Abstract
MicroRNAs (miRNAs) can function as tumor suppressors or oncogene promoters during tumor development. In this study, low levels of expression of miR-196b were detected in patients with chronic myeloid leukemia. Bisulfite genomic sequencing PCR and methylation-specific PCR were used to examine the methylation status of the CpG islands in the miR-196b promoter in K562 cells, patients with leukemia and healthy individuals. The CpG islands showed more methylation in patients with chronic myeloid leukemia compared with healthy individuals (P<0.05), which indicated that low expression of miR-196b may be associated with an increase in the methylation of CpG islands. The dual-luciferase reporter assay system demonstrated that BCR-ABL1 and HOXA9 are the target genes of miR-196b, which was consistent with predictions from bioinformatics software analyses. Further examination of cell function indicated that miR-196b acts to reduce BCR-ABL1 and HOXA9 protein levels, decrease cell proliferation rate and retard the cell cycle. A low level of expression of miR-196b can cause up-regulation of BCR-ABL1 and HOXA9 expression, which leads to the development of chronic myeloid leukemia. MiR-196b may represent an effective target for chronic myeloid leukemia therapy.
Introduction
MicroRNAs (miRNAs) are non-coding single-stranded RNAs of 19 to 25 nucleotides. They are involved in a variety of biological processes and function by binding to target mRNAs to cause degradation or inhibition of translation. Studies have shown that miRNAs are involved in tumorigenesis and can act as tumor suppressors or oncogene promoters [1]. MiRNA molecules can be regulated by epigenetic processes such as methylation, which can also be involved in tumor initiation and development [2].
The miRNA, miR-196b, is closely associated with some types of leukemia. Overexpression of this miRNA significantly delays mixed lineage leukemia-fusion-mediated leukemogenesis in primary bone marrow transplant patients [3]. In addition, miR-196b has been shown to be down-regulated in EB-3 cells and in patients with B-cell acute lymphocytic leukemia (ALL). These data indicate that miR-196b could be a potential therapeutic target in B-cell ALL [4]. In contrast, miR-196b was over-expressed in patients with acute myeloid leukemia (AML) and the carcinogenic NPM1 mutation [5].
Little is known of the role of miR-196b in chronic myeloid leukemia (CML). In this study, we have demonstrated that the expression of miR-196b is lower in CML patients than in healthy individuals. The BCR-ABL1 gene and HOXA9 gene have been identified as likely targets of miR-196b, using bioinformatics software [6]. Both BCR-ABL1 [7] and HOXA9 [8] have already been shown to be associated with CML. We therefore hypothesize that miR-196b acts as a tumor suppressor in CML, via the up-regulation of BCR-ABL1 and HOXA9. We have also investigated the role of epigenetic regulation in the decreased expression of miR-196b in CML.
Materials and Methods
Detection of miR-196b expression in clinical samples
The bone marrow from 16 patients with CML and 10 healthy age-matched controls, which obtained signed informed consent and approval by the institutional review board (Nanfang Hospital Medical Ethics Committee), was used for the preparation of mononuclear cells, using Ficoll-Paque PLUS (GE Healthcare, Piscataway, NJ, USA). Total RNA was extracted using TRIzol (Invitrogen, Carlsbad, CA, USA), in accordance with the manufacturer's instructions. Reverse transcription of cDNA was performed from total RNA samples, using specific miRNA primers from a Hairpin-it miRNAs qPCR Quantitation Kit (Genepharma, Shanghai, China). Real-time quantitative polymerase chain reaction (RT-qPCR) was used to quantify miR-196b expression, with a specific stem-loop primer from the Hairpin-it kit and an Applied Biosystems 7500 system (Applied Biosystems, Carlsbad, CA, USA). The small nuclear RNA, Cel-mir-39, was used as an internal reference for relative miRNA quantification. The primer sequences are shown in Table S1 in File S1.
CpG island detection
The CpG Island Searcher software (http://www.cpgislands.com/) [9] was used for the detection of CpG islands. The search parameters were as follows: GC content >55%; ratio of CpG observed versus CpG expected >0.65, length >500 base pairs.
Epigenetic drug treatment of K562 cells
K562 cells were purchased from the American Type Culture Collection and treated in three groups; 1. Treated with demethylation drug 5-Aza-2′-deoxycytidine (Aza, Sigma, St. Louis, MO USA), 2. Treated with histone deacetylase inhibitor 4-Phenylbutyric acid (PBA, Sigma), 3. Treated with both Aza and PBA (Aza+PBA). The experiment was performed as described previously, using Cell Counting Kit-8(CCK-8, Dojindo, Kumamoto, Japan) [10], [11]. DNA was extracted 72 h after treatment, using the DNA and Blood Mini Kit (Qiagen, Valencia, CA, USA), as described by the manufacturer.
Measurement of miR-196b promoter CpG island methylation status in K562 cells, by bisulfite genomic sequencing polymerase chain reaction (BSP)
Genomic DNA (1 μg) was treated with bisulfite, in accordance with the manufacturer's protocol (Qiagen EpiTect Bisulfite kit), and eluted in a total of 40 μl of elution buffer. The resulting DNA (2 μl) was used as the template in a 25-μl PCR reaction. Touchdown PCR was performed as follows: 98°C for 4 min; 20 cycles of 94°C for 45 s, 66–56°C for 45 s, with a 0.5°C reduction every cycle, and 72°C for 1 min; 20 cycles of 94°C for 45 s, 56°C for 45 s and 72°C for 1 min; a final extension at 72°C for 8 min. Bisulfite PCR products were gel purified (Promega, Fitchburg, WI, USA) and cloned into the pUC18-T plasmid (Sangon Biotech, Shanghai, China). Independent clones were sequenced using a BiQ analyzer (v2.00) [12]. Primer sequences are given in Table S1 in File S1.
