Abstract
Oxytocin (Oxt), produced in the hypothalamic paraventricular (PVN) and supraoptic nuclei (SON) for transport to and release from the posterior pituitary, was originally discovered through its role in lactation and parturition. Oxt also plays important roles in the central nervous system by influencing various behaviors. MicroRNAs (miRNAs), endogenous regulators of many genes, are a class of small noncoding RNAs that mediate post-transcriptional gene silencing. We performed miRNA expression profiling of the mouse hypothalamus by deep sequencing. Among the sequenced and cross-mapped small RNAs, expression of known miRNAs and unknown miRNAs candidates were analyzed. We investigated in detail one miRNA, miR-24, and found that it is a novel regulator of Oxt and controls both transcript and peptide levels of Oxt. These results provide insights into potential neurohypophysial hormone regulation mediated by miRNAs.
Keywords: microRNA, deep sequencing, hypothalamus, OXT, AVP
Introduction
The hypothalamo-neurohypophyseal system (HNS) consists of large magnocellular neurons (MCNs) of the supraoptic (SON) and paraventricular (PVN) hypothalamic nuclei that have axon projections to the posterior pituitary (Brownstein et al. 1980). The HNS is the major source of the neuropeptide hormones (and neurotransmitters) oxytocin (Oxt) and vasopressin (Avp) which have crucial roles in reproduction (lactation and parturition) and osmoregulation, respectively (Stoop 2012, Lohmeier 2003, Caldwell & Young 2006, Burbach et al. 2001, Young & Gainer 2003). In addition, Oxt and Avp mediate the confluence of complex processes related to homeostasis, and behaviors such as stress modulation, aggressive behavior and social recognition (Lee et al. 2009a, Caldwell et al. 2008). Expressions of Oxt and Avp in hypothalamus are specifically regulated (Caldwell & Young 2006, Burbach et al. 2001, Young & Gainer 2003). Lesser amounts of Oxt and Avp are made by smaller cells of the PVN and a few other forebrain nuclei and released into the central nervous system (CNS) to influence various other behaviors (Lee et al. 2009a, Caldwell et al. 2008). However, it is largely unclear what mechanisms are responsible for the highly selective regulation of the cell-type specific expression of Oxt and Avp (Ponzio et al. 2012). Therefore, it is important to understand how exactly the neurohypophysial hormones and neurotransmitters are regulated in the hypothalamus. Gene and peptide expressions in the HNS are the result of complex regulatory interactions involving cis-regulatory sequences, alternative splicing, post-translational regulation, and noncoding antisense RNAs (Murphy et al. 2012).
MicroRNAs (miRNAs) are evolutionally conserved short (18–23nt) noncoding RNAs that can exert multilevel inhibition or repression during formation of a double-stranded RNA duplex through the binding of the miRNA to a mRNA in the RNA-induced silencing complex (RISC). This usually triggers the inhibition of protein translation in the animal (Mattick & Makunin 2005, Eckstein 2005, Bartel 2004). Many miRNAs are closely involved in various functions of the HSN including circadian clock control (Alvarez-Saavedra et al. 2011), stress response (Nemoto et al. 2012), and osmotic regulation (Lee et al. 2006). In addition, the miR-7 family was found to be enriched in the hypothalamus of zebrafish (Tessmar-Raible et al. 2007), mouse (Lee et al. 2006, Herzer et al. 2012, Sanek & Young 2012), and human (Berezikov et al. 2006), suggesting they have a conserved role in gene regulation in hypothalamic neurons.
The recent use of deep sequencing (next generation sequencing) allows quantitative assessment of miRNAs. In this study, we investigated the miRNA expression profiles in the mouse hypothalamus using this technique. Furthermore, we found that an miRNA, mir-24, that in silico analysis suggested would interact with the Oxt mRNA, does indeed reduce transcript and peptide levels of Oxt in vitro.
Materials and Methods
Purification of small-sized RNA and construction of the small-sized RNA library
Mouse hypothalamus RNA was purified from two adult male C57Bl/6J mice (Jackson Laboratories, Bar Harbor, ME, USA). Mice were maintained on a 12-hour light/dark cycle (lights on at 4:00 AM) with food and water available ad libitum. All animal procedures were approved by the National Institute of Mental Health Animal Care and Use Committee and were in accordance with the National Institutes of Health guidelines on the care and use of animals. For each mouse, the brain was quickly removed from the skull after decapitation. The tissue was isolated by slicing the brain coronally between about 0.0 and −2.5 mm from the bregma, then sagittally at about 1.4 mm lateral to the midline, and finally separating the hypothalamus from remaining brain tissue with a cut just above the dorsal extent of the third ventricle. Small sized-RNA was extracted from the hypothalamus after pooling using the mirVana miRNA isolation kit (Ambion, Foster City, CA, USA) according to the manufacturer’s protocol. The small-sized RNA library was made as described previously (Lee & Hong 2012).
