterization of spin labeled Aβ42 fibrils.
(A) Chemical structure of spin label R1 used in this work. (B) Transmission electron microscopy images show similar morphologies for Aβ42 L17R1 and its mixture with either Aβ40 or Aβ42 wild type peptides.
Transmission electron microscopy shows that Aβ42 L17R1 forms short straight fibrils (Figure 2B). The fibrils have similar morphologies as previously studied Aβ40 fibrils under agitated conditions (Petkova et al. 2005, Kodali et al. 2010, Agopian & Guo 2012). We chose agitated condition because solid-state NMR studies show that agitation leads to highly homogeneous fibrils for Aβ40 (Bertini et al. 2011). Our previous study also reveals a more pronounced single-line feature for agitated Aβ40 fibrils when compared to quiescent fibrils (Agopian & Guo 2012), suggesting better packing interactions in agitated fibrils. When spin labeled Aβ42 is mixed with either Aβ42 WT or Aβ40 WT, the morphologies of the fibrils remain unchanged, suggesting that similar fibril structures in different samples (Figure 2B). Our results are consistent with previous findings that mixtures of Aβ42 and Aβ40 at different ratios form similar fibrils as Aβ42 or Aβ40 alone (Pauwels et al. 2012).
Effect of spin dilution on EPR spectral lineshape of spin-labeled Aβ42 fibrils
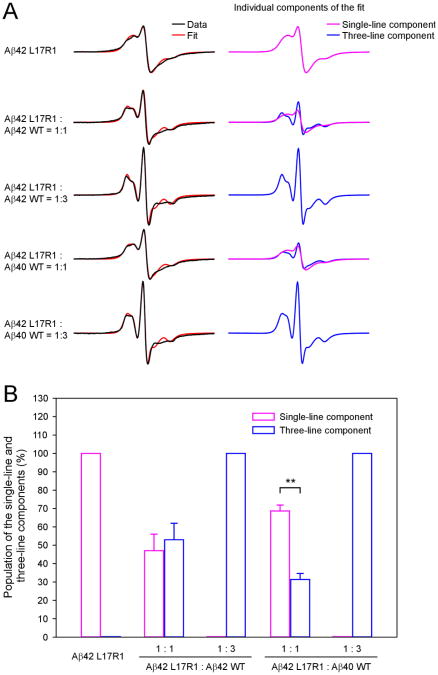
Aβ42 L17R1 fibrils gave rise to a single-line spectrum, suggesting a parallel in-register β structure in Aβ42 fibrils (Figure 3A, black traces). Spin dilution with Aβ42 WT at 1:1 and 1:3 molar ratios led to an evident three-line feature. Similar effects were observed when spin dilution was performed with Aβ40 WT. These results suggest that Aβ40 WT can be incorporated into Aβ42 fibrils. Meanwhile, the EPR lineshape for Aβ42 WT-diluted fibrils is not identical to Aβ40 WT-diluted fibrils, suggesting that Aβ40 WT is not equivalent to Aβ42 WT in terms of co-fibrillization with spin-labeled Aβ42.
Figure 3. EPR analysis of spin-labeled Aβ42 fibrils.
(A) EPR spectra of fully labeled Aβ42 fibrils and spin-diluted with either Aβ42 or Aβ40 wild-type proteins. The experimental spectra are shown in black and the best fits from spectral simulations are shown in red. Individual spectral components are shown in magenta and blue. (B) A bar graph showing the population of the single-line and three-line components from spectral simulations in the absence and presence of spin dilutions. For the 1:1 spin dilutions, results are expressed as mean ± SD (**p < 0.01, Student's t-test).
We performed spectral simulations to quantitatively analyze the population of single-line and three-line components in the spin-diluted spectra. The best non-linear least squares fits are shown in Figure 3A (red traces). The spectrum of fibrils formed by Aβ42 L17R1 can be fitted with only a single-line component. On the other hand, when spin dilution is performed at the 1:3 molar ratio, the spectrum of spin-diluted fibrils, whether with Aβ42 or Aβ40 WT, can be fitted with only a three-line component, suggesting that Aβ40 WT is capable of completely diluting out the spin-labeled Aβ42. Distributions of different fibril populations as a result of different Aβ42 and Aβ40 mixing ratios are summarized in Figure 4.
