Abstract
The polyketide synthase (PKS) biosynthetic code has recently expanded to include a newly recognized group of extender unit substrates derived from α,β-unsaturated acyl-CoA molecules that deliver diverse side chain chemistry to polyketide backbones. Herein we report the identification of a three-gene operon responsible for the biosynthesis of the PKS building block isobutyrylmalonyl-CoA associated with the macrolide ansalactam A from the marine bacterium Streptomyces sp. CNH189. Using a synthetic biology approach, we engineered the production of unnatural 36-methyl-FK506 in Streptomyces sp. KCTC 11604BP by incorporating the branched extender unit into FK506 biosynthesis in place of its natural C-21 allyl side chain, which has been shown to be critical for FK506’s potent immunosuppressant and neurite outgrowth activities.
Keywords: biosynthesis, crotonyl-CoA carboxylase, FK506, immunosuppressant, polyketide synthase
Introduction
FK506 (Tacrolimus, Prograf™) is a clinically relevant drug that blocks lymphocyte activation by forming a complex with the immunophilin FKBP12 and the protein phosphatase calcineurin.1 The bacterial natural product is assembled by the polyketide synthase (PKS) FkbA-C and nonribosomal peptide synthetase FkbP enzyme complex in Streptomyces sp. MA6548.2 This multi-modular enzymatic machinery condenses 4,5-dihydroxycyclohex-1-enecarboxylic acid, two malonyl-CoAs, two methoxymalonyl-acyl carrier proteins (-ACP), five methylmalonyl-CoAs, allylmalonyl-CoA and a pipecolate molecule through successive rounds of condensation reactions.3–5 A high degree in the complexity of FK506’s chemical structure is due to specific incorporation of these diverse precursors, which can be changed to alter its biological activity.6
Polyketide backbones have been engineered successfully in the past through building block substitutions. Precursor selectivity for starter and extender units can be predicted from modular acyltransferase (AT) domains, which perform gate keeping functions.7, 8 Substrate tolerance of the AT domain and promiscuity of downstream enzymes furthermore permit certain flexibility in the biosynthetic route.9 These features and an assorted repertoire of priming carboxylic acid substrates10 have afforded a genetic blueprint to precursor-directed combinatorial biosynthesis of rationally designed polyketide molecules.11 Complex polyketides can be further modified beyond starter unit substitutions as shown in an early study by Stassi et al. in which the methylmalonate-specific AT present in the erythromycin PKS was replaced by an ethylmalonate-specific AT to generate 6-desmethyl-6-ethyl-erythromycin. Production of the ethyl-substituted analog required supplementation with the requisite precursor in which the extender unit substrate ethylmalonyl-CoA was generated in vivo after expression of a crotonyl-CoA reductase/carboxylase (CCR).12 This example elegantly demonstrated a non-chemical complementation approach to provide an unnatural polyketide. However, the scope of polyketide engineering efforts has been restricted due to the combination of extender unit availability and AT domain specificity.13 Only six natural malonate derivatives with substitutions (e.g. alkyl, hydroxy, amino) at the C2 position were identified prior to 2009.14
Recently, we discovered a general pathway to a new group of extender units15, which are accessed from α,β-unsaturated acyl-CoA substrates in a CCR-dependent manner. This paradigm shift led to an increased appreciation of polyketide extender unit diversity and opened the door for new synthetic biology approaches to generate hybrid polyketide molecules. Genes for unusual precursor biosynthesis, such as allylmalonyl-CoA (per FK506 synthesis)3, 16 and chloroethylmalonyl-CoA (salinosporamide A)17, are associated with respective secondary metabolite gene clusters and in many cases are even organized in specialized operons. These gene cassettes are conveniently suited for plug and play engineering of polyketide biosynthetic pathways. Herein we report the engineered biosynthesis of 36-methyl-FK506 by replacing the endogenous allylmalonyl-CoA pathway with an exogenous pathway encoding isobutyrylmalonyl-CoA synthesis.