Measurement of miR-196b promoter CpG island methylation status in clinical samples, by methylation-specific polymerase chain reaction (MSP)
The DNA was extracted from the bone marrow of 45 leukemia patients and 10 healthy controls and then treated with bisulfite (EpitTect Bisulfite Kit, Qiagen). Promoter methylation of miR-196b was detected by PCR, using M- and U-PCR primers (Table S1 in File S1) to specifically recognize methylated or un-methylated versions of the promoter. The annealing temperature was 60°C for M-PCR, and 55°C for U-PCR, with 25 cycles used for each.
Bioinformatics prediction of miRNA binding sites
TargetScan (release 5.2) [13], PicTar [14], miRanda (August 2010 release) [15] and miRNA Viewer (April 2005 version) [16] were used to predict miR-196b binding sites.
Plasmid constructs and luciferase assay
Total RNA was isolated from K562 cells using TRIzol (Invitrogen). Reverse transcription-PCR was then used to amplify the 3′-UTR (untranslated region) of human BCR-ABL1 mRNA (1,988 base pairs). The M-MLV RTase cDNA Synthesis kit (Invitrogen) was used for this reaction. Next, Subcloning was performed by using gene splicing, with an overlap extension from the BCR-ABL1 mRNA 3′-UTR or the HOXA9 mRNA 3′-UTR. The resulting PCR product contained miR-196b, which was combined with the loci seed sequence mutation in the BCR-ABL1 3′-UTR and HOXA9 3′-UTR. The subcloned fragments were inserted into the reporter psiCHECK™-2 construct (harboring Renilla and firefly luciferase genes) [17] by double digestion with Xho I and Not I (New England Biolabs, Ipswich, MA, USA). The assembled constructs were then sequenced. Primers are displayed in Table S1 in File S1.
Co-transfection of 0.8 μg of the plasmids and 50 nM solutions of miR-196b mimics (Genepharma), into 293T cells (purchased from the American Type Culture Collection), was achieved using Lipofectamine 2000 (Invitrogen), in accordance with the manufacturer's instructions. The control supplied with the RNA mimics and transfection with Lipofectamine 2000, alone (no plasmid or miR-196b), were used as assay controls. Forty-eight hours after transfection the luciferase assay was performed using the dual-luciferase reporter assay system kit (Promega) and Infinite M200 (Tecan, Männedorf, Switzerland). A decrease in luciferase activity indicated degradation of the target (BCR-ABL1, HOXA9) mRNA. The experiments were performed in triplicate.
Plasmid constructs and lentivirus production
Healthy human bone marrow samples were used for DNA extraction. The human pre-miR-196b (284 base pairs) region was amplified and the PCR fragments were assembled with the plasmid pLVTHM, after double digestion with Mlu I and Cla I (New England Biolabs). The assembled constructs were then sequenced. The primers are shown in Table S1 in File S1.
The lentivirus vector was generated as described previously [18], with omission of the concentration step. The vector was classified as a 196b virus and a pLV virus. Cells that were infected with lentivirus were sorted by fluorescence-activated cell sorting (FACS). Green fluorescence indicated that cells were over-expressing miRNA-196b and pLV virus.
Transfection
Cells that were infected with lentivirus were grown in RPMI-1640 culture medium (Gibco, Grand Island, NY, USA), with 10% fetal bovine serum (Gibco), at 37°C in a 5% CO2 atmosphere. Transfection with 100 nM solutions of miR-196b inhibitors (Genepharma) was performed using Lipofectamine 2000 (Invitrogen), in accordance with the manufacturer's instructions, using 4×105 cells in six-well plates. The control supplied with the RNA inhibitor and transfection with Lipofectamine 2000, alone, served as controls for the assay.
The K562 cells were transfected with 50 nM solutions of BCR-ABL1 siRNA (ABL1-homo-265, Genepharma) and 50 nM solutions of HOXA9 siRNA (HOXA9-homo-452, Genepharma), using Lipofectamine 2000 (Invitrogen), in six-well plates (Table S2 in File S1). Control siRNA and transfection with Lipofectamine 2000, alone, were used as the controls for the assay. The cells were collected 48 h after transfection.
Cell proliferation and cell cycle assays
The proliferation rates of K562 cells and cells infected with lentivirus were quantified in 96-well plates, using the CCK-8 kit (Dojindo), in accordance with the manufacturer's instructions. Measurements were taken every 24 h for a total of 5 days. The proliferation rates of transfected cells were examined after 48 h. Proliferation under different conditions was assessed in triplicate. Cell cycle assays were performed as described in a previous study [19].
RT-qPCR analysis
Detection of miR-196b followed a similar method mentioned above, except that U6 was used as an internal reference. The primers are shown in Table S1 in File S1.
