Abstract
Background
Oxidative stress plays a key role in the etiology of male infertility. Significant alterations in the sperm proteome are associated with poor semen quality. The aim of the present study was to examine if elevated levels of reactive oxygen species cause an alteration in the proteomic profile of spermatozoa.
Methods
This prospective study consisted of 52 subjects: 32 infertile men and 20 normal donors. Seminal ejaculates were classified as ROS+ or ROS- and evaluated for their proteomic profile. Samples were pooled and subjected to LC-MS/MS analysis through in-solution digestion of proteins for peptide characterization. The expression profile of proteins present in human spermatozoa was examined using proteomic and bioinformatic analysis to elucidate the regulatory pathways of oxidative stress.
Results
Of the 74 proteins identified, 10 proteins with a 2-fold difference were overexpressed and 5 were underexpressed in the ROS+ group; energy metabolism and regulation, carbohydrate metabolic processes such as gluconeogenesis and glycolysis, protein modifications and oxidative stress regulation were some of the metabolic processes affected in ROS+ group.
Conclusions
We have identified proteins involved in a variety of functions associated with response and management of oxidative stress. In the present study we focused on proteins that showed a high degree of differential expression and thus, have a greater impact on the fertilizing potential of the spermatozoa. While proteomic analyses identified the potential biomarkers, further studies through Western Blot are necessary to validate the biomarker status of the proteins in pathological conditions.
Keywords: Spermatozoa, Reactive oxygen species, Oxidative stress, Proteomics, Male infertility
Background
Male-factor infertility contributes to 50% of infertile couples worldwide, and while the causes are multifactorial, current research has focused on oxidative stress. Specifically, oxidative stress may lead to alterations in protein expression levels in spermatozoa, causing molecular and genetic defects. Research has already shown that oxidative stress is associated with a variety of male infertility diagnosis such as varicocele, idiopathic infertility, spinal cord injury, prostatitis and leukocytospermia [1-8]. Varicocele is implicated in 35% of men with primary infertility and in 80% of infertile men with secondary infertility, and oxidative stress is one of the many causes of varicocele related infertility [9-11]. Oxidative stress in men with varicocele is also associated both with motility and grade of varicocele.
Understanding the protein profile of spermatozoa is essential for identifying the protein alterations that occur as a result of increased production of ROS and for better diagnosis of male infertility. The development of 2-dimensional-polyacrylamide gel electrophoresis (2-DE) coupled with Western blot have aided in the identification of proteins, in relation to sperm composition and function [12,13]. Furthermore advancements in mass spectrometry analyses of spermatozoa have broadened our knowledge and aided in the identification of pivotal sperm proteins [14,15]. Numerous studies have reported the presence of proteins in studies involving human spermatozoa through the implementation of proteomic techniques such as 2-DE, liquid chromatography tandem mass spectrometry (LC-MS/MS), matrix assisted laser desorption and ionization- time of flight tandem mass spectrometry (MALDI-TOF-MS/MS), and 2-dimensional difference in gel electrophoresis (DIGE) [16-21].
In this comparative study, we employed LC- MS/ MS and functional bioinformatic analyses to identify the relative abundance of proteins in spermatozoa obtained from seminal ejaculates of infertile men who were referred to the andrology clinical laboratory for advanced semen analysis (including parameters of oxidative stress such as levels of ROS in spermatozoa, concentration of antioxidant in the seminal plasma and the extent of DNA damage). We also included healthy men with high and low levels of ROS in their seminal ejaculates compared to those with low levels of ROS. Oxidative stress plays a major role in male infertility, but the crucial proteins that are differentially expressed in ROS are not known. Understanding the role of these proteins is therefore important in our understanding of the post-genomic events. It will help identify potential proteins that could serve as biomarkers to better manage and treat oxidative stress, and consequently, male infertility.
Methods
The study was approved by the Institutional Review Board of Cleveland Clinic and was conducted in accordance with national and international guidelines. A total of 52 subjects were enrolled in the study. Twenty healthy male volunteers of unproven fertility (normal semen analysis results but had not established a pregnancy) were selected on the basis of normal semen analysis. Thirty two infertile men attending our infertility clinic were referred to the andrology lab for advanced semen parameters of oxidative stress. The breakdown of the infertile subjects was as follows: primary infertility: n = 25 (25/ 32; 78.1%); Secondary infertility: n = 7 (7/25; 28%). Of the 25 men with primary infertility, 64% (16/ 25) had <2 years and 36% (9/ 25) >2 years. Clinical varicocele (grade 1-2) was present in 10 (62.5%) of the 16 men who presented with <2 years of primary infertility and in 5 (55.6%) of the 9 men with >2 years of primary infertility. In the 7 patients with secondary infertility, 6 (85.7%) had a clinical varicocele (grade 1-2).
