Abstract
Tyrosine depletion in metazoan proteins was recently explained to be due to the appearance of tyrosine kinases in Metazoa. Here, we present a complementary explanation for the depletion of tyrosine, stating the importance of tyrosine in signaling not only as a phosphorylation target, but also as a precursor for catecholamines and hormones. Molecules (dopamine, norepinephrine, and epinephrine, and to a lesser extent serotonin and melatonin) critical to metazoan multicellular signaling are also greatly dependent on a supply of tyrosine. These signaling molecules are synthesized in two highly linked pathways specific to metazoans. In addition, the shikimate pathway that non-metazoans use to synthesize the aromatic amino acids is not present in metazoans. These important pathway changes have occurred between Metazoa and other eukaryotes, causing significant changes to tyrosine metabolism and rendering tyrosine crucial for extracellular signaling. Additionally, the evolutionary and functional linkage between these two pathways and the resulting implications for neuropathology are discussed.
Keywords: tyrosine, dopamine, serotonin, Metazoa, pathway, evolution
Introduction
Recently, Tan et al. reported positive selection for tyrosine loss in metazoan proteins (Tan et al. 2009). The underlying reason for this was first speculated to be the concurrent emergence of tyrosine kinases in metazoan genomes, and indeed tyrosine phosphorylation is an important regulatory mechanism in metazoan signaling. However, Su and colleagues presented an alternative hypothesis, with increased GC content driving the reduction of the amount of tyrosine in metazoan proteins (Su et al. 2011). Both of these events may have contributed to the reduction of tyrosine in metazoan proteins to different extents, but there are additional important metazoan innovations missing from this discussion. One important change between metazoan and other organisms in the tree of life is the development of a nervous system accompanied by extracellular neurostransmitter signaling. Tyrosine is a key player in this system.
First, molecules originating with tyrosine include melanin and the catecholamine neurotransmitters: dopamine, norepinephrine, and epinephrine (Kanehisa et al. 2012; Kanehisa and Goto 2000). Tryptophan metabolism occurs in a parallel pathway to that of tyrosine, and it results in the production of serotonin and melatonin (Kanehisa et al. 2012; Kanehisa and Goto 2000), another monoamine neurotransmitter and a hormone, respectively. These molecules have significant roles in extracellular signaling. Second, the pathway to synthesize the aromatic amino acids in other eukaryotes is lost in Metazoa. In order to illuminate the importance of tyrosine in Metazoa, we complement the finding of positive selection of tyrosine loss in metazoan proteins by describing a concurrent biochemical revolution and the consequential evolution of the underlying systems biology.
The shikimate pathway used by prokaryotes, plants, and Protozoa is not present in Metazoa (Herrmann and Weaver 1999). Instead, an enzyme converting phenylalanine to tyrosine present in some protozoan species (Siltberg-Liberles et al. 2008) about 1200 mya underwent at least two gene duplications over the next 300 mya (Hedges et al. 2006). In Metazoa, this enzyme family, denoted as the aromatic amino acid hydroxylases (AAAHs), consists of a minimum of three members: phenylalanine hydroxylase (PAH), tyrosine hydroxylase (TH), and tryptophan hydroxylase (TPH). As their names imply, these enzymes hydroxylate their substrates: PAH converts L-Phe to L-Tyr, TH converts L-Tyr to L-Dopa, and TPH converts L-Trp to 5-hydroxytryptophan, a precursor of serotonin. The AAAHs are missing from most non-metazoan genomes, although the catalytic domain of AAAH has been found in some bacteria and in some non-metazoan eukaryotes (Cao et al. 2010; Siltberg-Liberles et al. 2008). The loss of the shikimate pathway rendered Metazoa unable to synthesize phenylalanine. Subsequently, phenylalanine has become an essential amino acid in most Metazoa (Fitzgerald and Szmant 1997). There are a few known exceptions in basal Metazoa, where a selection of corals have been found to synthesize phenylalanine (Fitzgerald and Szmant 1997), and components of the shikimate pathway were found in the genome of a close relative of the corals, Nematostella vectensis (Starcevic et al. 2008). However, phylogenetic analysis indicates that these components are of bacterial origin and horizontal gene transfer is a more likely explanation (Starcevic et al. 2008). Metazoan dependence on an intake of L-Phe in order to synthesize tyrosine is noteworthy for the positive selection of tyrosine loss in metazoan proteins.
Tyrosine to L-Dopa hydroxylation by TH is the beginning and rate limiting step of the pathway that generates L-Dopa, dopamine, norepinephrine, and epinephrine. TH is highly regulated by feedback inhibition by the downstream products in this pathway (Fitzpatrick 1999). The last AAAH, TPH, converts tryptophan into 5-hydroxytryptophan, a precursor of serotonin and melatonin. This is the initial step in the tryptophan metabolic pathway, which is separate from the tyrosine metabolic pathway where PAH and TH operate. However, these two pathways are connected on multiple levels. First, all AAAHs are dependent on tetrahydrobiopterin (BH4) as a cofactor. Km for BH4 is 2-3 μM for PAH and about 30 μM for both TH and TPH (Thöny et al. 2000). The higher affinity for the cofactor in PAH would allow the L-Phe to L-Tyr conversion to be more favorable than L-Tyr to L-Dopa or L-Trp to 5-hydroxytryptophan if the supply of BH4 is limited. Further linking the two pathways, the next step in both is catalyzed by the same enzyme, aromatic amino acid decarboxylase (AADC).