Figure 4. A schematic drawing shows distributions of different fibril populations resulting from spin dilution of spin-labeled Aβ42 with unlabeled Aβ40.
With 1:3 dilution, two fibril populations are present: interlaced fibrils and Aβ40 fibrils. With 1:1 dilution, all three fibril populations (labeled Aβ42, interlaced, and unlabeled Aβ40) are present.
When spin dilution is performed at 1:1 molar ratio, the EPR spectra contain both the single-line and three-line components (Figure 3A). Aβ42 WT-diluted spectrum consists of 47% single-line and 53% three-line components, whereas Aβ40 WT-diluted spectrum consists of 69% single-line and 31% three-line components (Figure 3B). Because the three-line component is a result of interlacing between unlabeled and labeled Aβ in the fibrils, the ratio of the three-line component populations in Aβ40- versus Aβ42-diluted samples can be used as an estimate for the relative efficiency of incorporating Aβ40 into Aβ42 fibrils. Using the data from Figure 3B, the efficiency of incorporating Aβ40 into Aβ42 fibrils, relative to incorporating Aβ42 into Aβ42 fibrils, is 58% (= 31% / 53%). In other words, when mixing Aβ40 and Aβ42 at 1:1 ratio, approximately 58% Aβ40 proteins are forming interlaced fibrils with Aβ42, and the remaining 42% forms Aβ40-only fibrils.
Overall, the EPR studies in this work reveal interactions between Aβ40 and Aβ42 in amyloid fibrils at a resolution of <10 Å. The strong spin exchange interaction that gives rise to the single-line EPR spectrum requires spin labels to be within ∼7 Å of each other, essentially the distance between adjacent β-strands in the fibril. The spin dilution experiments suggest that Aβ40 WT is able to disrupt the spin exchange interactions in spin-labeled Aβ42, suggesting Aβ40 must be incorporated in between spin-labeled Aβ42 molecules. Previous studies have suggested that Aβ42 and Aβ40, when mixed at different ratios, affect each other's aggregation behavior and the toxicities of the resulting aggregates (Jan et al. 2008, Yan & Wang 2007, Kuperstein et al. 2010, Pauwels et al. 2012). Sequestration of fluorescently-labeled Aβ42 with unlabeled Aβ40 (or vice versa) suggests that Aβ42 and Aβ40 form mixed aggregates (Frost et al. 2003), but the structural resolution for the mixed aggregates was low. A previous EPR study has shown that Aβ42 can be incorporated into spin-labeled Aβ40 fibrils (Török et al. 2002), but it was not clear whether Aβ40 can be incorporated into Aβ42 fibrils. Furthermore, Török et al. (2002) suggested that Aβ42 and Aβ40 co-mix equally well with spin-labeled Aβ40. Our quantitative analysis suggests that Aβ40 is less efficient in terms of being incorporated into Aβ42 fibrils. Our results are consistent with previous surface plasmon resonance studies by Pauwels et al. (2012) suggesting that monomeric Aβ42 and Aβ40 interact with each other, albeit less strongly than with themselves.
Implications for AD pathogenesis
Specific interactions between Aβ42 and Aβ40 as revealed by EPR provide insight on how Aβ42 to Aβ40 ratio plays an important role in AD. Our results suggest that coexistence of Aβ42 and Aβ40 in the extracellular space may generate aggregates containing three populations: Aβ42 alone, Aβ40 alone, and Aβ42/Aβ40 mix. Transgenic mice with over expression of Aβ40 alone do not develop amyloid pathology or form insoluble aggregates (McGowan et al. 2005), suggesting that the aggregation of Aβ40 is too slow to account for amyloid deposition in AD. The mixture of Aβ42 and Aβ40 also aggregates slowly compared to Aβ42 alone (Snyder et al. 1994, Yoshiike et al. 2003, Yan & Wang 2007, Jan et al. 2008, Kuperstein et al. 2010, Pauwels et al. 2012). Therefore, it is likely that the Aβ42-alone population is responsible for amyloid pathology in AD. The spin dilution experiments in this work show that the amount of Aβ42-alone population is determined by the Aβ42 to Aβ40 ratio. At Aβ42:Aβ40 ratio of 1:3, most of Aβ42 is in the Aβ42/Aβ40 mixture, while at Aβ42:Aβ40 ratio of 1:1, ∼69% of Aβ42 exists in Aβ42-alone population (Figure 3). As a result, increases in Aβ42/Aβ40 ratio lead to increased Aβ42-alone population, which causes AD pathology.