Semen samples were collected by masturbation after 2-3 days of sexual abstinence and analyzed according to WHO 2010 criteria [22]. A complete semen analysis was performed on both donors and patients and included both macroscopic (volume, pH, color, viscosity, liquefaction time, split or complete ejaculate) and microscopic parameters (concentration, motility, morphology, round cells and peroxidase or Endtz test). All subjects were also examined for levels of reactive oxygen species (ROS) in the seminal ejaculate. The remainder of the seminal ejaculate was frozen without any cryoprotectant and stored at -55°C for proteomic analysis.
Semen analysis
After liquefaction, complete semen analysis was performed to evaluate the sperm parameters (sperm count, percentages motility and morphology), according to WHO guidelines [22]. Semen analysis was done using a MicroCell counting chamber (Vitrolife, San Diego, CA). Smears of the raw semen were stained with a Diff-Quik kit (Baxter Healthcare Corporation, Inc., McGaw Park, IL) for assessment of sperm morphology according to WHO criteria.
Measurement of reactive oxygen species
In the freshly ejaculated and completely liquefied semen, ROS levels were measured by a chemiluminescence assay using luminol (5-amino-2, 3- dihydro-1, 4-phthalazinedione). Test samples consisted of luminol (10 μL, 5 mM) and 400 μL of semen. Negative controls were prepared by replacing the sperm suspension with phosphate buffered saline. Chemiluminescence was measured for 15 min using a Berthold luminometer (Autolumat Plus 953). Results were expressed as relative light units (RLU)/sec/ × 106 sperm. Reference value for ROS – group was: <20 RLU/s/×106 sperm. ROS levels >20 RLU/s/×106 sperm were considered positive [23].
Separation of spermatozoa and In-solution Digestion of Proteins
We were interested in examining the effect of ROS levels on the distribution / alteration of proteins in an effort to understand the underlying mechanism of infertility. Based on our published reference levels, all samples with ROS levels below 20 RLU/sec/ × 106 sperm were considered to be ROS – and those with ROS levels >20 RLU/sec/ × 106 sperm were considered to be ROS + [23]. For proteomic analysis, frozen seminal ejaculates from both the donors and patients were divided into 2 groups: ROS+ and ROS-, irrespective of the subject’s clinical diagnoses. Samples were centrifuge at 10,000 g for 10 minutes to pellet the spermatozoa and the clear seminal plasma was discarded. Samples were pooled into ROS+ and ROS – groups and solubilized in lysis buffer containing 2% octyl-β-glucopyranoside, 100 mM dithiothreitol (DTT), 9.8 M urea and protease inhibitors [12]. Spermatozoa samples were stored overnight in the refrigerator to allow for complete lysis of the spermatozoa. The samples were first precipitated in cold acetone, solubilized in 6 M urea, reduced with DTT, and alkylated with iodoacetamide. The samples were subsequently diluted to give a urea concentration <2 M and then digested with trypsin. The tryptic digestion was subjected to a C18 clean up and then brought up in 50 μL of 1% acetic acid [12].
Liquid chromatography- Mass Spectrometer analysis (LC-MS-MS)
The LC-MS/MS system was a Finnigan LTQ linear ion trap mass spectrometer system (Thermo Finnigan). Ten μL volumes of the extract were injected on a self-packed high performance liquid chromatography column (Phenomenex Jupiter C18 reversed-phase capillary chromatography column). Peptides were eluted from the column by an acetonitrile/0.1% formic acid gradient at a flow rate of 0.25 μL/min. They were introduced into the source of the mass spectrometer on-line. The microelectrospray ion source was operated at 2.5 kV. The digest was analyzed using the data dependent multitask capability of the instrument acquiring full scan mass spectra to determine peptide molecular weights and product ion spectra to determine amino acid sequence in successive instrument scans [12].