Here, we have surveyed selected eukaryotic genomes for the enzymes involved in the tyrosine and tryptophan pathways. The purpose of this investigation is to benchmark how tyrosine became an important signaling molecule in metazoans, not only as a phosphorylation target, but also as part of a metazoan-specific rewiring of the aromatic amino acids systems biology simultaneous with the appearance of tyrosine kinases.
Results
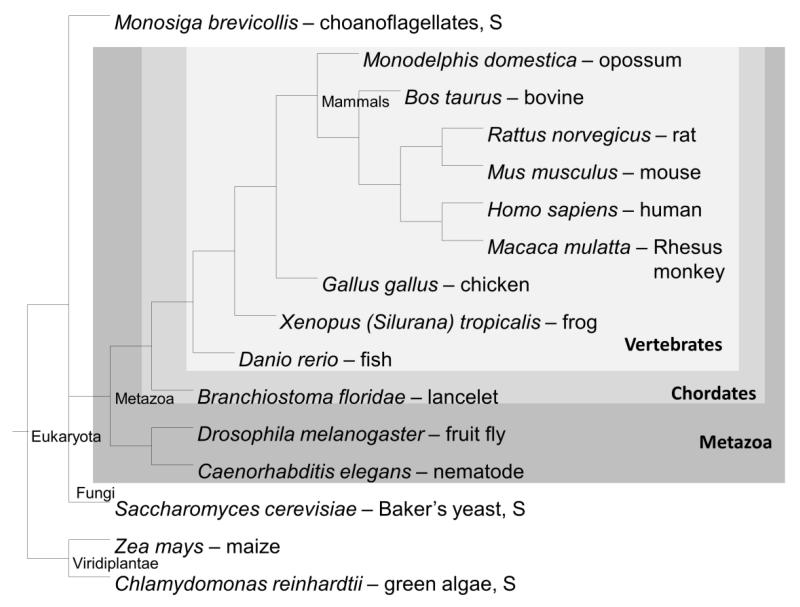
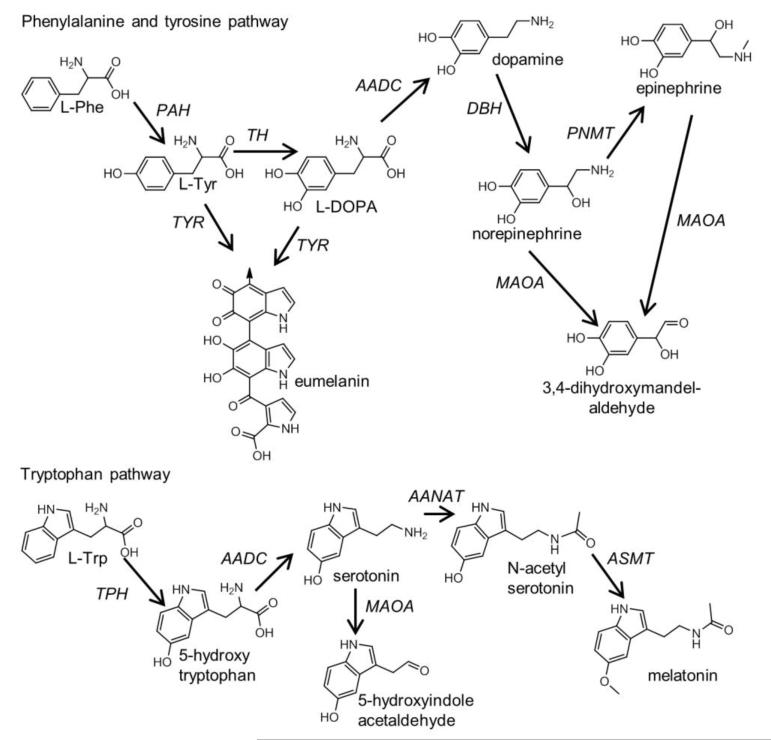
We investigated the selected eukaryotic species (Homo sapiens, Macaca mulatta, Rattus norvegicus, Mus musculus, Bos taurus, Monodelphis domestica, Gallus gallus, Xenopus tropicalis, Danio rerio, Branchiostoma floridae, Drosophila melanogaster, Caenorhabditis elegans, Saccharomyces cerevisiae, Monosiga brevicollis, Zea mays, and Chlamydomonas reinhardtii) (Figure 1) for homologs of the enzymes in the human tyrosine and tryptophan metabolic pathways. The enzymes in the tyrosine pathway convert the following reactions: L-Phe to L-Tyr (PAH), L-Tyr to L-Dopa (TH), L-Dopa to dopamine (aromatic amino acid decarboxylase [AADC]), dopamine to norepinephrine (dopamine beta hydroxylase [DBH]), and norepinephrine to epinephrine (phenylethanolamine N-methyltransferase [PNMT]) (Figure 2A). The enzymes in the tryptophan pathway convert the following reactions: L-Trp to 5-hydroxytryptophan (TPH), 5-hydroxytryptophan to serotonin (AADC), serotonin to N-acetylserotonin (arylalkylamine N-acetyltransferase [AANAT]), and N-acetylserotonin to melatonin (acetylserotonin O-metyltransferase [ASMT]) (Figure 2B). We also investigated two additional enzymes linked to these pathways: (1) monoamine oxidase A (MAOA), which degrades the monoamines norepinephrine, epinephrine, and serotonin, and (2) tyrosinase (TYR), which influences melanin production by converting L-Dopa to eumelanin, a form of melanin (Figure 2). We built all phylogenetic trees, besides PAH, TH, and TPH, using PhyML (Guindon et al. 2005). The phylogeny of AAAH (PAH, TH, and TPH) has recently been published (Siltberg-Liberles et al. 2008).