The finding that Aβ42 and Aβ40 form interlaced amyloid fibrils raises an intriguing question regarding in vivo Aβ aggregation. The amyloid plaques in AD brains contain mostly, sometimes only, Aβ42 (Iwatsubo et al. 1995, Iwatsubo et al. 1994, Gravina et al. 1995, Miller et al. 1993, Mak et al. 1994), even though the Aβ40 concentration in cerebrospinal fluid is several folds more than Aβ42. What is the explanation for this phenomenon when Aβ40 can be incorporated into Aβ42 fibrils in vitro? Two potential mechanisms may contribute to the preferential deposition of Aβ42 in amyloid plaques. First, the local concentration of Aβ42 at plaque deposition sites may be significantly higher than currently assumed. Aβ concentration in the extracellular space of human brain is difficult to analyze. An intracerebral microdialysis study of brain interstitial fluid Aβ in patients with acute brain injuries shows high Aβ40/Aβ42 ratios (∼14) for the pooled sample of 5 patients (Brody et al. 2008). However, microdialysis studies can only detect Aβ peptides in solution. Preferential binding of Aβ42 to extracellular matrix or cell membrane may lead to a high local Aβ42 concentration. Second, the aggregation of Aβ42 in vivo may be assisted by some other proteins or cell components such as lipids, which may preferentially promote the aggregation of Aβ42. Future investigations to achieve a better understanding of the discrepancy between in vitro and in vivo observations may shed new light on the pathogenesis of AD.
Acknowledgments
We thank Dr. Christian Altenbach and Dr. Wayne Hubbell for providing EPR analysis programs. This work was supported by the Alzheimer's Association (Grant NIRG-09-133555) and American Health Assistance Foundation (Grant A2010362). We acknowledge the use of instruments at the Electron Imaging Center for NanoMachines supported by NIH (1S10RR23057 to ZHZ) at the California NanoSystems Institute, UCLA. The authors declare no conflict of interest.
Abbreviations
- Aβ
amyloid β
- AD
Alzheimer's disease
- EPR
electron paramagnetic resonance
- WT
wild-type
References
- Agopian A, Guo Z. Structural origin of polymorphism for Alzheimer's amyloid-β fibrils. Biochem J. 2012;447:43–50. doi: 10.1042/BJ20120034. [DOI] [PubMed] [Google Scholar]
- Baker RT, Catanzariti AM, Karunasekara Y, Soboleva TA, Sharwood R, Whitney S, Board PG. Using deubiquitylating enzymes as research tools. Methods Enzymol. 2005;398:540–554. doi: 10.1016/S0076-6879(05)98044-0. [DOI] [PubMed] [Google Scholar]
- Bertini I, Gonnelli L, Luchinat C, Mao J, Nesi A. A new structural model of Aβ40 fibrils. J Am Chem Soc. 2011;133:16013–16022. doi: 10.1021/ja2035859. [DOI] [PubMed] [Google Scholar]
- Brody DL, Magnoni S, Schwetye KE, Spinner ML, Esparza TJ, Stocchetti N, Zipfel GJ, Holtzman DM. Amyloid-beta dynamics correlate with neurological status in the injured human brain. Science. 2008;321:1221–1224. doi: 10.1126/science.1161591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budil DE, Lee S, Saxena S, Freed JH. Nonlinear-least-squares analysis of slow-motion EPR spectra in one and two dimensions using a modified Levenberg-Marquardt algorithm. J Magn Reson, Ser A. 1996;120:155–189. [Google Scholar]