Data analysis
The data were analyzed using all collision- induced dissociation spectra collected in the experiment to search the National Center for Biotechnology Information (NCBI) human reference sequence database with the search program MASCOT (Matrix Science, Boston, MA). Mascot is a mass spectral search algorithm that uses mass spectrometry data to identify proteins from primary sequence databases. These searches were used to identify the proteins present in the in-solution digestions. After identification, a database consisting of all proteins identified in these searches was created and used for a second set of searches performed with SEQUEST (Thermo Finnigan). SEQUEST is a tandem mass spectrometry data analysis program used for protein identification. Sequest identifies collections of tandem mass spectra to peptide sequences that have been generated from databases of protein sequences. The results from these SEQUEST searches were used to determine the spectral counts. These spectral count values were normalized by the total number of spectral counts for all proteins in the sample and the number of amino acids present in the protein. As the precision of the proteomic analysis has an average error of 10-20%, a 2-fold change in protein expression was considered significant.
Furthermore functional bioinformatics analysis were done using publicly available (Gene Ontology (GO) annotations from GO Term Finder [24] and GO Term Mapper [25], UNIPROT [26], STRAP [27] and BioGPS [28] and proprietary software packages (Ingenuity Pathway Analysis (IPA) from Ingenuity® Systems [29] and Metacore™ from GeneGo Inc. [30] to identify the differentially affected processes, pathways, interactions and cellular distribution of the proteins in the two study groups.
Results
Semen parameters and oxidative stress parameters
Most of the infertile men were diagnosed with primary infertility and varicocele. The median and 25th and 75th percentile for sperm concentration (×106/ mL) in controls was 64.9 (45.4-74.9) versus patients 23.0 (13.0-41.4). The median and 25th and 75th percentile motility in controls was 58.3(50.0-65.7) compared to infertile men 50.5 (36.8-58.0). Among the patients, 46.8% (15/32) had > 1 × 106 round cells / mL compared to donors 35% (7/20) of the donors, of these 15 patients, 15.6% were positive for the Endtz test – a marker for granulocytes, which are the major contributors of ROS production. None of the donors was positive for the Endtz test. An overlap in morphology was seen in both the donors (3.5% ± 1.6%) and infertile men (3.6% ± 3.2%).
Among the patients 22 of 32 (68.8%) were positive for ROS compared to 9 of 20 (45%) in the controls. Levels of ROS were significantly lower (RLU/s/×106 sperm) median and 25th and 75th percentile were seen in donors [4.4(0, 300.2)] compared to infertile men [104(0, 1341)]. Among the primary infertility patients with <2 years of infertility, 80% were positive for ROS and 55.6% were positive in the patients with >2 years of primary infertility. A significantly larger percentage of men (83%) were positive for ROS in those with secondary infertility. Teratozoospermia was present in 48% of men with primary infertility.
When all subjects were categorized into ROS+ and ROS- groups, ROS levels (RLU/s/106sperm: median, 25th, 75th percentile) were significantly different and higher in the ROS+ group [2236(33, 4439)] group compared to the ROS – group [4 (0, 9); P < 0.01)].
MS identification of the differentially expressed proteins in ROS+ and ROS- samples
In this study, we analyzed the differential expression of proteins in spermatozoa obtained from seminal ejaculates with high levels of ROS+ and those with normal levels (ROS-). Tables 1 and 2 show the primary proteins with their NCBI database index number, calculated molecular weight, isoelectric point (pI), peptide coverage and Mascot scores for ROS+ and ROS- groups. Based on the SEQUEST score, a total of 74 proteins were identified and differential expression was calculated based on the normalized spectral count ratios between ROS+ and ROS-. In the ROS+ group, proteins that had >50% peptide coverage were: A-Kinase anchor protein 4 isoform 2; Lactotransferrin precursor; Tubulin, beta, 2; Tubulin, beta 4; Tubulin, alpha 3c; Ropporin, rhophilin associated protein 1B; Prolactin-induced protein; Histone cluster 1, H2aa; Beta actin and Triosephosphate isomerase 1 isoform 1. In the ROS- group, proteins that had >50% peptide coverage were: A-Kinase anchor protein 4 isoform 2; Lactotransferrin precursor; Spermatogenic Tubulin, beta, 2 precursor; Tubulin, beta 4; Tubulin, alpha 3c; Ropporin, rhophilin associated protein 1B; Prolactin-induced protein; Histone cluster 1, H2aa and Triosephosphate isomerase 1 isoform 1;
Table 1.