Figure 1.
The species tree. The phylogenetic relationships among the species included in this study according to the NCBI taxonomy, according to the ecdysozoa topology (Holton and Pisani 2010), is shown. The following abbreviations are used in the remainder of this text: (Homo sapiens: H. sapiens, Macaca mulatta: M. mulatta, Rattus norvegicus: R. norvegicus, Mus musculus: M. musculus, Bos taurus: B. taurus, Monodelphis domestica: M. domestica, Gallus gallus: G. gallus, Xenopus tropicalis: X. tropicalis, Danio rerio: D. rerio, Branchiostoma floridae: B. floridae, Drosophila melanogaster: D. melanogaster, Caenorhabditis elegans: C. elegans, Saccharomyces cerevisiae: S. cerevisiae, Monosiga brevicollis: M. brevicollis, Zea mays: Z. mays, and Chlamydomonas reinhardtii: C. reinhardtii). If a species is denoted ‘S’ it means ‘Single cell eukaryote,’ all others are multicellular eukaryotes.
Figure 2.
The tyrosine and tryptophan pathways in Metazoa.
The tyrosine pathway
AADC
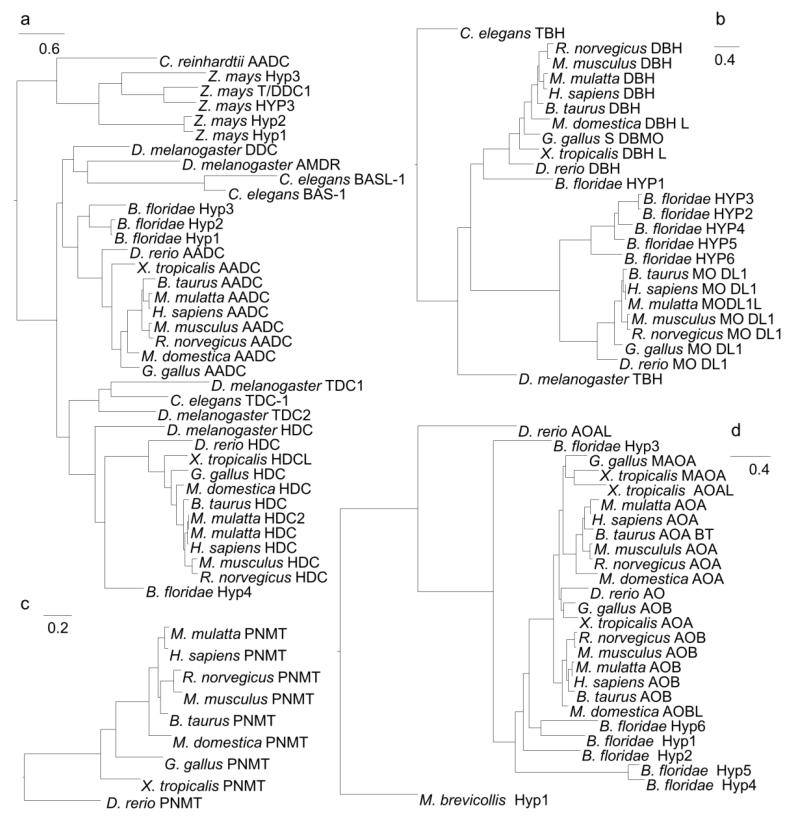
The phylogeny for AADC shows that homologs of AADC are present in both metazoans and plants (Figure 3A). The AADC protein is present with a single copy in vertebrates, while B. floridae, D. melanogaster, and C. elegans have at least two lineage-specific copies of this protein. Two paralogs, histidine decarboxylase (HDC) and tyrosine decarboxylase (TDC), are present in the tree. TDC is present in non-chordate metazoans but not in vertebrates, and HDC is present in all chordates and D. melanogaster, but not in C. elegans. The tree implies that TDC and HDC may have had a common ancestor in an ancestral metazoan and evolved different substrate specificities after the chordate-arthropod split. In addition, C. reinhardtii has a single copy classified as AADC that forms an outgroup to all metazoan AADC homologs. Zea mays also has five lineage-specific AADC homologs.
Figure 3.
Phylogenies for the different enzymes in the tyrosine and tryptophan pathways, AADC (A), DBH (B), PNMT (C), MAOA (D), AANAT (E), ASMT (F), and TYR (G).
DBH
The phylogeny for DBH shows two clades, DBH and its paralog DBH-like monooxygenase, with tyramine β-hydroxylase from C. elegans and D. melanogaster as outgroups (Figure 3B). This pattern is consistent with a chordate-specific gene duplication of tyramine β-hydroxylase, resulting in DBH and DBH-like monooxygenase. Both DBH and DBH-like are members of the copper monooxygenase family of proteins (Xin et al. 2004).