Deep sequencing data analysis
Preparation of the raw data and subsequent small-sized RNA mappings and predictions were performed with a proprietary software package, ACGT101-miR v3.5 (LC Sciences, Houston, TX, USA) as previously described (Ma et al. 2011). First, low-quality reads were removed from the raw reads. After removal of the adaptor sequences, and filtering of the low quality reads and simple artificial sequences, the mappable reads were extracted and the unique sequences generated by collapsing the identical sequences, with the occurrence count of each unique sequence as the unique sequence tag. These unique sequences were compared with the sequences of non-coding RNAs (rRNA, tRNA, snRNA, snoRNA) available in Rfam (http://www.sanger.ac.uk/software/Rfam) and in the GenBank non-coding RNA database (http://www.ncbi.nlm.nih.gov/) to clarify degradation fragments of non-coding RNA. Raw sequences were processed using Illumina’s (San Diego, CA, USA) Pipeline software and then subjected to a series of data filtration steps to analyze sequencing data using the ACGT101-miR software package (V3.5, LC Sciences). The reference database of M. musculus (ftp://ftp.ncbi.nih.gov/genomes/M musculus) was used for miRNA mapping. Hairpin RNA structures were predicted from the adjacent 60–80 nt sequences in either direction using mfold software (Zuker 2003). This study employed miRBase V17.0 (ftp://mirbase.org/pub/mirbase/CURRENT/) for sequence analysis to compare with miRNA and precursor miRNA (pre-miRNA) sequences. Oxt targeting candidate miRNAs were predicted from RegRNA (available at http://regrna.mbc.nctu.edu.tw/php/prediction.php).
Generation of DNA constructs
The complete murine Oxt (GenBank accession no. NM_011025.3) was amplified by RT-PCR from adult mouse hypothalamus total RNA. The PCR product was cloned into pSICHECK2 (Promega, Madison, WI) downstream from the Renilla luciferase coding sequence (pSICHECK2-Oxt) for luciferase assay. The Oxt cDNA was also cloned into pcDNA 3.1 vector (Life Technologies, Carlsbad, CA) to generate Oxt expressing cDNA (pcDNA 3.1-Oxt) for ELISA test.
Quantitative RT-PCR (qRT-PCR)
For quantitative analysis of miRNAs, two-step TaqMan real-time PCR was performed with reagents from Applied Biosystems (Foster City, CA, USA) according to the manufacturer’s instructions. SnoRNA-202 was used as an internal control and the expressions of miRNAs were compared to miR-7b. For Oxt qRT-PCR, one μg of total RNA was reverse transcribed using Omniscript Reverse Transcription (QIAGEN). Power SYBR Green PCR master mix (Applied Biosystems) was used for PCR reactions. The following primer sequences were used to assess Oxt and Gapdh mRNA expression. Oxt forward 5′-ATGGCCTGCCCCAGTCTCGC-3′ and Oxt reverse 5′-CCGGGCTGCAGCAGATGCCT-3′; Gapdh forward 5′-GGCATTGCTCTCAATGACAA-3′ and Gapdh reverse 5′-TGTGAGGGAGATGCTCAGTG-3′. For each treatment, all samples were measured in triplicate and the data are presented as ratio of Oxt /Gapdh.
Luciferase activity assay
For the reporter assay, COS-7 cells (2×105) were co-transfected in 24-well plates by using Lipofectamine 2000 (Life Technologies) with 25 nM of miRNA oligo (Bioneer, Deajeon, Korea) and 800 ng of pSICHECK2-Oxt DNA. Luciferase assays were performed 24 h later with the Dual-Luciferase reporter system (Promega) according to the manufacturer’s instructions. All experiments were performed in triplicate.
Enzyme-linked immunosorbent assay (ELISA)
For the reporter assay, AtT-20 cells (2×105, mouse pituitary cell line, ATCC No.CCL-89, American Type Culture Collection, Manassas, VA, USA) were seeded on 24-well plates and then 0.8 μg of pcDNA 3.1-Oxt DNA and 100 nM of miRNA mimic oligo (Bioneer, Deajeon, Korea) or nonspecific scrambled control siRNA (siControl, ON-TARGET plus Control Pool; D-001810-10, Dharmacon, Lafayette, CO, USA) were co-transfected using Lipofectamine (Life Technologies). Cell culture supernatants were collected 48 h after transfection and then measured using an Oxt ELISA kit (Assay Designs/Enzo Life Sciences, Ann Arbor, MI, USA) according to the manufacturer’s instructions.