Western blot analysis
Total protein was prepared and quantified as described in a previous study [20] and then transferred to a polyvinylidene difluoride membrane (Millipore, Bedford, MA, USA). The membranes were probed with antibodies for BCR-ABL1 (sc-23, 1∶500, Santa Cruz Biotechnology, CA, USA), HOXA9 (sc-81291, 1∶500 dilution, Santa Cruz Biotechnology) and β-actin (sc-130301, 1∶2,500 dilution, Santa Cruz Biotechnology, used as a gel loading control). An enhanced chemiluminescence fluorescence system (Thermo Scientific Pierce, Waltham, MA, USA) was used to detect antigen–antibody interactions.
Patient samples
Bone marrow samples were collected from 16 patients with CML, 14 patients with AML, 15 patients with ALL and 10 healthy, age-matched, controls (Table S3 in File S1). Informed consent was obtained from all individuals and approval was obtained by the institutional review board (Nanfang Hospital Medical Ethics Committee). The diagnosis of leukemia was based on clinical features and hematological characteristics, in accordance with the National Comprehensive Cancer Network Guidelines.
Statistical analysis
The results of MSP were analyzed by multi-sample rate χ2 tests, using SPSS 13.0 software. The results of the luciferase assay and cell proliferation detection assay were analyzed by one-way ANOVA, using SPSS 13.0 software.
Results
Methylation of CpG islands in the miR-196b promoter
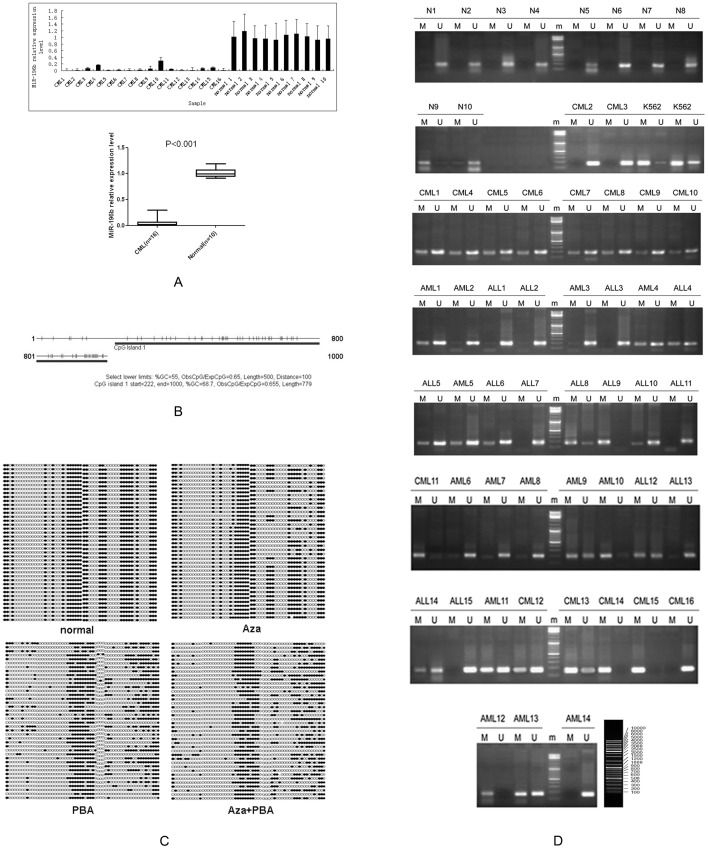
The average expression level of miR-196b was significantly lower (P<0.001) in the bone marrow samples from the 16 CML patients compared with the 10 healthy controls, as indicated by the RT-qPCR (Figure 1A). In light of this, we investigated the role of epigenetic mechanisms that may be involved in the silencing of miR-196b. A CpG island, similar to that present in many tumor suppressor genes, was found in the 1000 bases upstream from the transcription start site of miR-196b (Figure 1B).
Figure 1. MiR-196b expression and CpG island methylation.
(A) MiR-196b expression levels were significantly lower in CML patients than in healthy controls. (B) Predicted miR-196b promoter CpG islands, obtained from CpG Island Searcher software. (C) The BSP detection method demonstrated decreased methylation after treatment. (D) MSP test results for 45 patients with leukemia and 10 healthy controls. N represents the healthy controls.
The miR-196b CpG island methylation status, in K562 cells treated with 15.55 nM Aza, 1.5 nM PBA, and 15.55 nM Aza+1.5 nM PBA, was examined using BSP. The results indicated that methylation was decreased after all the treatments, particularly after the combined Aza + PBA treatment (Figure 1C).
MSP was used to detect methylation of the 423–591 bases upstream from the transcriptional start site of miR-196b in 45 patients with leukemia and 10 healthy individuals (Figure 1D). Multi-sample rate comparison χ2 tests showed sample to sample differences in CpG island methylation status (P = 0.005; Table 1). A significantly higher level of methylation was observed in CML patients compared with the healthy controls (P = 0.001).
Table 1. Analysis of CpG island methylation status by MSP.
| Typing | Methylation + | Methylation − | Total | ?2 | P(two sides) |
| CML | 13(81.3%) | 3(18.8%) | 16 | 12.652 | 0.005 |
| AML | 8(57.1%) | 6(42.9%) | 14 | ||
| ALL | 8(53.3%) | 7(46.7%) | 15 | ||
| healthy controls | 1(10%) | 9(90%) | 10 |
Six pairwise comparisons were performed, and the significance level was adjusted to 0.0083, in accordance with the Bonferroni correction. CML vs. healthy controls, Fisher's exact test, P = 0.001; AML vs. healthy controls, Fisher's exact test, P = 0.033; ALL vs. healthy controls, Fisher's exact test, P = 0.040; CML vs. AML, Fisher's exact test, P = 0.236; CML vs. ALL, Fisher's exact test, P = 0.135; AML vs. ALL, P = 0.837.