PNMT
The phylogeny for PNMT reveals a vertebrate-specific enzyme with no paralogs (Figure 3C). No related proteins were found in vertebrates or other organisms.
MAOA
The phylogeny for MAOA is chordate-specific, but a hypothetical protein in M. brevicollis forms an outgroup (Figure 3D). The phylogenies for both the MAOA and the MAOB clades follow the correct species taxonomy for mammals. Outside the mammalian clades the species taxonomy is less accurately reconstructed. MAOA from G. gallus and X. tropicalis are found as outgroups to the MAOA clade, but additional copies from these species are grouped with the AO copy from D. rerio in the MAOA clade. For the MAOB clade, there are no copies from non-mammalian species. B. floridae has six hypothetical copies; all form outgroups to the MAOA and MAOB clades, but a divergent AOA-like from D. rerio in turn forms an outgroup to these six copies. Setini, Pierucci, Senatori, and Nicotra found evidence for only one copy of MAO in D. rerio (Setini et al. 2005), indicating that the distant AOA-like copy from D. rerio may no longer perform the MAO function. MAOA and MAOB perform highly similar functions with different specificities. MAOA has a higher affinity than MAOB for norepinephrine, epinephrine, and serotonin (Haavik et al. 2008; Wells and Bjorksten 1989). The pattern we observe suggests that the principal MAOA and MAOB functions are likely conserved in mammals but may vary in non-mammals.
The tryptophan pathway
As described above, AADC is found in all metazoans surveyed except for C. elegans. The ancestor of AADC appears to predate Metazoa as homologs are found in plants.
AANAT
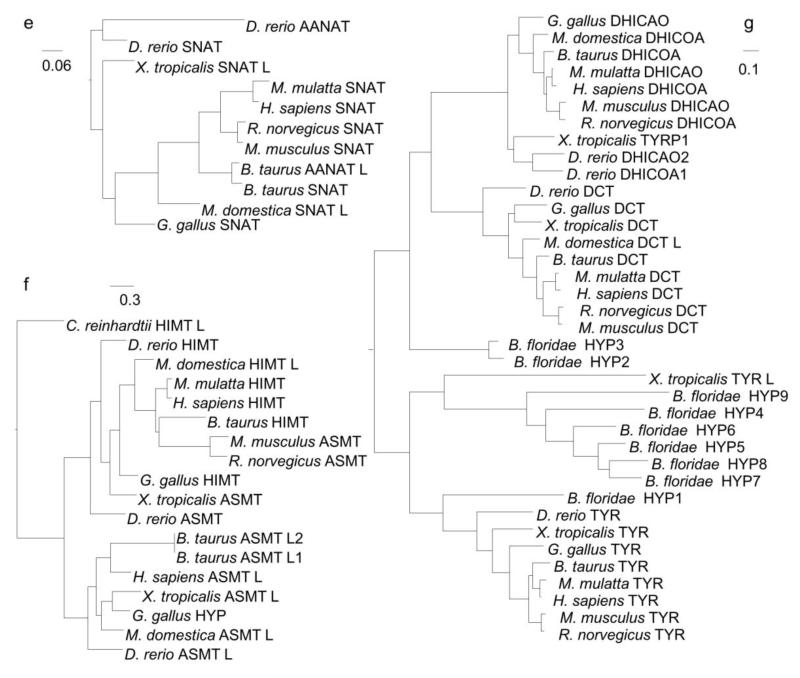
The phylogeny for AANAT demonstrates a vertebrate-specific enzyme family (Figure 3E). Coon, Bégay, Deurloo, Falcón, and Klein found two copies of AANAT in bony fishes (Coon et al. 1999), which is consistent with the two zebrafish genes present in this tree.
ASMT
The phylogeny for ASMT reveals it is another vertebrate-specific enzyme (Figure 3F). ASMT has a paralog, ASMT-like, in some of the vertebrate species investigated here, but it appears to be absent from many of the mammals investigated in this study. Finally, it should be noted that although no ASMT or ASMT-like enzyme was found in non-vertebrate metazoans, a remote homolog is present in C. reinhardtii. This outgroup suggests that ASMT may have evolved from a protein in an ancestral eukaryote and/or a horizontal gene transfer event occurred.
TYR
The phylogeny for TYR revealed two paralogs in other vertebrates, l-dopachrome tautomerase (DCT) and tyrosinase-related protein 1 (TYRP1), while TYR itself is chordate specific (Figure 3G). DCT and TYRP1 belong to a different part of the tyrosine metabolic pathway (Kanehisa et al. 2012). Similar to TYR, together they assist in converting l-DOPA into eumelanin. As observed for previous phylogenies, B. floridae has several lineage-specific hypothetical proteins.
To summarize the phylogenies for the enzymes in the tyrosine and tryptophan metabolic pathways generated here, it is clear that these pathways have continuously evolved, adding novel enzymes, functions, and signaling molecules as organismal complexity increased and effective population size decreased.