Results
Identification of the isobutyrylmalonyl-CoA biosynthetic gene cassette
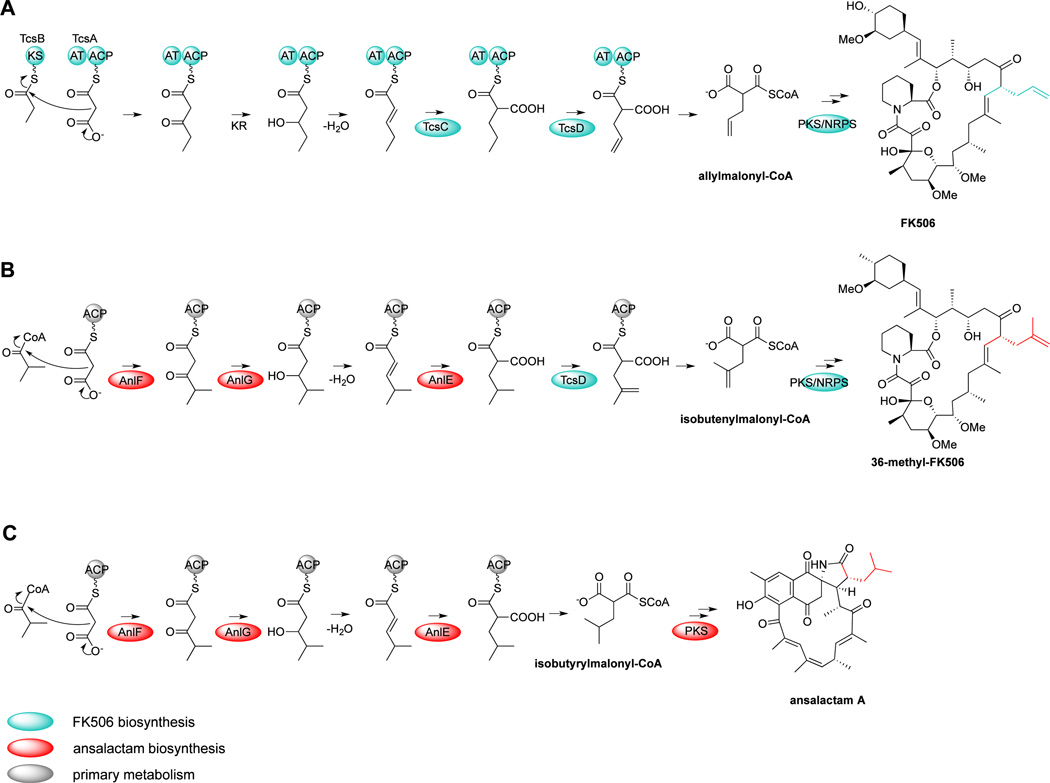
Recently the pathway to the allyl side chain in FK506 was independently characterized by Petkovic3 and our group16 through a series of chemical, biochemical and genetic experiments in which the dedicated ketosynthase TcsB initiates a discrete, multi-step enzymatic pathway to allylmalonyl-CoA (Figure 1a). Using the tcsB deletion mutant strain, we produced two novel analogs, 36-fluoro-FK520 and 36-methyl-FK506, through chemical complementation with 4-fluorocrotonate and 4-methylpentanoate, respectively.16 The exogenously added precursors were converted in vivo to their respective α-substituted malonyl-CoAs with the help of the allylmalonyl-CoA CCR TcsC. In the case of 4-methylpentanoic acid, we hypothesize that the resultant isobutyrylmalonyl-CoA (ibMCoA) is further acted upon by the dehydrogenase TcsD to ultimately provide isobut-2-enylmalonyl-CoA for enzymatic assimilation into 36-methyl-FK506. Since the 36-methyl derivative exhibits improved neurite outgrowth activity in comparison with the parent natural product FK506, we set about to genetically engineer its construction without the requirement of chemical complementation.
Figure 1. Genetic engineering approach towards 36-methyl-FK506 production.
(a) The FK506 producer S. sp. KCTC 11604BP synthesizes allylmalonyl-CoA, which derives from a multi-step pathway involving the ketosynthase TcsB, the cognate acyltransferase and acyl-carrier protein TcsA, the crotonyl-CoA carboxylase TcsC, and the dehydrogenase TcsD. (b) The enzymatic machineries for FK506 (a) and isobutyrylmalonyl-CoA (c) biosynthesis were combined from two different streptomycetes to generate the hybrid pathway of 36-methyl-FK506 in Streptomyces sp. KCTC 11604BP ΔtcsB. (c) The ansalactam producer S. sp. CNH189 assembles isobutyrylmalony-CoA using the FabH-like β-ketoacyl-ACP-synthase AnlF, the dehydrogenase AnlG, and the crotonyl-CoA carboxylase AnlE.