Prediction of target genes
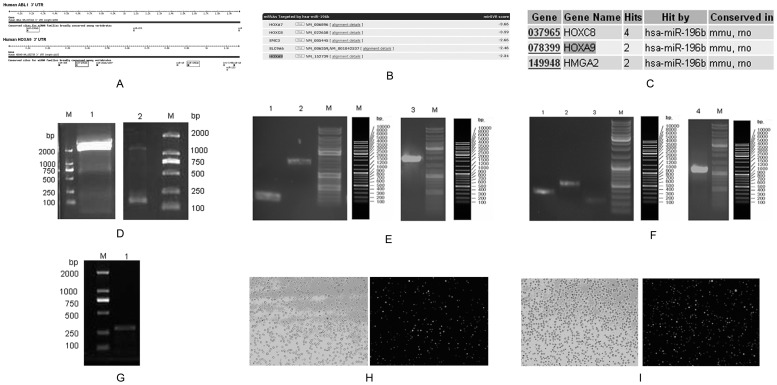
The TargetScan software predicted that BCR-ABL1 and HOXA9, which are closely associated with CML, are target genes of miR-196b (Figure 2A). Both miRanda (Figure 2B) and miRNA Viewer (Figure 2C) software predicted that the target genes of miR-196b included HOXA9. In contrast, the PicTar software failed to identify any target genes of miR-196b. Overall, these results indicated that BCR-ABL1 and HOXA9 are likely to be target genes of miR-196b.
Figure 2. Target gene prediction, plasmid constructs and lentivirus infection.
(A) Target genes of miR-196b, predicted by TargetScan. (B) Target genes of miR-196b, predicted by miRanda. (C) Target genes of miR-196b predicted by miRNA Viewer. (D1): BCR-ABL1-3′-UTR, (D2): HOXA9-3′-UTR. (E1): BCR-ABL1-3′-UTR-mut-1, (E2): BCR-ABL1-3′-UTR-mut-2, (E3): BCR-ABL1-3′-UTR-mutant. (F1): HOXA9-3′-UTR-mut-1, (F2): HOXA9-3′-UTR-mut-2, (F3): HOXA9-3′-UTR-mut-3, (F4): HOXA9-3′-UTR-mutant. (G): pre-miR-196b. (H): K562 cells infected by 196b virus (10×). (I): K562 cells infected by pLV virus (10×).
Validation of miR-196b target genes by the dual-luciferase reporter assay
The 3′-UTR of human BCR-ABL1 mRNA (Figure 2D1) and HOXA9 mRNA (Figure 2D2) were amplified from the total RNA of K562 cells. The BCR-ABL1 3′-UTR-mutant (Figure 2E3), which contained one combined loci seed sequence, was subcloned by division into two segments (Figure 2E1, 2E2). The HOXA9 3′-UTR-mutant (Figure 2F4), which contained two combined loci seed sequences, was subcloned by division into three segments (Figures 2F1, 2FC2, 2F3). The data from the assay were analyzed using one-way ANOVA, which revealed unequal variances between treatment factors in the miR-196b mimics plus BCR-ABL1 3′-UTR group (P<0.001). The results of multiple comparisons showed that miR-196b mimics plus BCR-ABL1 3′-UTR significantly decreased the fluorescence value (P<0.05), which indicated the inclusion BCR-ABL1 in the target genes of miR-196b. Unequal variances were also observed between treatment factors in the miR-196b mimics plus HOXA9 3′-UTR group (P<0.001). Again, the results of multiple comparisons showed that miR-196b mimics plus HOXA9 3′-UTR caused a decrease in the fluorescence value (P<0.05), which supports the hypothesis that HOXA9 is a target gene of miR-196b (Table 2).
Table 2. Dual-luciferase reporter gene assay.
| n |
 ±s ±s
|
|
| miR-196b mimics+BCR-ABL1-3′-UTR | 3 | 532.793±111.245* |
| miR-196b mimicscontrol+BCR-ABL1-3′-UTR | 3 | 1497.843±204.271 |
| miR-196b mimics+BCR-ABL1-3′-UTR -mutant | 3 | 1523.627±109.590 |
| lipo2000+BCR-ABL1-3′-UTR | 3 | 1488.233±94.823 |
| lipo2000+BCR-ABL1-3′-UTR -mutant | 3 | 1473.147±158.225 |
| F | 45.234 | (welch) |
| P | <0.001 | |
| miR-196b mimics+HOXA9-3′-UTR | 3 | 507.629±32.345# |
| miR-196b mimics contro+HOXA9-3′-UTR | 3 | 1160.171±60.471 |
| miR-196b mimics+ HOXA9-3′-UTR -mutant | 3 | 1080.374±121.249 |
| lipo2000+ HOXA9-3′-UTR | 3 | 1197.352±162.872 |
| lipo2000+HOXA9-3′-UTR-mutant | 3 | 1107.123±30.490 |
| F | 168.622 | (welch) |
| P | <0.001 |
vs. miR-196b mimics control+BCR-ABL1-3′-UTR, P = 0.006; * vs. miR-196b mimics+ BCR-ABL1-3′-UTR -mutant, P<0.001; * vs. lipo2000+ BCR-ABL1-3′-UTR, P<0.001; * vs. lipo2000+BCR-ABL1-3′-UTR -mutant, P = 0.001 (missing case, pairwise comparisons). # vs. miR-196b mimics contro+HOXA9-3′-UTR, P<0.001; # vs. miR-196b mimics+ HOXA9-3′-UTR -mutant, P = 0.016; # vs. lipo2000+ HOXA9-3′-UTR, P = 0.027; # vs. lipo2000+HOXA9-3′-UTR -mutant, P<0.001 (missing case, pairwise comparisons).