Discussion
Overall, it was found that enzymes involved in tyrosine and tryptophan metabolism narrow in phyletic distribution further down the pathways, consistent with more recent expansions of these pathways in vertebrates. The tyrosine pathway begins with PAH converting phenylalanine into tyrosine, which TH then converts into L-Dopa. PAH and TH are both found throughout Metazoa (Siltberg-Liberles et al. 2008). The next step in the pathway is L-Dopa to dopamine, catalyzed by AADC. AADC is found across Metazoa. Next, DBH, which catalyzes dopamine to norepinephrine, is chordate-specific. DBH has homologs in C. elegans and D. melanogaster that predate the chordate-specific gene duplication which yielded DBH and DBH-like monooxygenase. It is possible that these distant copies are able to perform similar functions. PNMT, which catalyzes norepinephrine to epinephrine, is only found in vertebrates and no other homologs were detected. MAOA, which is involved in catabolism of these catecholamines is chordate-specific with a distant homolog found in M. brevicollis.
The tryptophan metabolic pathway is structured in a similar manner to the tyrosine pathway. TPH is present in Metazoa (Siltberg-Liberles et al. 2008). However, a gene duplication in chordates or vertebrates resulted in TPH1 and TPH2, with TPH2 being specific to the central nervous system (Walther et al. 2003). The next enzyme in the pathway, AADC, is shared with the tyrosine pathway. Here it converts 5-hydroxytryptophan to serotonin. The two last enzymes in the tryptophan pathway that convert serotonin to N-actetylserotonin and N-acetylserotonin to melatonin are AANAT and ASMT, respectively. These are both vertebrate-specific (Figure 3E,F).
TYR and its homologs, DCT and TYRP1, are all tyrosinases. It is still unclear whether tyrosinase is expressed in the central nervous system (CNS; 19, 20) and the mechanism for neuromelanin formation in the CNS is unknown.
The reported positive selection of tyrosine loss in protein during metazoan evolution shows a positive correlation between tyrosine loss and number of cell types, but it should be noted that these results are derived from proteins longer than 200 residues (Tan et al. 2009). There may be differences in shorter proteins. As metazoan proteins underwent a depletion of tyrosine content, the metabolic pathways used for tyrosine biosynthesis changed as well. These results provide further insight into the evolution of these systems. Based on these results, the specific enzymes in tyrosine and tryptophan pathways are all unique to metazoans, chordates, or vertebrates, although some have remote homologs in other organisms. Overall, the distribution of these enzymes across Metazoa indicates that most components of these metabolic pathways arose only recently in evolutionary history.
Monoamines have modulatory functions in both vertebrates and invertebrates. Invertebrates synthesize dopamine and serotonin through the same enzymes as vertebrates: TH, TPH2, and AADC (Blenau and Baumann 2001). Since a homologous yet distant AADC sequence was found in D. melanogaster and dopamine is found in invertebrates, the AADC protein in the fly likely serves the same function as the vertebrate protein: catalyzation of L-Dopa into dopamine. However, it does not appear that most invertebrates synthesize norepinephrine or epinephrine (Blenau and Baumann 2001). Instead, they create a compound called octopamine, similar to norepinephrine, by converting tyrosine into tyramine and then into octopamine.
The octopaminergic system in invertebrates is considered to be homologous to the adrenergic system in vertebrates, involving modulation of most physiological processes including those of the CNS (Roeder 1999). The AADC homolog tyrosine decarboxylase converts tyrosine into tyramine, which tyramine β-hydroxylase (a homolog of DBH) then catalyzes into octopamine. Because AADC and DBH catalyze the production of norepinephrine, this supports the homology of the octopaminergic and adrenergic pathways.
In invertebrates, octopamine acts as a modulatory neurotransmitter. It serves several functions in the invertebrate nervous system, including regulation of neural plasticity and reward responses (Koon et al. 2011; Schwaerzel et al. 2003). Tyramine may also be a neurotransmitter in invertebrates, but the evidence is inconclusive (Lange 2009). Although octopamine is a monoamine, it is not metabolized by monoamine oxidase (Roeder 1999). Instead, it is primarily inactivated by addition of an acetyl group by N-acetyltransferases, similar to the metabolism of serotonin by AANAT. The phylogenetic origin of these N-acetyltransferases is unclear and there appears to be no evidence from the results or the literature indicating homology with monoamine oxidases or AANAT.
In addition, both serotonin and melatonin have biological functions outside of chordates. Melatonin, which regulates mammalian circadian rhythms, also serves a protective function in both plants and animals (Hardeland et al. 2011; Tan et al. 2012). Although melatonin has similar functions in plants and Metazoa, plants produce melatonin through a different pathway than vertebrates, with tryptamine as an intermediate between tryptophan and serotonin instead of 5-hydroxytryptophan (Fujiwara et al. 2010). In the present study, AADC homologs were found in invertebrates and in plants. As AADC converts 5-hydroxytryptophan to serotonin, these homologs support shared evolutionary origins for the enzymes involved in parts of the tryptophan metabolism for plants and animals. Since serotonin is a melatonin precursor, it is present in organisms that produce melatonin, including plants. A modulatory neurotransmitter for vertebrates, serotonin also modulates behavior in invertebrates such as honeybees, sea slugs, and moths (Barbas et al.; Gatellier et al. 2004; Scheiner et al. 2006). While serotonin’s function in plants is unclear, it may support plant defense and immune responses (Fujiwara et al. 2010; Ishihara et al. 2008).