Polyketides with isobutyryl side chains are rare and include two recently characterized ansamycin compound families, the ansalactams18 and divergolides19. Isotope labeling experiments showed that this biosynthetic unit is derived from isobutyrate and acetate rather than L-leucine. We postulated in the context of ansalactam A biosynthesis that condensation of malonyl-CoA and isobutyryl-CoA with subsequent dehydrogenation and dehydration would yield 4-methyl-2-pentenoyl-CoA, which is then reductively carboxylated by a crotonyl-CoA carboxylase homolog affording ibMCoA (Figure 1c). To probe for the molecular mechanism of ibMCoA biosynthesis, we analyzed the draft genome of the ansalactam producer Streptomyces sp. CNH189, which we previously reported in the context of co-produced merochlorin antibiotics.20 Bioinformatic analysis of the genome sequence identified a type I PKS gene locus containing biosynthetic genes consistent with the ansamycin typical starter unit 3,5-aminohydroxybenzoic acid.21 The gene cluster also included a small operon coding for the crotonyl-CoA carboxylase (CCR) AnlE, the FabH-like β-ketoacylsynthase (KSIII) AnlF, and the dehydrogenase (DH) AnlG, which is consistent with the recently reported ibMCoA gene set associated with the divergolide macrolides.19 Phylogenetic analysis revealed that AnlE is closely related to CCRs that reductively carboxylate α,β-unsaturated acyl-CoA substrates to generate C2-substituted malonyl units.15 Therefore anlE, F and G are predicted to encode enzymes for a CCR-dependent pathway to the isobutyrylmalonyl-CoA PKS substrate in ansalactam A biosynthesis.
Biosynthesis of 36-methyl-FK506 through a genetic approach
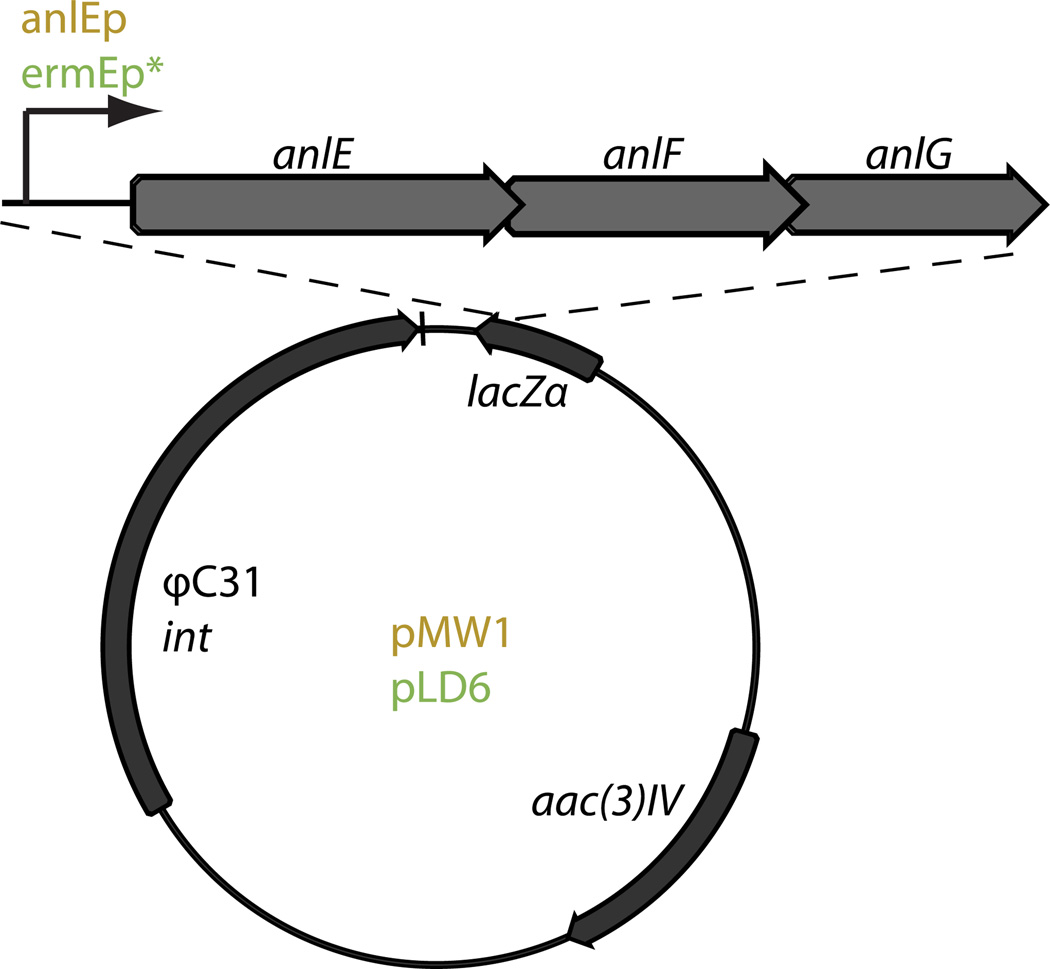
To introduce the ibMCoA pathway into the FK506 producer for the engineered production of 36-methyl-FK506 (Figure 1b), we cloned the three-gene cassette anlE–G from the ansalactam producer strain S. sp. CNH189 and created constructs based on the integrative vector pSET152. Gene expression was placed under control of the native promoter in vector pMW1 and the constitutive ermE* promoter in vector pLD6 (Figure 2). The plasmids were conjugated into the mutant Streptomyces sp. KCTC 11604BP ΔtcsB, in which background production of FK506 was eliminated due to the deletion of the KS TcsB (Figure 3a,b). Prior to plasmid introduction into the ΔtcsB mutant, we observed trace levels of the target molecule, 36-methyl-FK506, at the limit of detection. This observation suggested that endogenous 3-keto-4-methylpentyl thioesters from branched chain fatty acid synthesis may be synthesized and diverted to ibMCoA through the TcsC-catalyzed reductive carboxylation of 4-methyl-2-pentenoyl-CoA.