Protein name, NCBI database index number, calculated MW, pI, peptide coverage and Mascot score in ROS+ group
| No. | Protein name | NCBI database index number | Calculated MW (kDa) | pI |
LTQ |
|
|---|---|---|---|---|---|---|
|
Peptides |
Mascot |
|||||
| (Coverage) | score | |||||
| 1 |
A-Kinase anchor protein 4 isoform 2 |
21493039 |
94 |
6.7 |
34 (54%) |
10988 |
| 2 |
lactotransferrin precursor |
54607120 |
80 |
8.5 |
36 (64%) |
7287 |
| 3 |
tubulin, beta, 2 |
5174735 |
50 |
4.7 |
24 (68%) |
6702 |
| 4 |
tubulin, beta 4 |
21361322 |
50 |
4.8 |
19 (57%) |
5071 |
| 5 |
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
7657116 |
44 |
8.4 |
12 (48%) |
5711 |
| 6 |
tubulin, alpha 3c |
17921993 |
50 |
4.9 |
15 (51%) |
2942 |
| 7 |
ropporin, rhophilin associated protein 1B |
59891409 |
24 |
5.1 |
7 (56%) |
2587 |
| 8 |
semenogelin II precursor |
4506885 |
65 |
9.1 |
13 (22%) |
2277 |
| 9 |
outer dense fiber of sperm tails 2 isoform 2 |
24430183 |
73 |
7.2 |
16 (31%) |
2051 |
| 10 |
semenogelin I isoform a preproprotein |
4506883 |
52 |
9.3 |
12 (31%) |
1983 |
| 11 |
sperm protein associated with the nucleus, X chromosome, family member C |
13435137 |
11 |
5.0 |
3 (45%) |
1892 |
| 12 |
tubulin, alpha, ubiquitous |
57013276 |
50 |
4.9 |
13 (46%) |
1616 |
| 13 |
glutathione S-transferase mu 3 |
23065552 |
26 |
5.3 |
7 (37%) |
1595 |
| 14 |
prolactin-induced protein |
4505821 |
16 |
8.2 |
8 (62%) |
1559 |
| 15 |
A Kinase anchor protein 3 |
21493041 |
95 |
5.8 |
20 (35%) |
1416 |
| 16 |
histone cluster 1, H2ba |
24586679 |
14 |
10.3 |
1 (11%) |
1371 |
| 17 |
mitochondrial malate dehydrogenase precursor |
21735621 |
35 |
8.9 |
8 (33%) |
1342 |
| 18 |
histone cluster 1, H2aa |
25092737 |
14 |
10.8 |
6 (58%) |
1334 |
| 19 |
clusterin isoform 1 |
42716297 |
58 |
6.2 |
6 (16%) |
1204 |
| 20 |
ropporin |
21359920 |
24 |
5.5 |
6 (43%) |
1184 |
| 21 |
sorbitol dehydrogenase |
156627571 |
38 |
8.2 |
7 (35%) |
1095 |
| 22 |
mitochondrial ATP synthase beta subunit precursor |
32189394 |
56 |
5.2 |
16 (53%) |
976 |
| 23 |
beta actin |
4501885 |
42 |
5.2 |
9 (54%) |
909 |
| 24 |
triosephosphate isomerase 1 isoform 1 |
4507645 |
26 |
6.4 |
9 (55%) |
761 |
| 25 |
glyceraldehyde-3-phosphate dehydrogenase |
7669492 |
36 |
8.5 |
6 (32%) |
706 |
| 26 |
L-lactate dehydrogenase C– 1 peptide |
4504973 |
36 |
7.0 |
1 (5%) |
674 |
| 27 |
fatty acid synthase |
41872631 |
275 |
6.0 |
3 (8%) |
669 |
| 28 |
sperm autoantigenic protein 17 |
8394343 |
17 |
4.7 |
2 (22%) |
592 |
| 29 |
pyruvate kinase, muscle isoform M2 |
33286418 |
58 |
7.9 |
11 (38%) |
560 |
| 30 |
ATP synthase, H + transporting, mitochondrial F1 complex, alpha subunit precursor |
4757810 |
59 |
9.1 |
8 (23%) |
542 |
| 31 |
brain creatine kinase |
21536286 |
42 |
5.3 |
7 (32%) |
511 |
| 32 |
heat shock protein 90 kDa beta, member 1 |
4507677 |
92 |
4.7 |
6 (10%) |
502 |
| 33 |
heat shock 70 kDa protein 2 |
13676857 |
70 |
5.5 |
10 (21%) |