Further research should be conducted regarding connections between diseases involving enzymes in these pathways. Variants in genes for both tyrosinase and tyrosinase-related protein 1 have been associated with melanoma (Gudbjartsson et al. 2008). Similarly, Nan, Kraft, Hunter, and Han found that mutations in the tyrosinase-related protein 1 gene were associated with melanoma, while mutations in the gene for tyrosinase were associated with squamous cell carcinoma, a different type of skin cancer (Nan et al. 2009). These pathways are therefore relevant to the etiology of skin cancer as well as other diseases. Melanoma is associated with mortality both from Parkinson’s disease and amyotrophic lateral sclerosis (ALS), a motor neuron disease (Baade et al. 2007; Freedman et al. 2005). Although the cause of the association between ALS and melanoma is unknown, it may relate to the tyrosine metabolic pathway. Polymorphisms in the promoter region of the gene coding for PNMT, another gene in the tyrosine pathway, have been associated with both sporadic (non-familial) early-onset Alzheimer’s disease and multiple sclerosis (MS), a disease involving the immune system and neuron demyelination (Mann et al. 2002; Mann et al. 2001). Additional research regarding connections among these diseases may provide insight into the mechanisms behind their pathologies.
When searching for the origin of these genes in chordates and vertebrates, the possibility of horizontal gene transfer should also be investigated. Iyer, Aravind, Coon, Klein, and Koonin found homologs in bacteria of all the studied enzymes except for DBH and TYR, and suggested that the origin of most of these genes in chordates and vertebrates is horizontal gene transfer from bacteria (Iyer et al. 2004). As no single bacterial species is known to contain all enzymes in either pathway, more research is needed to discover if these genes stem from a bacterial ancestor or if multiple horizontal gene transfers from metazoans to different bacteria is a plausible explanation.
As demonstrated by Parkinson’s disease and melanoma, connections between seemingly unrelated diseases can be found through the tyrosine and tryptophan metabolic pathways. This study found the phylogenies of enzymes in these pathways, which relate to many human diseases. The enzymes investigated are common to chordates and vertebrates, suggesting a recent origin for these specific metabolic pathways. The compounds created by these pathways are biologically important, and this study provides a basis for further investigation into the evolution of their functions in vertebrates. With these findings, metazoan evolution, and more specifically vertebrate evolution, can be investigated with consideration to the catecholamine neurotransmitters, serotonin, melanin, and melatonin.
The evolution of enzymes involved in tyrosine metabolism along new trajectories in Metazoa suggests that the novel metazoan tyrosine metabolic pathway is an integral element for the organismal complexity seen in animals, and as such, tyrosine metabolism should be included as a complement to the current theories aiming to explain the loss of tyrosine in metazoan proteins. While tyrosine kinases are irrefutably of vast importance for signaling and regulation in metazoa, the neurotransmitter- and hormone-based systems founded on the aromatic amino acids in general, and perhaps of tyrosine specifically, cannot be forgotten. The pathway involving tyrosine metabolism is different between metazoan and other organisms and this pathway has continued to expand in vertebrates. Vertebrate gene duplication has facilitated finer regulatory finesse, and tyrosine has become even more imperative as the precursor of epinephrine. Epinephrine, the last addition to the tyrosine pathway, is perhaps most famous for its role in the flight or fight response in animals, but it is also a main coordinator of cell signaling in vertebrates. As new parts are added onto the pathway, these components alter the balance of tyrosine and all other substrates and products. The fraction of tyrosine residues is decreasing with organismal complexity (Tan et al. 2009), and based on these signaling mechanisms, it is not surprising. Instead, we might have to ask: is there enough tyrosine to go around?
Methods
Homologous sequences for each enzyme in the pathways were identified using BLAST in selected eukaryotic species: Homo sapiens, Macaca mulatta, Rattus norvegicus, Mus musculus, Bos taurus, Monodelphis domestica, Gallus gallus, Xenopus tropicalis, Danio rerio, Brachiostoma floridae, Drosophila melanogaster, Caenorhabditis elegans, Saccharomyces cerevisiae, Monosiga brevicollis, Zea mays, and Chlamydomonas reinhardtii.
Multiple sequence alignments were constructed using MAFFT (Katoh et al. 2002) and model testing for the aligned data was performed using default settings in ProtTest3 (Darriba et al. 2011). ProtTest3 suggested the following best models for the different datasets according to the Akaike Information Criteria: AADC, LG+I+G+F; DBH, LG+I+G; MAOA, LG+G+F; TYR, JTT+G; PNMT, JTT+G; AANAT, JTT+G+F; and ASMT, LG+I+G+F. The models suggested by ProtTest3 (Darriba et al. 2011) were used to reconstruct the phylogenetic tree for each enzyme respectively using PhyML (Guindon et al. 2005) (Table 1). The protein sequence trees were rooted based on the NIH common tree for the selected species.
Table 1. The model of evolution used for phylogenetic reconstructions.
| Dataset | Model of evolution |
|---|---|
| AADC | LG+G+F |
| DBH | LG+I+G |
| PNMT | JTT+G |
| AANAT | JTT+G+F |
| ASMT | LG+I+G+F |
| MAOA | LG+G+F |
| TYR | JTT+G |
Supplementary Material
Acknowledgements
Special thanks to John Horner for assistance with manuscript preparation. This work was supported by Award Number P20RR016474 from the National Center for Research Resources. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Research Resources or the National Institutes of Health.