Statistical analysis
All data are presented as the means ± standard deviations (SD). Significant variation analysis was used to calculate the parametric two-tailed non-paired t-test (for miRNA qRT-PCR and ELISA) or the non-parametric Kruskal-Wallis ANOVA analysis (for Oxt qRT-PCR) was used as the data was not in a normal distribution. All analyses were performed using Origin 8.0 (OriginLab, MA, USA), and p-values of ≤ 0.05 were considered statistically significant.
Results
Deep sequencing data analysis and verification
11.5 million sequences were isolated and read from the Mus musculus small RNA samples. The collected sequences of 15–26 nt were extracted as valid small RNAs and were compared with various RNA databases. Sequencing of small RNA libraries from hypothalamus resulted in 27.62×106 raw sequences (counts). After removal of 6.02×106 (21.1%) raw sequences that did not meet the acceptance criteria of our filters, 21.60×106 (78.2%) were considered as “mappable” sequences. Of these mappable sequences, the majority of the small RNAs were 20–24 nt in size (77.7% of mappable sequences), which is typical of mature miRNA products (Fig. 1A). The mature miRNAs identified from the predicted miRNAs were divided into several groups. In one group, 914 unique miRNAs (total 61.90%) were mapped to mmu-miRNAs/pre-miRNAs in the miRbase. A second group included 54 unique miRNAs (total 0.10%) that mapped to Mammalia (novel miRNA in M. musculas) miRNAs/pre-miRNAs in miRBase. A final group contained approximately 3.3% of the small-sized RNA clones that were mapped to genomic locations (within hairpins) not previously annotated in miRBase 17 as mRNA, rRNA, tRNA, or non-coding RNA; we classified then as novel miRNA candidates (Fig. 1B). However, most of them had small clone counts. All classified miRNAs are summarized in Table S1 and the raw sequencing data were deposited in the NCBI Sequence Read Archive (SRA) database.
Figure 1. Distribution and analysis of sequenced small RNAs from mouse hypothalamus.
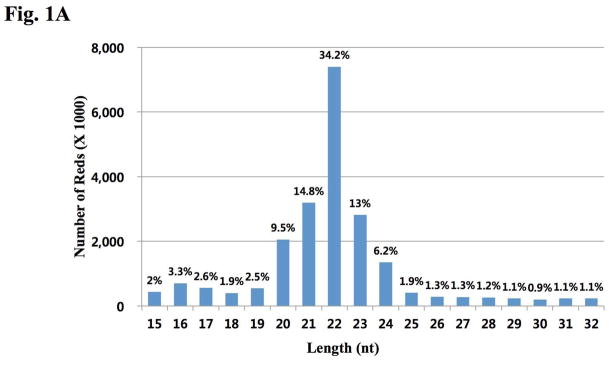
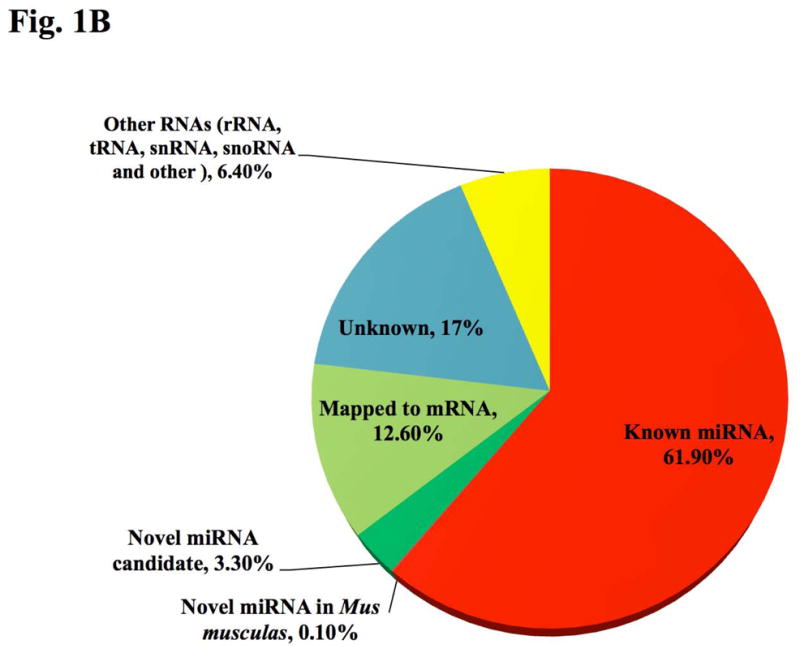
There were 27628729 Sequence reads generated. (A) Size distribution of sequenced miRNAs is shown. The nucleotide (nt) lengths of cloned miRNAs are shown on the X-axis; the number of total reads by deep sequencing are shown on the Y-axis. Reads with length >32 nt were not chosen for mapping. (B) Categorical distribution of sequenced small RNAs from mouse hypothalamus.