Modulation of BCR-ABL1 and HOXA9 expression by miR-196b small interfering (si)RNA and epigenetic drugs
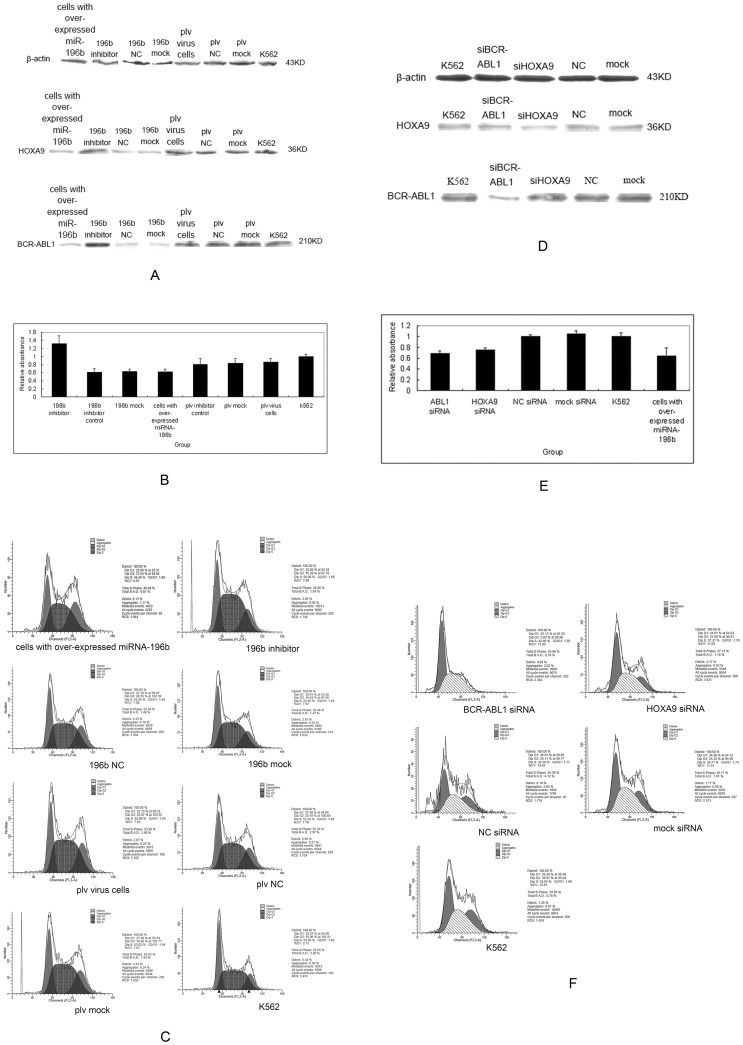
Human pre-miR-196b was amplified from normal human bone marrow (Figure 2G). Follow up testing was performed on K562 cells infected by 196b virus (Figure 2H) and pLV virus (Figure 2I). A restoration of the expression of miR-196b reduced BCR-ABL1 and HOXA9 protein levels (Figure 3A). This was accompanied by a dramatic decrease in the cell proliferation rate (Figure 3B, Table 3) and retardation of the G2 stage (Figure 3C). A reduction in the expression of miRNA-196b, in the cells where it had been overexpressed, restored BCR-ABL1 and HOXA9 protein levels (Figure 3A), enhanced cell proliferation (P<0.05) (Figure 3B, Table 3), and retarded the S stage of the cell cycle (Figure 3C). In addition, down-regulation of BCR-ABL1, by specific siRNAs, reduced BCR-ABL1 protein levels (Figure 3D) and inhibited proliferation (P<0.05) (Figure 3E, Table 3), as also observed in cells that showed over-expression of miRNA-196b and a retarded G1 stage (Figure 3F). Down-regulation of HOXA9, by specific siRNAs, reduced HOXA9 protein levels (Figure 3D) and induced proliferation arrest (P<0.05) (Figure 3E, Table 3), as observed in cells that showed over-expression of miRNA-196b. This also resulted in the retardation of the G1 stage of the cell cycle (Figure 3F).
Figure 3. Cell functions after transfection and epigenetic drugs.
(A) Protein levels after reintroduction of miR-196b into K562 cells. (B) Proliferation rates of K562 cells after reintroduction or reduction of miR-196b. (C) Cell cycle analysis after reintroduction or reduction of miR-196b. (D) Protein levels after down-regulation of BCR-ABL1 and HOXA9 by specific siRNAs. (E) Proliferation rates after down-regulation of BCR-ABL1 and HOXA9 by specific siRNAs. (F) Cell cycle analysis after down-regulation of BCR-ABL1 and HOXA9 by specific siRNAs. (G) MiR-196b expression in K562 cells after treatment with Aza and PBA separately, or Aza + PBA. (H) BCR-ABL1 and HOXA9 protein levels after treatment with Aza, PBA or Aza + PBA.