448 |
| 34 |
heat shock 90 kDa protein 1, alpha isoform 1 |
153792590 |
98 |
5.0 |
8 (13%) |
447 |
| 35 |
saccharopine dehydrogenase (putative) |
55770836 |
47 |
9.2 |
5 (29%) |
395 |
| 36 |
actin, alpha 1, skeletal muscle |
4501881 |
42 |
5.2 |
6 (36%) |
374 |
| 37 |
glutamine synthetase |
19923206 |
42 |
6.4 |
3 (9%) |
374 |
| 38 |
fructose-bisphosphate aldolase A |
4557305 |
39 |
8.3 |
5 (29%) |
366 |
| 39 |
eukaryotic translation elongation factor 1 alpha 1 |
4503471 |
50 |
9.1 |
8 (36%) |
347 |
| 40 |
fibronectin 1 isoform 3 preproprotein |
16933542 |
262 |
5.4 |
6 (4%) |
321 |
| 41 |
prostate, ovary, testis expressed protein on chromosome 2 |
153791352 |
123 |
5.8 |
4 (8%) |
317 |
| 42 |
acetyl-Coenzyme A acetyltransferase 1 precursor |
4557237 |
45 |
8.9 |
9 (33%) |
316 |
| 43 |
voltage-dependent anion channel 2 |
42476281 |
32 |
7.4 |
4 (22%) |
293 |
| 44 |
outer dense fiber of sperm tails 1 |
194248060 |
30 |
8.4 |
4 (26%) |
283 |
| 45 |
phosphoglycerate kinase 2 |
31543397 |
45 |
8.7 |
5 (30%) |
253 |
| 46 |
histone cluster 1, H2ae |
10645195 |
14 |
11.0 |
4 (49%) |
251 |
| 47 |
protein disulfide – isomerase A3 precursor |
21361657 |
57 |
5.9 |
4 (13%) |
243 |
| 48 |
acid phosphatase, prostate short isoform precursor |
6382064 |
44 |
5.8 |
5 (16%) |
240 |
| 49 |
voltage-dependent anion channel 3 isoform b |
25188179 |
30 |
8.8 |
5 (27%) |
226 |
| 50 |
phospholipase A2, group IIA precursor |
4505849 |
16 |
9.4 |
2 (23%) |
202 |
| 51 |
transglutaminase 4 (prostate) |
156627577 |
77 |
6.3 |
8 (17%) |
187 |
| 52 |
glutathione peroxidase 4 isoform A precursor |
75709200 |
22 |
8.6 |
4 (32%) |
181 |
| 53 |
heat shock 70 kDa protein 5 |
16507237 |
72 |
5.0 |
4 (8%) |
161 |
| 54 |
heat shock protein beta-1 |
4504517 |
22 |
5.9 |
3 (31%) |
160 |
| 55 |
angiotensin I converting enzyme 1 isoform 1 precursor |
4503273 |
150 |
5.9 |
3 (4%) |
160 |
| 56 |
peroxiredoxin 6 |
4758638 |
25 |
6.0 |
2 (16%) |
145 |
| 57 |
chaperonin containing TCP1, subunit 5 (epsilon) |
24307939 |
60 |
5.4 |
4 (12%) |
142 |
| 58 |
valosin-containing protein |
6005942 |
89 |
5.1 |
3 (7%) |
142 |
| 59 |
sperm acrosomal membrane protein 14 – 1 peptide |
19424138 |
13 |
5.4 |
1 (11%) |
132 |
| 60 |
chaperonin containing TCP1, subunit 8 (theta) |
48762932 |
60 |
5.4 |
4 (14%) |
131 |
| 61 |
L-lactate dehydrogenase A isoform 1 |
5031857 |
36 |
8.4 |
2 (13%) |
124 |
| 62 |
phosphoglycerate dehydrogenase |
23308577 |
57 |
6.2 |
3 (13%) |
112 |
| 63 |
clathrin heavy chain 1 |
4758012 |
193 |
5.4 |
4 (5%) |
108 |
| 64 |
chaperonin containing TCP1, subunit 4 (delta) |
38455427 |
58 |
7.9 |
5 (21%) |
99 |
| 65 |
peptidylprolyl isomerase A |
10863927 |
18 |
7.6 |
2 (23%) |
96 |
| 66 |
fumarate hydratase precursor |
19743875 |
54 |
8.8 |
8 (31%) |
92 |
| 67 |
RAB2A, member RAS oncogene family |
4506365 |
23 |
6.0 |
3 (18%) |
92 |
| 68 | olfactomedin 4 precursor | 32313593 | 57 | 5.5 | 2 (5%) | 90 |
Table 2.