Figure 2. Expression constructs containing the ibMCoA gene cassette.
The three-gene operon anlE–G was amplified from the ansalactam producer strain S. sp. CNH189 and cloned into pSET152 containing the native upstream region (orange) or the ermEp sequence (green) affording the expression constructs pMW1 and pLD6, respectively.
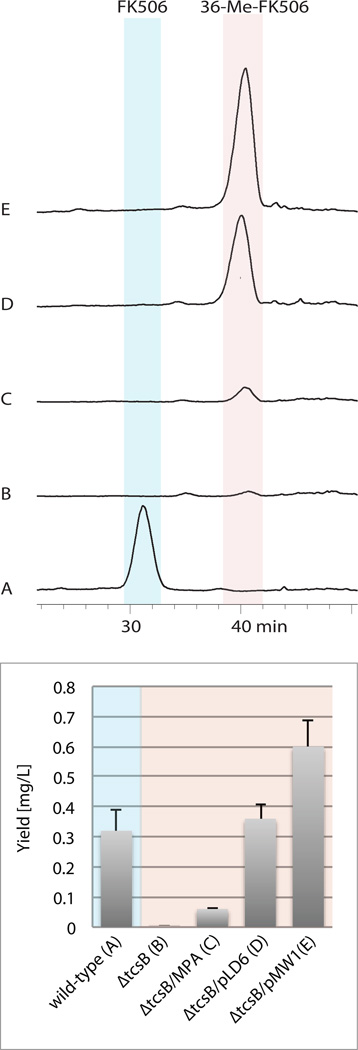
Figure 3. Production of 36-methyl-FK506 via heterologous expression of the ibMCoA pathway in a S. sp. KCTC 11604BP ΔtcbB mutant strain.
Representative HPLC-ESI-MS chromatograms visualized by selected ion monitoring for FK506 (m/z 821 [M+NH4]+; in blue) and 36-Me-FK506 (m/z 835 [M+NH4]+; in red) in top panel. Results were obtained from five independent cultivation experiments of (a) the FK506 producing wild-type strain, (b) Streptomyces sp. KCTC 11604BP ΔtcbB, (c) the ΔtcbB mutant strain supplemented with 4-methylpentanoic acid (MPA), (d) the tcbB deletion strain with ermE*p-ibMCoA biosynthetic genes (pLD6), and (e) with the native promoter-ibMCoA biosynthetic genes (pMW1). Production levels of FK50 (in blue) and 36-Me-FK506 (in red) are graphically displayed in the bottom panel.
We previously reported the mutasynthetic production of 36-methyl-FK506 upon chemical supplementation of the culture with 4-methylpentanoic acid,16 however, the in vivo production levels at 0.06 ± 0.009 mg/L were modest (Figure 3c). Upon chromosomal integration of vectors pLD6 and pMW1, the production of 36-methyl-FK506 was significantly increased 6-fold to 0.36 ± 0.047 mg/L and 10-fold to 0.60 ± 0.089 mg/L, respectively. (Figure 3d,e). Highest production levels were measured when the ibMCoA gene cassette remained under control of the native pathway promoter, which resulted in 2-fold higher yields compared to FK506 production in the native host when similarly grown on solid media (0.32 ± 0.069 mg/L). The industrially optimized strain Streptomyces sp. TST10, which overproduces FK506 at approximately 900 mg/L in liquid culture,22 could provide the chassis for optimized 36-methyl-FK506 biosynthesis.