- Columbus L, Kálai T, Jeko J, Hideg K, Hubbell WL. Molecular motion of spin labeled side chains in α-helices: analysis by variation of side chain structure. Biochemistry. 2001;40:3828–3846. doi: 10.1021/bi002645h. [DOI] [PubMed] [Google Scholar]
- Frost D, Gorman PM, Yip CM, Chakrabartty A. Co-incorporation of Aβ40 and Aβ42 to form mixed pre-fibrillar aggregates. Eur J Biochem. 2003;270:654–663. doi: 10.1046/j.1432-1033.2003.03415.x. [DOI] [PubMed] [Google Scholar]
- Gravina SA, Ho LB, Eckman CB, Long KE, Otvos L, Younkin LH, Suzuki N, Younkin SG. Amyloid β protein (Aβ) in Alzheimer's disease brain: Biochemical and immunocytochemical analysis with antibodies specific for forms ending at Aβ40 or Aβ42(43) J Biol Chem. 1995;270:7013–7016. doi: 10.1074/jbc.270.13.7013. [DOI] [PubMed] [Google Scholar]
- Iwatsubo T, Mann DMA, Odaka A, Suzuki N, Ihara Y. Amyloid β protein (Aβ) deposition: Aβ42(43) precedes Aβ40 in Down syndrome. Ann Neurol. 1995;37:294–299. doi: 10.1002/ana.410370305. [DOI] [PubMed] [Google Scholar]
- Iwatsubo T, Odaka A, Suzuki N, Mizusawa H, Nukina N, Ihara Y. Visualization of Aβ42(43) and Aβ40 in senile plaques with end-specific Aβ monoclonals: Evidence that an initially deposited species is Aβ42(43) Neuron. 1994;13:45–53. doi: 10.1016/0896-6273(94)90458-8. [DOI] [PubMed] [Google Scholar]
- Jan A, Gokce O, Luthi-Carter R, Lashuel HA. The ratio of monomeric to aggregated forms of Abeta40 and Abeta42 is an important determinant of amyloid-beta aggregation, fibrillogenesis, and toxicity. J Biol Chem. 2008;283:28176–28189. doi: 10.1074/jbc.M803159200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Onstead L, Randle S, Price R, Smithson L, Zwizinski C, Dickson DW, Golde T, McGowan E. Abeta40 inhibits amyloid deposition in vivo. J Neurosci. 2007;27:627–633. doi: 10.1523/JNEUROSCI.4849-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodali R, Williams AD, Chemuru S, Wetzel R. Aβ(1-40) forms five distinct amyloid structures whose β-sheet contents and fibril stabilities are correlated. J Mol Biol. 2010;401:503–517. doi: 10.1016/j.jmb.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar-Singh S, Theuns J, Van Broeck B, et al. Mean age-of-onset of familial Alzheimer disease caused by presenilin mutations correlates with both increased Aβ42 and decreased Aβ40. Hum Mutat. 2006;27:686–695. doi: 10.1002/humu.20336. [DOI] [PubMed] [Google Scholar]
- Kuperstein I, Broersen K, Benilova I, et al. Neurotoxicity of Alzheimer's disease Abeta peptides is induced by small changes in the Aβ42 to Aβ40 ratio. EMBO J. 2010;29:3408–3420. doi: 10.1038/emboj.2010.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lührs T, Ritter C, Adrian M, Riek-Loher D, Bohrmann B, Doeli H, Schubert D, Riek R. 3D structure of Alzheimer's amyloid-β(1-42) fibrils. Proc Natl Acad Sci USA. 2005;102:17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak K, Yang FS, Vinters HV, Frautschy SA, Cole GM. Polyclonals to beta-amyloid(1-42) identify most plaque and vascular deposits in Alzheimer cortex, but not striatum. Brain Res. 1994;667:138–142. doi: 10.1016/0006-8993(94)91725-6. [DOI] [PubMed] [Google Scholar]
- Margittai M, Langen R. Fibrils with parallel in-register structure constitute a major class of amyloid fibrils: molecular insights from electron paramagnetic resonance spectroscopy. Q Rev Biophys. 2008;41:265–297. doi: 10.1017/S0033583508004733. [DOI] [PubMed] [Google Scholar]