Table 3. Cell proliferation assessed by CCK-8 assay after miR-196b and siRNA transfection.
| n |
 ±s ±s
|
|
| miR-196b inhibitor | 3 | 3.674±0.080* |
| miR-196b inhibitorcontrol | 3 | 1.690±0.035 |
| miR-196b mock | 3 | 1.703±0.018 |
| cells with over-expressedmiRNA-196b | 3 | 1.716±0.079 |
| K562 | 3 | 2.611±0.103 |
| F | 336.464 | (welch) |
| P | <0.001 | |
| BCR-ABL1 siRNA | 3 | 1.821±0.063** |
| HOXA9 siRNA | 3 | 1.939±0.079# |
| NC siRNA | 3 | 2.717±0.126 |
| mock siRNA | 3 | 2.654±0.075 |
| K562 | 3 | 2.675±0.129 |
| cells with over-expressedmiRNA-196b | 3 | 1.676±0.060 |
| F | 69.425 | (welch) |
| P | <0.001 |
vs. miR-196b inhibitor control, P = 0.001; * vs. miR-196b mock, P = 0.003; * vs. cells with over-expressed miRNA-196b, P<0.001; * vs. K562, P = 0.002; cells with over-expressed miRNA-196b vs. K562, P = 0.004 (missing case, pairwise comparisons). ** vs. HOXA9 siRNA, P = 0.851; ** vs. K562, P = 0.032; **vs. cells with over-expressed miRNA-196b, P = 0.495; # vs. K562, P = 0.034; # vs. cells with over-expressed miRNA-196b, P = 0.166 (missing case, pairwise comparisons).
Promoter methylation can be reversed by specific epigenetic drugs. We treated K562 cells with Aza, PBA and Aza + PBA, which resulted in partial but efficient demethylation of the miR-196b promoter. The miR-196b expression was restored in K562 cells that were treated with Aza + PBA but not in those treated with Aza and PBA, separately (Figure 3G). The three treatment groups showed a significant reduction in both BCR-ABL1 and HOXA9 protein levels (Figure 3H).
Discussion
The levels of expression of miR-196b were significantly lower in CML patients than in healthy controls. The methylation of the miR-196b promoter CpG islands was also increased in CML patients, which implied that expression levels and promoter methylation status could be linked. The differences observed between CML patients and healthy individuals correlate to the mechanisms of action of many tumor suppressor genes [21], [22], which indicates that miR-196b could be involved in the mechanism that underlies the development of CML. The regulation of miRNA-196b by DNA methylation has been shown to be involved in the development of many other cancers and may be significant in CML. For example, Tsai KW et al. demonstrated that abnormal DNA hypomethylation induces the overexpression of miR-196b in gastric cancer [23]. Hulf T et al. found that miR-196b was repressed by epigenetic activity in prostate cancer cells [24]. Abe W et al. suggested that the expression of miR-196b in endometriotic cyst stromal cells is repressed by hypermethylation of the DNA of the miR-196b gene [25]. Schotte D et al. demonstrated that up-regulation of miR-196b coincides with reduced DNA methylation at CpG islands in the promoter regions of miR-196b in pediatric ALL [26]. The differences in levels of miR-196b and methylation that were observed between ALL and CML require further studies to identify whether the discrepancy was related to different sub-types of cancer, different stages of cancer or differences between adults and children.
Statistical analyses also indicated that, unlike CML, there were no significant differences in the methylation status of the miR-196b promoter CpG islands between healthy controls and patients with other types of leukemia. The differences between types of leukemia were also not significant. Further studies are needed to determine if this difference is related to the fact that CML involves the increased proliferation of completely differentiated cells, and results from abnormal signal transduction or incontrollable cell proliferation [27], rather than representing a cell-maturity disorder, as in ALL.
The BCR-ABL1 and HOXA9 protein levels were reduced in K562 cells that were treated with demethylating agents. These results indicated that a reduction in the methylation of the miR-196b promoter could inhibit BCR-ABL1 and HOXA9 protein expression. The use of demethylating agents, including Aza and PBA, could provide a potential treatment for CML. However, miR-196b expression was not restored in cells treated with Aza and PBA, separately, whilst it was restored in cells treated with a combination of Aza + PBA. This may indicate that the mechanism of change of miR-196b expression involved not only hypermethylation, but also excessive deacetylation [28].
The luciferase assay confirmed that BCR-ABL1 and HOXA9 are target genes of miR-196b. Western blotting indicated that the reintroduction of miR-196b, into K562 cells, reduced BCR-ABL1 and HOXA9 protein levels. The reduction of miRNA-196b expression restored BCR-ABL1 and HOXA9 protein levels. These data confirmed the roles of BCR-ABL1 and HOXA9 as miR-196b target genes. There are no published studies of BCR-ABL1 as a target for miR-196b, but there are several papers that have shown that HOXA9 is a target gene. For example, Li Z et al. reported that miR-196b directly targets HOXA9/MEIS1 oncogenes in MLL-rearranged leukemia [29]. Shen J et al. showed that the host gene of miR-196b was HOXA9 in hepatocellular carcinoma [30]. Tsai KW et al. reported that miR-196b is located in the HOXA cluster, within exon AB of HOXA9 (chromosome7), in gastric cancer [22].