Nine selected miRNAs were verified by qRT-PCR using specific TaqMan probes and their expression levels were normalized to a highly expressed miRNA (miR-7b) in the hypothalamus (Lee et al. 2006, Herzer et al. 2012, Sanek & Young 2012)(Figure 2). These data showed that relative expression of selected miRNAs by qRT-PCR were roughly co-related with the deep sequencing data.
Figure 2. Verification of miRNA expression from deep sequencing data.
Real-time PCR analysis of miRNAs in hypothalamus total RNA and sno-202 was used for the reference control gene. miRNA fold expression data were normalized to expression levels of miR-7b in the hypothalamus. Data show the means from three independent experiments (error bars indicate standard deviations).
MiR-24 directly targets Oxt by interaction with the exon/3′ UTR boundary
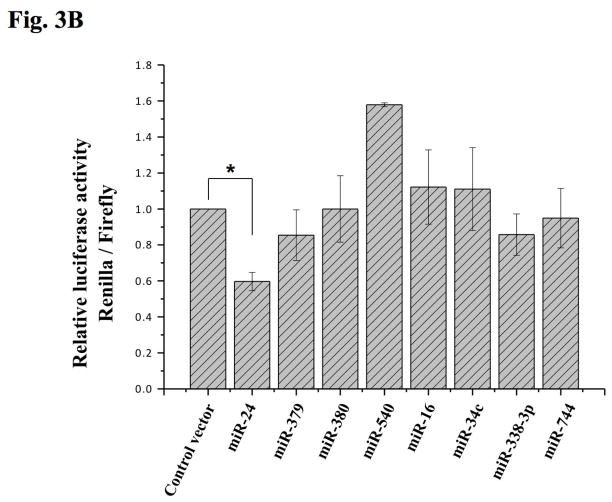
To find target miRNAs of Oxt, we analyzed candidates by using the bioinformatic tools at the RegRNA database. The analysis revealed that eight miRNAs that were highly expressed in the mouse hypothalamus from our deep sequencing, harbored complementary sites to portions of the Oxt mRNA, not only in the 3′ UTR but also in coding regions of the mRNA (Fig. 3A). We examined these other regions because recent studies suggested that miRNAs can target them (Lee et al. 2009b, Qin et al. 2010). To investigate the potential interactions experimentally, the full length mouse Oxt cDNA was subcloned after the Renilla luciferase coding sequence and co-transfected into Cos-7 cells along with the eight miRNA oligonucleotides. Only miR-24 oligo produced a significant (approximately 40%) decrease in luciferase activity compared with control empty vector whereas the others showed not more than a 20% reduction (Fig 3B). These findings show that miR-24 can interact with the Oxt mRNA and inhibit translation from the chimeric transcript.
Figure 3. Direct targeting of Oxt by miR-24.
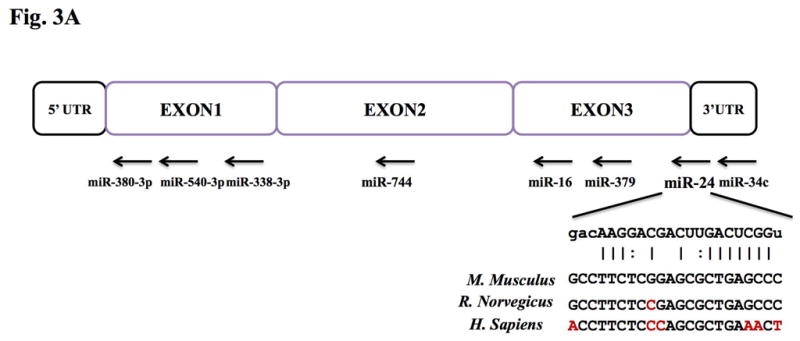
(A) Schematic diagram of putative miRNA binding sites within the Oxt gene. Each box represents exons and UTRs. Also shown are the predicted duplex formation between miR-24 and the targeted Oxt gene and the comparisons of conserved targeting regions between mouse and other species (non-conserved regions are shown in red). (B) Luciferase activity of the Oxt cDNA reporter gene in the presence of the miRNA oligos. COS-7 cells were cotransfected with both the reporter gene and miRNA mimic oligos; miR-24, miR-379, miR-380, miR-540, miR-16, miR-34c, miR-338-3p and miR-744. Renilla luciferase data were normalized to firefly luciferase data and the fold value was compared to control vector transfection. Data show the means from three independent transfections (error bars indicate standard deviations; *, p≤0.05 for between control vector and miR-24).