Footnotes
Current affiliation: University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA.
References
- Baade PD, Fritschi L, Freedman DM. Mortality due to amyotrophic lateral sclerosis and Parkinson’s disease among melanoma patients. Neuroepidemiology. 2007;28:16–20. doi: 10.1159/000097851. [DOI] [PubMed] [Google Scholar]
- Barbas D, DesGroseillers L, Castellucci VF, et al. Multiple serotonergic mechanisms contributing to sensitization in aplysia: evidence of diverse serotonin receptor subtypes. Learning & memory (Cold Spring Harbor, NY) 10:373–86. doi: 10.1101/lm.66103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blenau W, Baumann A. Molecular and pharmacological properties of insect biogenic amine receptors: lessons from Drosophila melanogaster and Apis mellifera. Archives of insect biochemistry and physiology. 2001;48:13–38. doi: 10.1002/arch.1055. [DOI] [PubMed] [Google Scholar]
- Cao J, Shi F, Liu X, et al. Phylogenetic analysis and evolution of aromatic amino acid hydroxylase. FEBS letters. 2010;584:4775–82. doi: 10.1016/j.febslet.2010.11.005. [DOI] [PubMed] [Google Scholar]
- Coon SL, Bégay V, Deurloo D, et al. Two arylalkylamine N-acetyltransferase genes mediate melatonin synthesis in fish. The Journal of biological chemistry. 1999;274:9076–82. doi: 10.1074/jbc.274.13.9076. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics (Oxford, England) 2011;27:1164–5. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald LM, Szmant AM. Biosynthesis of “essential” amino acids by scleractinian corals. The Biochemical journal. 1997;322(Pt 1):213–21. doi: 10.1042/bj3220213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick PF. Tetrahydropterin-dependent amino acid hydroxylases. Annual review of biochemistry. 1999;68:355–81. doi: 10.1146/annurev.biochem.68.1.355. [DOI] [PubMed] [Google Scholar]
- Freedman DM, Travis LB, Gridley G, Kuncl RW. Amyotrophic lateral sclerosis mortality in 1.9 million US cancer survivors. Neuroepidemiology. 2005;25:176–80. doi: 10.1159/000087447. [DOI] [PubMed] [Google Scholar]
- Fujiwara T, Maisonneuve S, Isshiki M, et al. Sekiguchi lesion gene encodes a cytochrome P450 monooxygenase that catalyzes conversion of tryptamine to serotonin in rice. The Journal of biological chemistry. 2010;285:11308–13. doi: 10.1074/jbc.M109.091371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatellier L, Nagao T, Kanzaki R. Serotonin modifies the sensitivity of the male silkmoth to pheromone. The Journal of experimental biology. 2004;207:2487–96. doi: 10.1242/jeb.01035. [DOI] [PubMed] [Google Scholar]
- Gudbjartsson DF, Sulem P, Stacey SN, et al. ASIP and TYR pigmentation variants associate with cutaneous melanoma and basal cell carcinoma. Nature genetics. 2008;40:886–91. doi: 10.1038/ng.161. [DOI] [PubMed] [Google Scholar]
- Guindon S, Lethiec F, Duroux P, Gascuel O. PHYML Online--a web server for fast maximum likelihood-based phylogenetic inference. Nucleic acids research. 2005;33:W557–9. doi: 10.1093/nar/gki352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haavik J, Blau N, Thöny B. Mutations in human monoamine-related neurotransmitter pathway genes. Human mutation. 2008;29:891–902. doi: 10.1002/humu.20700. [DOI] [PubMed] [Google Scholar]
- Hardeland R, Cardinali DP, Srinivasan V, et al. Melatonin--a pleiotropic, orchestrating regulator molecule. Progress in neurobiology. 2011;93:350–84. doi: 10.1016/j.pneurobio.2010.12.004. [DOI] [PubMed] [Google Scholar]
- Hedges SB, Dudley J, Kumar S. TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics (Oxford, England) 2006;22:2971–2. doi: 10.1093/bioinformatics/btl505. [DOI] [PubMed] [Google Scholar]
- Herrmann KM, Weaver LM. THE SHIKIMATE PATHWAY. Annual review of plant physiology and plant molecular biology. 1999;50:473–503. doi: 10.1146/annurev.arplant.50.1.473. [DOI] [PubMed] [Google Scholar]
- Holton TA, Pisani D. Deep genomic-scale analyses of the metazoa reject Coelomata: evidence from single- and multigene families analyzed under a supertree and supermatrix paradigm. Genome biology and evolution. 2010;2:310–24. doi: 10.1093/gbe/evq016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara A, Hashimoto Y, Tanaka C, et al. The tryptophan pathway is involved in the defense responses of rice against pathogenic infection via serotonin production. The Plant. 2008;54:481–95. doi: 10.1111/j.1365-313X.2008.03441.x. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Aravind L, Coon SL, et al. Evolution of cell-cell signaling in animals: did late horizontal gene transfer from bacteria have a role? Trends in genetics: TIG. 2004;20:292–9. doi: 10.1016/j.tig.2004.05.007. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic acids research. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, et al. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic acids research. 2012;40:D109–14. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic acids research. 2002;30:3059–66. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koon AC, Ashley J, Barria R, et al. Autoregulatory and paracrine control of synaptic and behavioral plasticity by octopaminergic signaling. Nature neuroscience. 2011;14:190–9. doi: 10.1038/nn.2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange AB. Tyramine: from octopamine precursor to neuroactive chemical in insects. General and comparative endocrinology. 2009;162:18–26. doi: 10.1016/j.ygcen.2008.05.021. [DOI] [PubMed] [Google Scholar]