To date, this is the first report on the effects of miR-196b on cell proliferation in K562 cells. The overexpression of miR-196b was associated with decreased cell proliferation, whilst a reduction of expression enhanced cell proliferation. This indicated that miR-196b is involved in the proliferation of K562 cells. Recent studies have indicated that miR-196b also inhibits cell proliferation and promotes apoptosis in B-cell ALL cells [4], but promotes proliferation and improves survival in gastric cancer cells [23]. Hence, the effects of miR-196b on cell proliferation differ between different cell lines. The BCR-ABL1 gene is important for regulating cell proliferation in CML [31]. It is translated to produce a fusion protein with abnormal tyrosine kinase activity, which results in hematopoietic stem cell dysregulation, myeloid progenitor cell proliferation and reduced apoptosis. The HOXA9 gene is necessary for the normal differentiation of hematopoietic cells and HOXA9 deficiency can result in reduced cell proliferation, differentiation and apoptosis induction [32]. The reintroduction of miR-196b was associated with inhibition of BCR-ABL1 and HOXA9 expression and the consequent reduction of their regulatory effects on proliferation. The expression and function of the genes could be restored by reducing the expression of miRNA-196b. In addition, down-regulation of BCR-ABL1 and HOXA9, by specific siRNAs, resulted in the inhibition of cell proliferation, compared with cells that over-expressed miRNA-196b. This demonstrated that the inhibition of proliferation by miR-196b occurred by the inhibition of both BCR-ABL1 and HOXA9.
In conclusion, this study demonstrated that low expression levels of the tumor suppressor, miR-196b, were associated with up-regulation of the oncogenes BCR-ABL1 and HOXA9, ultimately leading to the development of CML. These results verified our previous hypothesis and indicated that miR-196b is a potential target for therapeutic intervention in CML.
Supporting Information
Supporting file containing Tables S1, S2, S3. Table S1. Primers used for detection of miRNAs and for cloning and mutagenesis of the human BCR-ABL1 3′-UTR and HOXA9 3′-UTR. Table S2. BCR-ABL1 and HOXA9 target sequences for RNA interference. The siRNA ID includes the start nucleotide of the targeted sequence in the reference transcript for the human BCR-ABL1 gene and HOXA9 gene. Table S3. Patient samples.
(DOC)
Acknowledgments
We would like to thank Qiang Li (Department of Clinical Laboratory of Nanfang Hospital) and Jinfang Zhang (Department of Hematology of Nanfang Hospital) for providing bone marrow samples, Dr. Avram Hershko and Dr. Yiqiang Cai of Yale University for helpful discussions and comments on the manuscript, and Xuhui Tan (Statistics Department of Southern Medical University) for assistance with statistical analysis.
Funding Statement
This work was supported by Guangdong Medical Research Foundation (B2013220), and Youth Science and technology personnel training project of “Scientific research plan” of Southern Medical University, and an “Outstanding Leadership Grant” from Guangdong Province (C1030925), and Key Project of Science and Technology Research grants from the Ministry of Education (B1000409). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Lujambio A, Lowe SW (2012) The microcosmos of cancer. Nature 482: 347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Caffarelli E, Filetici P (2012) Epigenetic regulation in cancer development. Front Biosci 17: 2682–2694. [DOI] [PubMed] [Google Scholar]
- 3. Li Z, Huang H, Chen P, He M, Li Y, et al. (2012) miR-196b directly targets both HOXA9/MEIS1 oncogenes and FAS tumour suppressor in MLL-rearranged leukaemia. Nat Commun 3: 688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bhatia S, Kaul D, Varma N (2010) Potential tumor suppressive function of miR-196b in B-cell lineage acute lymphoblastic leukemia. Mol Cell Biochem 340: 97–106. [DOI] [PubMed] [Google Scholar]
- 5. Coskun E, von der Heide EK, Schlee C, Kuhnl A, Gokbuget N, et al. (2011) The role of microRNA-196a and microRNA-196b as ERG regulators in acute myeloid leukemia and acute T-lymphoblastic leukemia. Leuk Res 35: 208–213. [DOI] [PubMed] [Google Scholar]
- 6. Thomas M, Lieberman J, Lal A (2010) Desperately seeking microRNA targets. Nat Struct Mol Biol 17: 1169–1174. [DOI] [PubMed] [Google Scholar]
- 7. Quintas-Cardama A, Cortes J (2009) Molecular biology of bcr-abl1-positive chronic myeloid leukemia. Blood 113: 1619–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tedeschi FA, Cardozo MA, Valentini R, Zalazar FE (2010) Co-expression of HoxA9 and bcr-abl genes in chronic myeloid leukemia. Leuk Lymphoma 51: 892–896. [DOI] [PubMed] [Google Scholar]
- 9. Takai D, Jones PA (2003) The CpG island searcher: a new WWW resource. In Silico Biol 3: 235–240. [PubMed] [Google Scholar]
- 10. Du X, Nagata S, Ise T, Stetler-Stevenson M, Pastan I (2008) FCRL1 on chronic lymphocytic leukemia, hairy cell leukemia, and B-cell non-Hodgkin lymphoma as a target of immunotoxins. Blood 111: 338–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bueno MJ, Perez de Castro I, Gomez de Cedron M, Santos J, Calin GA, et al. (2008) Genetic and epigenetic silencing of microRNA-203 enhances ABL1 and BCR-ABL1 oncogene expression. Cancer Cell 13: 496–506. [DOI] [PubMed] [Google Scholar]