Mir-24 regulates both transcript and peptide levels of Oxt
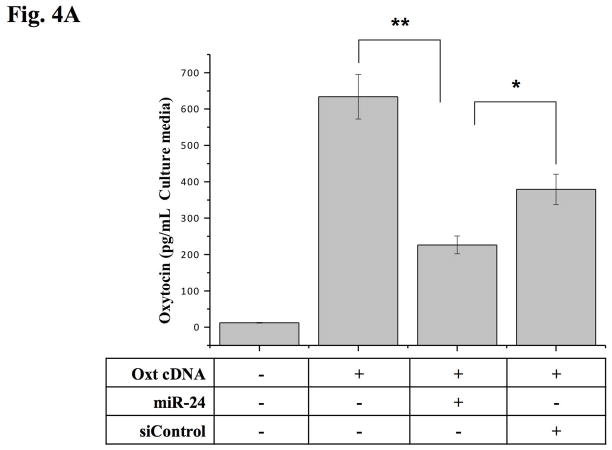
To learn whether miR-24 can affect Oxt peptide levels, we next examined the effect of this miRNA in cultured tissue cells co-transfected with the Oxt cDNA. ELISA from cultured media obtained from AtT-20 cells 48 hr after Oxt transfection revealed dramatically reduced levels of Oxt peptide after miR-24 co-transfection (Fig. 4A). In contrast, siControl transfected cells showed small reduction in Oxt protein expression, possibly the result of an off-target effect. Because inhibition of expression by miRNAs may also be mediated by mRNA degradation (Behm-Ansmant et al. 2006b, Behm-Ansmant et al. 2006a), we examined whether Oxt mRNA levels might be affected by miR-24. Interestingly, mRNA levels were also lower (Fig. 4B), perhaps by reduced transcription and/or mRNA degradation.
Figure 4. Translational and transcriptional repression of mouse Oxt by miR-24.
(A) ELISA assay showed Oxt peptide expression was specifically reduced following transfection of the miR-24. Data show the means from three independent experiments (error bars indicate standard deviations; **, p≤0.001 between control vector and miR-24 and *; p≤0.05 between miR-24 and siControl). (B) qRT-PCR of Oxt after Oxt cDNA transfection with or without miRNA-transfection (miR-24 or siControl) into AtT-20 cells. (Inserted box) Visualized final qRT-PCR products. Exogenous Oxt expression was reduced approximately 50% by miR-24 compare to siControl transfection (error bars indicate standard deviations; *, p≤0.05 between control vector and miR-24 and between miR-24 and siControl).
Discussion
It is well known that miRNA levels are tightly controlled and deregulation of miRNAs is a hallmark of impaired brain development and nervous system diseases (Kosik 2006). In addition, miRNAs modulate almost all biological events and signaling cascades (Bartel 2009, Ouellet et al. 2006). miRNA expression profiles of the PVN and SON of the hypothalamus or various region of the mouse brain including hypothalamus by miRNA microarray methods were previously reported (Lee et al. 2006, Bak et al. 2008). Despite its convenient and reliable use, microarray techniques have limitations such as reliance on known sequences, relatively low sensitivity and specificity, and poor range of quantification. Deep sequencing enables the identification and quantification of small-sized RNAs such as miRNAs, including novel ones (Landgraf et al. 2007, Taft et al. 2009). Using this technique, we analyzed detailed miRNA profiles from adult mouse hypothalamus. The most highly expressed miRNA in the mouse hypothalamus was miR-127, which is known as one of the highest expressed miRNAs in the brain (Lu et al. 2005), though its function there is still unclear. miR-7b (Table S1), abundant in the hypothalamus (Lee et al. 2006, Herzer et al. 2012, Sanek & Young 2012), is up-regulated in response to osmotic stress and inhibits c-Fos (known as marker for neuronal activity) by binding 3′ UTR of the c-Fos gene (Lee et al. 2006). For this reason, we verified the expression of various hypothalamus miRNAs by qRT-PCR and normalized to miR-7b. Many of the miRNAs that were reported to be influential in brain function were also expressed in relatively high amounts as revealed by our profiling analysis. For example, the miR-9 gene is conserved in vertebrates where it regulates the midbrain-hindbrain boundary formation and, with miR-124, plays a role in neuronal differentiation (Leucht et al. 2008). The expression of miR-874 is dramatically reduced in glioblastoma tissues, compared to normal brain tissues. miR-124, another miRNA that is down-regulated in glioblastoma stem cells, also exhibits reduced expression in glioblastoma tissues by deep sequencing (Cheng et al. 2012). Among the highly expressed miRNAs, we used bioinformatic tools to find possible miRNAs that could regulate Oxt expression. As originally described, targeting to inhibit mRNAs by miRNAs is accomplished by base-pairing between the 3′ UTRs of the target genes and miRNAs (Robins et al. 2005). However, recently, many studies have found other sites of mRNA interaction including the coding sequence (Qin et al., Tay et al. 2008, Forman et al. 2008) and 5′ UTR (Lee et al. 2009b). Interestingly, the miR-24 target site in Oxt spans the boundary between the coding sequence and 3′ UTR (Fig. 3A) and miR-24 was the only miRNA that showed significant binding activity by luciferase assay among the miRNAs predicted from various programs (Fig. 3B). Interestingly, one of the Oxt-targeting candidate miRNAs, miR-540 showed up-regulated luciferase activity rather than down-regulation (Fig. 3B). This can be explained by RNA activation (RNAa) (Pushparaj et al. 2008, Lee in press, Huang et al. 2010), i.e., miR-540 interacted with promoter of the Renilla luciferase gene and thereby, resulted the elevated luciferase activity. However, whether miR-540 can also interact with Oxt needs investigation.