The successful engineering of 36-methyl-FK506 can be attributed to relaxed specificity of the native dehydrogenase TcsD and the AT4 domain in FkbB toward ibMCoA. The catalytic machinery of FK506 effectively accepts the unnatural building block ibMCoA in place of the native propylmalonyl-CoA, converts it to an unsaturated C4-methylated substrate, and incorporates it into the growing polyketide chain. These experiments confirm the metabolic function of the anlE–G operon and even demonstrate the plug and play character of this gene cassette in polyketide redesign.
Establishing a tool box for future engineering efforts
Tacrolimus and structural analogs have therapeutic potential beyond their current application as immunomodulators, e.g. as drugs in inflammatory diseases and neurological disorders, and as antimalarial agents, which indicates that the highly functionalized FK506 pharmacophore exhibits multiple modes of action.23, 24 To harvest their full medicinal potential, large chemistry efforts have been devoted to generating FK506 analogs,25 while biosynthetic engineering efforts have been mostly limited to the structurally related immunosuppressants FK520 (ascomycin) and rapamycin.26, 27 FK506 biosynthesis has only recently been subjected to genetic engineering efforts, owning to the successful pathway elucidation of the unusual extender unit allylmalonyl-CoA.3, 16, 28, 29
This rare natural PKS substrate belongs to the newly established CCR group of α-substituted malonates that are produced from specialized biosynthesis gene operons that accompany the cognate PKS pathway genes.15 A complementary system to a broad variety of extender units was recently established with variants of the promiscuous acyl-CoA synthetase MatB to engineer natural and non-natural PKS substrates.30 This study reported that trans-ATs are tolerant toward non-natural extender units. To further open the door for polyketide bioengineering via the incorporation of tailored extender units, which can impact biological function as in FK506 and salinosporamide A or introduce chemoselective ligation handles, will require the tailoring of native AT domains that select the substrate for incorporation into the growing polyketide molecule.
Methods
DNA sequencing and computer-assisted sequence analysis
S. sp CNH189 genomic DNA was sequenced using paired-end Illumina DNA sequencing as reported previously.20 The software program Geneious was used for bioinformatic analysis.31
Accession codes
The DNA sequences were deposited in GenBank under accession number JX867614 for the crotonyl-CoA carboxylase gene anlE, JX867615 for the 3-oxoacyl-(acyl-carrier-protein)-synthase gene anlF and JX867616 for the 3-hydroxybutyryl-CoA dehydrogenase gene anlG.
Construction of expression vectors pLD06 and pMW1
For heterologous expression of the ibMCoA biosynthetic gene cassette, we employed the integrative vector pSET152. Two constructs were designed containing either the constitutively expressed Streptomyces promoter ermE*p (pLD06) or the native ibMCoA promoter fragment (pMW1). The ermE*p promoter was isolated by PCR from pUWL201 using primers GCATAACTAGTGCGAGTGTCCGTTCGAG (SpeI) and CTTGATCTAGAGGATCCTACCAACCGGCAC (XbaI). The fragment (191 bp) was digested and cloned into the XbaI restriction site of pSET152 to give vector pLD02. Subsequently the ibMCoA gene cassette (3382 bp) including the native ribosome binding site was PCR amplified from Streptomyces sp. CNH189 genomic DNA using primers GTCGTCTAGAAGAACCGATCGTTTGACCAG (XbaI) and CTAGGAATTCATCAGTCGGTCAGGTCGTTC (EcoRI). The fragment was then cloned into pLD02 to yield expression vector pLD06. For construction of vector pMW1, the same gene cassette including its native promoter region (169 bp) and ribosome binding site were amplified from genomic DNA using primers GTCGTCTAGAGGTCTCTTTCGCTCAAGACC (XbaI) and CTAGGAATTCATCAGTCGGTCAGGTCGTTC (EcoRI). The fragment was subsequently cloned into the XbaI and EcoRI restriction sites of pSET152. Both inserts were sequenced to confirm their integrity.