- 12. Lutsik P, Feuerbach L, Arand J, Lengauer T, Walter J, et al. (2011) BiQ Analyzer HT: locus-specific analysis of DNA methylation by high-throughput bisulfite sequencing. Nucleic Acids Res 39: W551–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20. [DOI] [PubMed] [Google Scholar]
- 14. Lall S, Grun D, Krek A, Chen K, Wang YL, et al. (2006) A genome-wide map of conserved microRNA targets in C. elegans. Curr Biol 16: 460–471. [DOI] [PubMed] [Google Scholar]
- 15. John B, Enright AJ, Aravin A, Tuschl T, Sander C, et al. (2004) Human MicroRNA targets. PLoS Biol 2: e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kiezun A, Artzi S, Modai S, Volk N, Isakov O, et al. (2012) miRviewer: a multispecies microRNA homologous viewer. BMC Res Notes 5: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pottier N, Maurin T, Chevalier B, Puissegur MP, Lebrigand K, et al. (2009) Identification of keratinocyte growth factor as a target of microRNA-155 in lung fibroblasts: implication in epithelial-mesenchymal interactions. PLoS One 4: e6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Frka K, Facchinello N, Del Vecchio C, Carpi A, Curtarello M, et al. (2009) Lentiviral-mediated RNAi in vivo silencing of Col6a1, a gene with complex tissue specific expression pattern. J Biotechnol 141: 8–17. [DOI] [PubMed] [Google Scholar]
- 19. Ballabeni A, Park IH, Zhao R, Wang W, Lerou PH, et al. (2011) Cell cycle adaptations of embryonic stem cells. Proc Natl Acad Sci U S A 108: 19252–19257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu Y, Song Y, Ma W, Zheng W, Yin H (2013) Decreased microRNA-30a levels are associated with enhanced ABL1 and BCR-ABL1 expression in chronic myeloid leukemia. Leuk Res 37: 349–356. [DOI] [PubMed] [Google Scholar]
- 21. Kucuk C, Iqbal J, Hu X, Gaulard P, De Leval L, et al. (2011) PRDM1 is a tumor suppressor gene in natural killer cell malignancies. Proc Natl Acad Sci U S A 108: 20119–20124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Agrawal-Singh S, Isken F, Agelopoulos K, Klein HU, Thoennissen NH, et al. (2012) Genome-wide analysis of histone H3 acetylation patterns in AML identifies PRDX2 as an epigenetically silenced tumor suppressor gene. Blood 119: 2346–2357. [DOI] [PubMed] [Google Scholar]
- 23. Tsai KW, Hu LY, Wu CW, Li SC, Lai CH, et al. (2010) Epigenetic regulation of miR-196b expression in gastric cancer. Genes Chromosomes Cancer 49: 969–980. [DOI] [PubMed] [Google Scholar]
- 24. Hulf T, Sibbritt T, Wiklund ED, Bert S, Strbenac D, et al. (2011) Discovery pipeline for epigenetically deregulated miRNAs in cancer: integration of primary miRNA transcription. BMC Genomics 12: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Abe W, Nasu K, Nakada C, Kawano Y, Moriyama M, et al. (2013) miR-196b targets c-myc and Bcl-2 expression, inhibits proliferation and induces apoptosis in endometriotic stromal cells. Hum Reprod 28: 750–761. [DOI] [PubMed] [Google Scholar]
- 26. Schotte D, Lange-Turenhout EA, Stumpel DJ, Stam RW, Buijs-Gladdines JG, et al. (2010) Expression of miR-196b is not exclusively MLL-driven but is especially linked to activation of HOXA genes in pediatric acute lymphoblastic leukemia. Haematologica 95: 1675–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Helgason GV, Karvela M, Holyoake TL (2011) Kill one bird with two stones: potential efficacy of BCR-ABL and autophagy inhibition in CML. Blood 118: 2035–2043. [DOI] [PubMed] [Google Scholar]
- 28. Kobayashi T, Matsuoka K, Sheikh SZ, Russo SM, Mishima Y, et al. (2012) IL-10 regulates Il12b expression via histone deacetylation: implications for intestinal macrophage homeostasis. J Immunol 189: 1792–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li Z, Huang H, Chen P, He M, Li Y, et al. (2012) miR-196b directly targets both HOXA9/MEIS1 oncogenes and FAS tumour suppressor in MLL-rearranged leukaemia. Nat Commun 3: 688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shen J, Wang S, Zhang YJ, Kappil MA, Chen Wu H, et al. (2012) Genome-wide aberrant DNA methylation of microRNA host genes in hepatocellular carcinoma. Epigenetics 7: 1230–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Notta F, Mullighan CG, Wang JC, Poeppl A, Doulatov S, et al. (2011) Evolution of human BCR-ABL1 lymphoblastic leukaemia-initiating cells. Nature 469: 362–367. [DOI] [PubMed] [Google Scholar]
- 32. Bach C, Buhl S, Mueller D, Garcia-Cuellar MP, Maethner E, et al. (2010) Leukemogenic transformation by HOXA cluster genes. Blood 115: 2910–2918. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting file containing Tables S1, S2, S3. Table S1. Primers used for detection of miRNAs and for cloning and mutagenesis of the human BCR-ABL1 3′-UTR and HOXA9 3′-UTR. Table S2. BCR-ABL1 and HOXA9 target sequences for RNA interference. The siRNA ID includes the start nucleotide of the targeted sequence in the reference transcript for the human BCR-ABL1 gene and HOXA9 gene. Table S3. Patient samples.
(DOC)