One study has shown that miR-24 can regulate apoptosis by targeting the coding sequence of the FAF1 gene (Qin et al. 2010). Targeting other than the 3′ UTR could be one mechanism by which miRNAs could regulate genes that have short 3′ UTRs, like Oxt. Furthermore, miR-24 reduced both transcript and peptide levels of Oxt (Fig. 4A and B), suggesting its noncanonical regulation (Lee in press). We also tried to find an miRNA candidate form the profiled sequences that may be able to target Oxtr and Avp but failed to verify it at both the transcriptional and translational level; although miR-873 could reduce luciferase activity by binding to the Oxtr 3′ UTR (data not shown) and needs further investigation of any relevance between Oxtr and miR-873.
Deep sequencing profile analysis of annotated clones resulted in certain novel miRNA candidates although the read counts were extremely low. However, we cannot exclude the possibility that those novel miRNA candidates could be induced by neuronal stimuli and therefore functionally activated. The structure and biological significance of these candidates need to be elucidated in future studies.
It is well known that steroid hormones can regulate Oxt and Oxtr (Lee et al. 2009a, Choleris et al. 2003). Oxt and Oxtr promoters of many species harbor binding sites for the estrogen receptor (estrogen-response elements) and are stimulated by estrogen and thyroid hormones (Mohr & Schmitz 1991, Richard & Zingg 1990, Bale & Dorsa 1995, Lee et al. 2009a). The promoter region of the Oxt gene is important for its cell-specific expression but the precise cis-domains responsible for gene expression are unclear (Gainer 2012). However, no regulatory mechanism for Oxt expression has been fully characterized.
We propose here that miR-24, an abundant miRNA in hypothalamus, could be a possible regulator of Oxt. Our data showed that miR-24 could target the boundary region between the coding sequence and 3′ UTR and inhibit Oxt production. Deep sequencing analysis can be used to study miRNA gene profiling from selected brain regions and can help collect useful data to predict region-specific miRNAs that regulate neural gene expression.
Supplementary Material
All classified miRNAs in this study by deep sequencing.
Acknowledgments
H.L. conceived the project, designed the experiments, and analyzed the data. J.C and H.L. drafted the manuscript. J.C., N.A.S., and S.M.K. performed the experiments. Y.L. and S.H. assisted experimentally and intellectually. W.S.Y. supervised the project and revised the manuscript.
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2011-0028240 and 2011-0014086). W.S.Y. and N.A.S. were supported by the NIMH Intramural Research Program (Z01-MH-002498-22).
Footnotes
NCBI sequence read archive accession number. Raw sequencing data are available under Accession Number SRS396791.
The authors declare no conflict of interest.
Additional supporting information may be found in the online version of this article at the publisher’s web site.
References
- Alvarez-Saavedra M, Antoun G, Yanagiya A, Oliva-Hernandez R, Cornejo-Palma D, Perez-Iratxeta C, Sonenberg N, Cheng HY. miRNA-132 orchestrates chromatin remodeling and translational control of the circadian clock. Hum Mol Genet. 2011;20:731–751. doi: 10.1093/hmg/ddq519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak M, Silahtaroglu A, Moller M, Christensen M, Rath MF, Skryabin B, Tommerup N, Kauppinen S. MicroRNA expression in the adult mouse central nervous system. RNA. 2008;14:432–444. doi: 10.1261/rna.783108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bale TL, Dorsa DM. Regulation of oxytocin receptor messenger ribonucleic acid in the ventromedial hypothalamus by testosterone and its metabolites. Endocrinology. 1995;136:5135–5138. doi: 10.1210/endo.136.11.7588251. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006a;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant I, Rehwinkel J, Izaurralde E. MicroRNAs silence gene expression by repressing protein expression and/or by promoting mRNA decay. Cold Spring Harb Symp Quant Biol. 2006b;71:523–530. doi: 10.1101/sqb.2006.71.013. [DOI] [PubMed] [Google Scholar]