Generation of exconjugants
The constructs pLD06 and pMW1 were transferred into Escherichia coli S17-1 and introduced into S. sp. KCTC 11604BP ΔtcsB by intergeneric conjugation.32 Apramycin resistance mutants were selected and designated as S. sp. KCTC 11604BP ΔtcsB/pLD06 or pMW1.
Analysis of FK506 congeners
For production of 36-methyl FK506, the S. sp. KCTC 11604BP Δ tcsB strain and mutants expressing isobutyrylmalonyl-CoA biosynthetic genes were cultivated at 28°C for 5 days on R2YE agar.16 In case of the S. sp. KCTC 11604BP Δ tcsB mutant, 4-methylpentanoic acid (Sigma) was added to R2YE solid medium at a final concentration of 10 mM at the time of inoculation. The grown cultures (50 ml) of the mutant strains were mixed with 1 volume of MeOH and shaken at room temperature for 12 h. The cell debris was removed by centrifugation before rotary evaporation. The MeOH extract residues were dissolved in 1 volume of water, and then extracted with 1 volume of EtOAc. The organic extract was evaporated to dryness under reduced pressure and then dissolved in 0.2 ml MeOH for HPLC–ESI–MS/MS analysis. Samples were separated on an ACQUITY UPLC BEH C18 column (50×2.1 mm, 1.7 µm; Waters) interfaced with a Waters/Micromass Quattro micro/MS instrument tracing by MS/MS using a gradient of MeCN at a flow rate of 0.08 ml/min over 70 min starting with 80% (v/v) aqueous MeCN containing 10 mM ammonium acetate and 0.1% acetic acid. Tracing was done by MS/MS operated in multiple reactions monitoring mode choosing mass pairs specific for the selected analytes to detect the transition from parent ion as an ammonium adduct to product ion: 835 > 590 for 36-methyl FK506.33 Quantification of 36-methyl-FK506 was measured by comparison with authentic FK506 using HPLC-ESI-MS analysis in which the detection limit was established at 0.2 µg/L. Five independent experiments were carried out under identical conditions.
Acknowledgments
This work was generously supported by the National Institutes of Health (CA127622 to B.S.M), the National Research Foundation of Korea (20100018430 to Y.J.Y), and the Intelligent Synthetic Biology Center of Global Frontier Project funded by MEST (2011-0031961 to Y.J.Y). We thank Dr. L. Kaysser for experimental advice.
Footnotes
The authors declare no competing financial interest.
References
- 1.Kissinger CR, Parge HE, Knighton DR, Lewis CT, Pelletier LA, Tempczyk A, Kalish VJ, Tucker KD, Showalter RE, Moomaw EW, et al. Crystal structures of human calcineurin and the human FKBP12-FK506-calcineurin complex. Nature. 1995;378:641–644. doi: 10.1038/378641a0. [DOI] [PubMed] [Google Scholar]
- 2.Motamedi H, Shafiee A. The biosynthetic gene cluster for the macrolactone ring of the immunosuppressant FK506. Eur. J. Biochem. 1998;256:528–534. doi: 10.1046/j.1432-1327.1998.2560528.x. [DOI] [PubMed] [Google Scholar]
- 3.Goranovic D, Kosec G, Mrak P, Fujs S, Horvat J, Kuscer E, Kopitar G, Petkovic H. Origin of the allyl group in FK506 biosynthesis. J. Biol. Chem. 2010;285:14292–14300. doi: 10.1074/jbc.M109.059600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andexer JN, Kendrew SG, Nur-e-Alam M, Lazos O, Foster TA, Zimmermann AS, Warneck TD, Suthar D, Coates NJ, Koehn FE, Skotnicki JS, Carter GT, Gregory MA, Martin CJ, Moss SJ, Leadlay PF, Wilkinson B. Biosynthesis of the immunosuppressants FK506, FK520, and rapamycin involves a previously undescribed family of enzymes acting on chorismate. Proc. Natl. Acad. Sci. U.S.A. 2011;108:4776–4781. doi: 10.1073/pnas.1015773108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu K, Chung L, Revill WP, Katz L, Reeves CD. The FK520 gene cluster of Streptomyces hygroscopicus var ascomyceticus (ATCC 14891) contains genes for biosynthesis of unusual polyketide extender units. Gene. 2000;251:81–90. doi: 10.1016/s0378-1119(00)00171-2. [DOI] [PubMed] [Google Scholar]