- McGowan E, Pickford F, Kim J, et al. Abeta42 is essential for parenchymal and vascular amyloid deposition in mice. Neuron. 2005;47:191–199. doi: 10.1016/j.neuron.2005.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DL, Papayannopoulos IA, Styles J, Bobin SA, Lin YY, Biemann K, Iqbal K. Peptide compositions of the cerebrovascular and senile plaque core amyloid deposits of Alzheimer's disease. Arch Biochem Biophys. 1993;301:41–52. doi: 10.1006/abbi.1993.1112. [DOI] [PubMed] [Google Scholar]
- Ngo S, Guo Z. Key residues for the oligomerization of Aβ42 protein in Alzheimer's disease. Biochem Biophys Res Commun. 2011;414:512–516. doi: 10.1016/j.bbrc.2011.09.097. [DOI] [PubMed] [Google Scholar]
- Olofsson A, Sauer-Eriksson AE, Ohman A. The solvent protection of Alzheimer amyloid-β-(1-42) fibrils as determined by solution NMR spectroscopy. J Biol Chem. 2006;281:477–483. doi: 10.1074/jbc.M508962200. [DOI] [PubMed] [Google Scholar]
- Pauwels K, Williams TL, Morris KL, et al. The structural basis for increased toxicity of pathological Aβ42:Aβ40 ratios in Alzheimer's disease. J Biol Chem. 2012;287:5650–5660. doi: 10.1074/jbc.M111.264473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkova AT, Leapman RD, Guo ZH, Yau WM, Mattson MP, Tycko R. Self-propagating, molecular-level polymorphism in Alzheimer's β-amyloid fibrils. Science. 2005;307:262–265. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
- Schneider DJ, Freed JH. Calculating slow motional magnetic resonance specta. In: Berliner LJ, Reuben J, editors. Spin Labeling: Theory and Applications. Plenum Press; New York: 1989. p. 1.p. 76. [Google Scholar]
- Shahnawaz M, Thapa A, Park IS. Stable activity of a deubiquitylating enzyme (Usp2-cc) in the presence of high concentrations of urea and its application to purify aggregation-prone peptides. Biochem Biophys Res Commun. 2007;359:801–805. doi: 10.1016/j.bbrc.2007.05.186. [DOI] [PubMed] [Google Scholar]
- Snyder SW, Ladror US, Wade WS, Wang GT, Barrett LW, Matayoshi ED, Huffaker HJ, Krafft GA, Holzman TF. Amyloid-beta aggregation: Selective inhibition of aggregation in mixtures of amyloid with different chain lengths. Biophys J. 1994;67:1216–1228. doi: 10.1016/S0006-3495(94)80591-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Török M, Milton S, Kayed R, Wu P, McIntire T, Glabe CG, Langen R. Structural and dynamic features of Alzheimer's Aβ peptide in amyloid fibrils studied by site-directed spin labeling. J Biol Chem. 2002;277:40810–40815. doi: 10.1074/jbc.M205659200. [DOI] [PubMed] [Google Scholar]
- Tycko R. Molecular structure of amyloid fibrils: insights from solid-state NMR. Q Rev Biophys. 2006;39:1–55. doi: 10.1017/S0033583506004173. [DOI] [PubMed] [Google Scholar]
- Yan Y, Wang C. Aβ40 protects non-toxic Aβ42 monomer from aggregation. J Mol Biol. 2007;369:909–916. doi: 10.1016/j.jmb.2007.04.014. [DOI] [PubMed] [Google Scholar]
- Yoshiike Y, Chui DH, Akagi T, Tanaka N, Takashima A. Specific compositions of amyloid-beta peptides as the determinant of toxic beta-aggregation. J Biol Chem. 2003;278:23648–23655. doi: 10.1074/jbc.M212785200. [DOI] [PubMed] [Google Scholar]