- Berezikov E, Thuemmler F, van Laake LW, Kondova I, Bontrop R, Cuppen E, Plasterk RH. Diversity of microRNAs in human and chimpanzee brain. Nat Genet. 2006;38:1375–1377. doi: 10.1038/ng1914. [DOI] [PubMed] [Google Scholar]
- Brownstein MJ, Russell JT, Gainer H. Synthesis, transport, and release of posterior pituitary hormones. Science. 1980;207:373–378. doi: 10.1126/science.6153132. [DOI] [PubMed] [Google Scholar]
- Burbach JP, Luckman SM, Murphy D, Gainer H. Gene regulation in the magnocellular hypothalamo-neurohypophysial system. Physiol Rev. 2001;81:1197–1267. doi: 10.1152/physrev.2001.81.3.1197. [DOI] [PubMed] [Google Scholar]
- Caldwell HK, Lee HJ, Macbeth AH, Young WS., 3rd Vasopressin: behavioral roles of an “original” neuropeptide. Prog Neurobiol. 2008;84:1–24. doi: 10.1016/j.pneurobio.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell HK, Young WS., 3rd . Oxytocin and Vasopressin: Genetics and Behavioral Impolications. In: Lim R, editor. Neuroactive Proteins and Peptides. Springer; New York: 2006. pp. 573–607. [Google Scholar]
- Cheng JQ, Lang MF, Yang S, et al. Genome-Wide Profiling Identified a Set of miRNAs that Are Differentially Expressed in Glioblastoma Stem Cells and Normal Neural Stem Cells. PLoS ONE. 2012;7:e36248. doi: 10.1371/journal.pone.0036248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choleris E, Gustafsson JA, Korach KS, Muglia LJ, Pfaff DW, Ogawa S. An estrogen-dependent four-gene micronet regulating social recognition: a study with oxytocin and estrogen receptor-alpha and -beta knockout mice. Proc Natl Acad Sci USA. 2003;100:6192–6197. doi: 10.1073/pnas.0631699100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckstein F. Small non-coding RNAs as magic bullets. Trends Biochem Sci. 2005;30:445–452. doi: 10.1016/j.tibs.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Forman JJ, Legesse-Miller A, Coller HA. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc Natl Acad Sci USA. 2008;105:14879–14884. doi: 10.1073/pnas.0803230105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gainer H. Cell-type specific expression of oxytocin and vasopressin genes: an experimental odyssey. J Neuroendocrinol. 2012;24:528–538. doi: 10.1111/j.1365-2826.2011.02236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzer S, Silahtaroglu A, Meister B. Locked nucleic acid-based in situ hybridisation reveals miR-7a as a hypothalamus-enriched microRNA with a distinct expression pattern. J Neuroendocrinol. 2012;24:1492–1504. doi: 10.1111/j.1365-2826.2012.02358.x. [DOI] [PubMed] [Google Scholar]
- Huang V, Qin Y, Wang J, Wang X, Place RF, Lin G, Lue TF, Li LC. RNAa is conserved in mammalian cells. PLoS One. 2010;5:e8848. doi: 10.1371/journal.pone.0008848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosik KS. The neuronal microRNA system. Nat Rev Neurosci. 2006;7:911–920. doi: 10.1038/nrn2037. [DOI] [PubMed] [Google Scholar]
- Landgraf R, Kessler MS, Bunck M, et al. Candidate genes of anxiety-related behavior in HAB/LAB rats and mice: focus on vasopressin and glyoxalase-I. Neurosci Biobehav Rev. 2007;31:89–102. doi: 10.1016/j.neubiorev.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Lee HJ. Exceptional storeis of microRNAs. Exp Biol Med. :1–6. (in press) [Google Scholar]
- Lee HJ, Hong SH. Analysis of microRNA-size, small RNAs in Streptococcus mutans by deep sequencing. FEMS Microbiol lett. 2012;326:131–136. doi: 10.1111/j.1574-6968.2011.02441.x. [DOI] [PubMed] [Google Scholar]
- Lee HJ, Macbeth AH, Pagani JH, Young WS., 3rd Oxytocin: the great facilitator of life. Prog Neurobiol. 2009a;88:127–151. doi: 10.1016/j.pneurobio.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Palkovits M, Young WS., 3rd miR-7b, a microRNA up-regulated in the hypothalamus after chronic hyperosmolar stimulation, inhibits Fos translation. Proc Natl Acad Sci USA. 2006;103:15669–15674. doi: 10.1073/pnas.0605781103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Ajay SS, Yook JI, Kim HS, Hong SH, Kim NH, Dhanasekaran SM, Chinnaiyan AM, Athey BD. New class of microRNA targets containing simultaneous 5′-UTR and 3′-UTR interaction sites. Genome Res. 2009b;19:1175–1183. doi: 10.1101/gr.089367.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leucht C, Stigloher C, Wizenmann A, Klafke R, Folchert A, Bally-Cuif L. MicroRNA-9 directs late organizer activity of the midbrain-hindbrain boundary. Nat Neurosci. 2008;11:641–648. doi: 10.1038/nn.2115. [DOI] [PubMed] [Google Scholar]