- 6.Koehn FE. Biosynthetic medicinal chemistry of natural product drugs. Med. Chem. Comm. 2012;3:854–865. [Google Scholar]
- 7.Haydock SF, Aparicio JF, Molnar I, Schwecke T, Khaw LE, Konig A, Marsden AF, Galloway IS, Staunton J, Leadlay PF. Divergent sequence motifs correlated with the substrate specificity of (methyl)malonyl-CoA:acyl carrier protein transacylase domains in modular polyketide synthases. FEBS Lett. 1995;374:246–248. doi: 10.1016/0014-5793(95)01119-y. [DOI] [PubMed] [Google Scholar]
- 8.Petkovic H, Lill RE, Sheridan RM, Wilkinson B, McCormick EL, McArthur HA, Staunton J, Leadlay PF, Kendrew SG. A novel erythromycin, 6-desmethyl erythromycin D, made by substituting an acyltransferase domain of the erythromycin polyketide synthase. J. Antibiot. 2003;56:543–551. doi: 10.7164/antibiotics.56.543. [DOI] [PubMed] [Google Scholar]
- 9.Fischbach MA, Clardy J. One pathway, many products. Nat. Chem. Biol. 2007;3:353–355. doi: 10.1038/nchembio0707-353. [DOI] [PubMed] [Google Scholar]
- 10.Moore BS, Hertweck C. Biosynthesis and attachment of novel bacterial polyketide synthase starter units. Nat. Prod. Rep. 2002;19:70–99. doi: 10.1039/b003939j. [DOI] [PubMed] [Google Scholar]
- 11.Goss RJ, Shankar S, Fayad AA. The generation of "unnatural" products: synthetic biology meets synthetic chemistry. Nat Prod Rep. 2012;29(8):870–889. doi: 10.1039/c2np00001f. [DOI] [PubMed] [Google Scholar]
- 12.Stassi DL, Kakavas SJ, Reynolds KA, Gunawardana G, Swanson S, Zeidner D, Jackson M, Liu H, Buko A, Katz L. Ethyl-substituted erythromycin derivatives produced by directed metabolic engineering. Proc. Natl. Acad. Sci. U.S.A. 1998;95:7305–7309. doi: 10.1073/pnas.95.13.7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pohl NL, Hans M, Lee HY, Kim YS, Cane DE, Khosla C. Remarkably broad substrate tolerance of malonyl-CoA synthetase, an enzyme capable of intracellular synthesis of polyketide precursors. J. Am. Chem. Soc. 2001;123:5822–5823. doi: 10.1021/ja0028368. [DOI] [PubMed] [Google Scholar]
- 14.Chan YA, Podevels AM, Kevany BM, Thomas MG. Biosynthesis of polyketide synthase extender units. Nat. Prod. Rep. 2009;26:90–114. doi: 10.1039/b801658p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilson MC, Moore BS. Beyond ethylmalonyl-CoA: The functional role of crotonyl-CoA carboxylase/reductase homologs in expanding polyketide diversity. Nat. Prod. Rep. 2012;29:72–86. doi: 10.1039/c1np00082a. [DOI] [PubMed] [Google Scholar]
- 16.Mo S, Kim DH, Lee JH, Park JW, Basnet DB, Ban YH, Yoo YJ, Chen S-w, Park SR, Choi EA, Kim E, Jin Y-Y, Lee S-K, Park JY, Liu Y, Lee MO, Lee KS, Kim SJ, Kim D, Park BC, Lee S-g, Kwon HJ, Suh J-W, Moore BS, Lim S-K, Yoon YJ. Biosynthesis of the allylmalonyl-CoA extender unit for the FK506 polyketide synthase proceeds through a dedicated polyketide synthase and facilitates the mutasynthesis of analogues. J. Am. Chem. Soc. 2011;133:976–985. doi: 10.1021/ja108399b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eustáquio AS, McGlinchey RP, Liu Y, Hazzard C, Beer LL, Florova G, Alhamadsheh MM, Lechner A, Kale AJ, Kobayashi Y, Reynolds KA, Moore BS. Biosynthesis of the salinosporamide A polyketide synthase substrate chloroethylmalonyl-CoA from S-adenosyl-L-methionine. Proc. Natl. Acad. Sci. U.S.A. 2009;106:12295–12300. doi: 10.1073/pnas.0901237106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilson MC, Nam S-J, Gulder TAM, Kauffman CA, Jensen PR, Fenical W, Moore BS. Structure and biosynthesis of the marine streptomycete ansamycin ansalactam A and its distinctive branched chain polyketide extender unit. J. Am. Chem. Soc. 2011;133:1971–1977. doi: 10.1021/ja109226s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu Z, Ding L, Hertweck C. A branched extender unit shared between two orthogonal polyketide pathways in an endophyte. Angew. Chem. Int. Ed. 2011;50:4667–4670. doi: 10.1002/anie.201008265. [DOI] [PubMed] [Google Scholar]