- Mann MB, Wu S, Rostamkhani M, et al. Phenylethanolamine N-methyltransferase (PNMT) gene and early-onset Alzheimer disease. American journal of medical genetics. 2001;105:312–6. doi: 10.1002/ajmg.1363. [DOI] [PubMed] [Google Scholar]
- Mann MB, Wu S, Rostamkhani M, et al. Association between the phenylethanolamine N-methyltransferase gene and multiple sclerosis. Journal of neuroimmunology. 2002;124:101–5. doi: 10.1016/s0165-5728(02)00009-7. [DOI] [PubMed] [Google Scholar]
- Nan H, Kraft P, Hunter DJ, Han J. Genetic variants in pigmentation genes, pigmentary phenotypes, and risk of skin cancer in Caucasians. International journal of cancer Journal international du cancer. 2009;125:909–17. doi: 10.1002/ijc.24327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder T. Octopamine in invertebrates. Progress in neurobiology. 1999;59:533–61. doi: 10.1016/s0301-0082(99)00016-7. [DOI] [PubMed] [Google Scholar]
- Scheiner R, Baumann A, Blenau W. Aminergic control and modulation of honeybee behaviour. Current neuropharmacology. 2006;4:259–76. doi: 10.2174/157015906778520791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaerzel M, Monastirioti M, Scholz H, et al. Dopamine and octopamine differentiate between aversive and appetitive olfactory memories in Drosophila. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2003;23:10495–502. doi: 10.1523/JNEUROSCI.23-33-10495.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setini A, Pierucci F, Senatori O, Nicotra A. Molecular characterization of monoamine oxidase in zebrafish (Danio rerio) Comparative biochemistry and physiology Part B, Biochemistry & molecular biology. 2005;140:153–61. doi: 10.1016/j.cbpc.2004.10.002. [DOI] [PubMed] [Google Scholar]
- Siltberg-Liberles J, Steen IH, Svebak RM, Martinez A. The phylogeny of the aromatic amino acid hydroxylases revisited by characterizing phenylalanine hydroxylase from Dictyostelium discoideum. Gene. 2008;427:86–92. doi: 10.1016/j.gene.2008.09.005. [DOI] [PubMed] [Google Scholar]
- Starcevic A, Akthar S, Dunlap WC, et al. Enzymes of the shikimic acid pathway encoded in the genome of a basal metazoan, Nematostella vectensis, have microbial origins. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:2533–7. doi: 10.1073/pnas.0707388105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z, Huang W, Gu X. Comment on “Positive selection of tyrosine loss in metazoan evolution”. Science (New York, NY) 2011;332:917. doi: 10.1126/science.1187374. author reply 917. [DOI] [PubMed] [Google Scholar]
- Tan CSH, Pasculescu A, Lim WA, et al. Positive selection of tyrosine loss in metazoan evolution. Science (New York, NY) 2009;325:1686–8. doi: 10.1126/science.1174301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan D-X, Hardeland R, Manchester LC, et al. Functional roles of melatonin in plants, and perspectives in nutritional and agricultural science. Journal of experimental botany. 2012;63:577–97. doi: 10.1093/jxb/err256. [DOI] [PubMed] [Google Scholar]
- Thöny B, Auerbach G, Blau N. Tetrahydrobiopterin biosynthesis, regeneration and functions. The Biochemical journal 347 Pt. 2000;1:1–16. [PMC free article] [PubMed] [Google Scholar]
- Tribl F, Arzberger T, Riederer P, Gerlach M. Tyrosinase is not detected in human catecholaminergic neurons by immunohistochemistry and Western blot analysis. Journal of neural transmission Supplementum. 2007:51–5. doi: 10.1007/978-3-211-73574-9_8. [DOI] [PubMed] [Google Scholar]
- Walther DJ, Peter J-U, Bashammakh S, et al. Synthesis of serotonin by a second tryptophan hydroxylase isoform. Science (New York, NY) 2003;299:76. doi: 10.1126/science.1078197. [DOI] [PubMed] [Google Scholar]
- Wells DG, Bjorksten AR. Monoamine oxidase inhibitors revisited. Canadian journal of anaesthesia = Journal canadien d’anesthésie. 1989;36:64–74. doi: 10.1007/BF03010890. [DOI] [PubMed] [Google Scholar]
- Xin X, Mains RE, Eipper BA. Monooxygenase X, a member of the copper-dependent monooxygenase family localized to the endoplasmic reticulum. The Journal of biological chemistry. 2004;279:48159–67. doi: 10.1074/jbc.M407486200. [DOI] [PubMed] [Google Scholar]
- Xu Y, Stokes AH, Freeman WM, et al. Tyrosinase mRNA is expressed in human substantia nigra. Brain research Molecular brain research. 1997;45:159–62. doi: 10.1016/s0169-328x(96)00308-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.