- Lohmeier TE. Neurohypophysial hormones. Am J Physiol Regul Integr Comp Physiol. 2003;285:R715–717. doi: 10.1152/ajpregu.00390.2003. [DOI] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Ma M, Huang Y, Gong Z, Zhuang L, Li C, Yang H, Tong Y, Liu W, Cao W. Discovery of DNA viruses in wild-caught mosquitoes using small RNA high throughput sequencing. PLoS One. 2011;6:e24758. doi: 10.1371/journal.pone.0024758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS, Makunin IV. Small regulatory RNAs in mammals. Hum Mol Genet. 2005;14(Spec No 1):R121–132. doi: 10.1093/hmg/ddi101. [DOI] [PubMed] [Google Scholar]
- Mohr E, Schmitz E. Functional characterization of estrogen and glucocorticoid responsive elements in the rat oxytocin gene. Brain Res Mol Brain Res. 1991;9:293–298. doi: 10.1016/0169-328x(91)90075-9. [DOI] [PubMed] [Google Scholar]
- Murphy D, Konopacka A, Hindmarch C, et al. The hypothalamic-neurohypophyseal system: from genome to physiology. J Neuroendocrinol. 2012;24:539–553. doi: 10.1111/j.1365-2826.2011.02241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto T, Mano A, Shibasaki T. Increased expression of miR-325-3p by urocortin 2 and its involvement in stress-induced suppression of LH secretion in rat pituitary. Am J Physiol Endocrinol Metab. 2012;302:E781–787. doi: 10.1152/ajpendo.00616.2011. [DOI] [PubMed] [Google Scholar]
- Ouellet DL, Perron MP, Gobeil LA, Plante P, Provost P. MicroRNAs in gene regulation: when the smallest governs it all. J Biomed Biotechnol. 2006;2006:69616. doi: 10.1155/JBB/2006/69616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponzio TA, Fields RL, Rashid OM, Salinas YD, Lubelski D, Gainer H. Cell-type specific expression of the vasopressin gene analyzed by AAV mediated gene delivery of promoter deletion constructs into the rat SON in vivo. PLoS One. 2012;7:e48860. doi: 10.1371/journal.pone.0048860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pushparaj PN, Aarthi JJ, Kumar SD, Manikandan J. RNAi and RNAa - The Yin and Yang of RNAome. Bioinformation. 2008;2:235–237. doi: 10.6026/97320630002235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin W, Shi Y, Zhao B, Yao C, Jin L, Ma J, Jin Y. miR-24 regulates apoptosis by targeting the open reading frame (ORF) region of FAF1 in cancer cells. PLoS One. 2010;5:e9429. doi: 10.1371/journal.pone.0009429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Z, Kearney P, Plaisance K, Parsons CH. Pivotal advance: Kaposi’s sarcoma-associated herpesvirus (KSHV)-encoded microRNA specifically induce IL-6 and IL-10 secretion by macrophages and monocytes. J Leukoc Biol. 87:25–34. doi: 10.1189/jlb.0409251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard S, Zingg HH. The human oxytocin gene promoter is regulated by estrogens. J Biol Chem. 1990;265:6098–6103. [PubMed] [Google Scholar]
- Robins H, Li Y, Padgett RW. Incorporating structure to predict microRNA targets. Proc Natl Acad Sci USA. 2005;102:4006–4009. doi: 10.1073/pnas.0500775102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanek NA, Young WS. Investigating the in vivo expression patterns of miR-7 microRNA family members in the adult mouse brain. MicroRNA. 2012;1:11–18. doi: 10.2174/2211536611201010011. [DOI] [PubMed] [Google Scholar]
- Stoop R. Neuromodulation by oxytocin and vasopressin. Neuron. 2012;76:142–159. doi: 10.1016/j.neuron.2012.09.025. [DOI] [PubMed] [Google Scholar]
- Taft RJ, Glazov EA, Lassmann T, Hayashizaki Y, Carninci P, Mattick JS. Small RNAs derived from snoRNAs. RNA. 2009;15:1233–1240. doi: 10.1261/rna.1528909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- Tessmar-Raible K, Raible F, Christodoulou F, Guy K, Rembold M, Hausen H, Arendt D. Conserved sensory-neurosecretory cell types in annelid and fish forebrain: insights into hypothalamus evolution. Cell. 2007;129:1389–1400. doi: 10.1016/j.cell.2007.04.041. [DOI] [PubMed] [Google Scholar]
- Young WS, 3rd, Gainer H. Transgenesis and the study of expression, cellular targeting and function of oxytocin, vasopressin and their receptors. Neuroendocrinology. 2003;78:185–203. doi: 10.1159/000073702. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
All classified miRNAs in this study by deep sequencing.