- 20.Kaysser L, Bernhardt P, Nam S-J, Loesgen S, Ruby JG, Skewes-Cox P, Jensen PR, Fenical W, Moore BS. Merochlorins A–D, cyclic meroterpenoid antibiotics biosynthesized in divergent pathways with vanadium-dependent chloroperoxidases. J. Am. Chem. Soc. 2012;134:11988–11991. doi: 10.1021/ja305665f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang Q, Shen Y, Bai L. Biosynthesis of 3,5-AHBA-derived natural products. Nat. Prod. Rep. 2012;29:243–263. doi: 10.1039/c2np00019a. [DOI] [PubMed] [Google Scholar]
- 22.Jung S, Moon S, Lee K, Park YJ, Yoon S, Yoo YJ. Strain development of Streptomyces sp. for tacrolimus production using sequential adaptation. J. Ind. Microbiol. Biotechnol. 2009;36:1467–1471. doi: 10.1007/s10295-009-0634-8. [DOI] [PubMed] [Google Scholar]
- 23.Chattopadhaya S, Harikishore A, Yoon HS. Role of FK506 binding proteins in neurodegenerative disorders. Curr. Med. Chem. 2011;18:5380–5397. doi: 10.2174/092986711798194441. [DOI] [PubMed] [Google Scholar]
- 24.Monaghan P, Fardis M, Revill WP, Bell A. Antimalarial effects of macrolactones related to FK520 (ascomycin) are independent of the immunosuppressive properties of the compounds. J. Infect. Dis. 2005;191:1342–1349. doi: 10.1086/428454. [DOI] [PubMed] [Google Scholar]
- 25.Maddess ML, Tackett MN, Ley SV. Total synthesis studies on macrocyclic pipecolic acid natural products: FK506, the antascomicins and rapamycin. Prog. Drug Res. 2008;66:13, 15–186. doi: 10.1007/978-3-7643-8595-8_2. [DOI] [PubMed] [Google Scholar]
- 26.Revill WP, Voda J, Reeves CR, Chung L, Schirmer A, Ashley G, Carney JR, Fardis M, Carreras CW, Zhou Y, Feng L, Tucker E, Robinson D, Gold BG. Genetically engineered analogs of ascomycin for nerve regeneration. J. Pharmacol. Exp. Ther. 2002;302:1278–1285. doi: 10.1124/jpet.102.034264. [DOI] [PubMed] [Google Scholar]
- 27.Ritacco FV, Graziani EI, Summers MY, Zabriskie TM, Yu K, Bernan VS, Carter GT, Greenstein M. Production of novel rapamycin analogs by precursor-directed biosynthesis. Appl. Environ. Microbiol. 2005;71:1971–1976. doi: 10.1128/AEM.71.4.1971-1976.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen D, Zhang Q, Cen P, Xu Z, Liu W. Improvement of FK506 production in Streptomyces tsukubaensis by genetic enhancement of the supply of unusual polyketide extender units via utilization of two distinct site-specific recombination systems. Appl. Environ. Microbiol. 2012;78:5093–5103. doi: 10.1128/AEM.00450-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kosec G, Goranovic D, Mrak P, Fujs S, Kuscer E, Horvat J, Kopitar G, Petkovic H. Novel chemobiosynthetic approach for exclusive production of FK506. Metab Eng. 2012;14:39–46. doi: 10.1016/j.ymben.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 30.Koryakina I, McArthur J, Randall S, Draelos M, Musiol EM, Muddiman DC, Weber T, Williams GJ. Poly-specific trans-acyltransferase machinery revealed via engineered acyl-CoA synthetases. ACS Chem Biol. 2012 doi: 10.1021/cb3003489. [DOI] [PubMed] [Google Scholar]
- 31.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat. Biotech. 1983;1:784–791. [Google Scholar]
- 33.Park JW, Mo SJ, Park SR, Ban YH, Yoo YJ, Yoon YJ. Liquid chromatography-mass spectrometry characterization of FK506 biosynthetic intermediates in Streptomyces clavuligerus KCTC 10561BP. Anal. Biochem. 2009;393:1–7. doi: 10.1016/j.ab.2009.06.021. [DOI] [PubMed] [Google Scholar]