Protein name, NCBI database index number, calculated MW, pI, peptide coverage and Mascot score in ROS- group
| No. | Protein name | NCBI database index number | Calculated MW (kDa) | pI |
LTQ |
|
|---|---|---|---|---|---|---|
| Peptides (Coverage) | Mascot score | |||||
| 1 |
A-Kinase anchor protein 4 isoform 2 |
21493039 |
94 |
6.7 |
34 (54%) |
11284 |
| 2 |
lactotransferrin precursor |
54607120 |
80 |
8.5 |
32 (56%) |
6049 |
| 3 |
gyceraldehyde-3-phosphate dehydrogenase, |
7657116 |
44 |
8.4 |
12 (48%) |
5959 |
| 4 |
spermatogenic tubulin, beta, 2 precursor |
5174735 |
50 |
4.7 |
25 (68%) |
5842 |
| 5 |
semenogelin I isoform a preproprotein |
4506883 |
52 |
9.3 |
11(24%) |
4965 |
| 6 |
tubulin, beta 4 |
21361322 |
50 |
4.8 |
19 (58%) |
4879 |
| 7 |
semenogelin II precursor |
4506885 |
65 |
9.1 |
15 (25%) |
4205 |
| 8 |
tubulin, alpha 3c |
17921993 |
50 |
4.9 |
15 (56%) |
2645 |
| 9 |
ropporin, rhophilin associated protein 1B |
59891409 |
24 |
5.1 |
6 (53%) |
2427 |
| 10 |
prolactin-induced protein |
4505821 |
16 |
8.2 |
8 (62%) |
2164 |
| 11 |
A-kinase anchor protein 3 |
21493041 |
95 |
5.8 |
16 (28%) |
1800 |
| 12 |
outer dense fiber of sperm tails 2 Isoform 2 |
24430183 |
73 |
7.2 |
16 (31%) |
1722 |
| 13 |
sperm protein associated with the Nucleus, X chromosome, family Member C |
13435137 |
11 |
5.0 |
3 (45%) |
1437 |
| 14 |
tubulin alpha 6 |
14389309 |
50 |
4.9 |
11 (45%) |
1374 |
| 15 |
ropporin |
21359920 |
24 |
5.5 |
6 (43%) |
1280 |
| 16 |
clusterin isoform 1 |
42716297 |
58 |
6.2 |
5 (15%) |
1257 |
| 17 |
sorbitol dehydrogenase |
156627571 |
38 |
8.2 |
4 (23%) |
1249 |
| 18 |
beta actin |
4501885 |
42 |
5.2 |
6 (34%) |
1232 |
| 19 |
pyruvate kinase, muscle isoform M1 |
3328642 |
58 |
7.6 |
13 (47%) |
1133 |
| 20 |
histone cluster 1, H2aa |
25092737 |
14 |
10.8 |
4 (51%) |
1121 |
| 21 |
mitochondrial ATP synthase beta subunit precursor |
32189394 |
56 |
5.2 |
13 (38%) |
1031 |
| 22 |
fatty acid synthase |
41872631 |
275 |
6.0 |
10 (9%) |
935 |
| 23 |
glutathione S-transferase mu 3 |
23065552 |
26 |
5.3 |
7 (32%) |
929 |
| 24 |
histone cluster 1, H2ba -1 peptide |
24586679 |
14 |
10.3 |
1 (11%) |
823 |
| 25 |
fibronectin 1 isoform 3 preproprotein |
16933542 |
262 |
5.4 |
16 (11%) |
790 |
| 26 |
L-lactate dehydrogenase C |
4504973 |
36 |
7.0 |
2 (7%) |
693 |
| 27 |
heat shock 90 kDa protein 1, alpha isoform 2 |
154146191 |
85 |
4.9 |
10 (18%) |
682 |
| 28 |
glyceraldehyde-3-phosphate dehydrogenase |
7669492 |
36 |
8.5 |
3 (15%) |
621 |
| 29 |
sperm autoantigenic protein 17 |
8394343 |
17 |
4.7 |
2 (22%) |
552 |
| 30 |
triosephosphate isomerase 1 isoform 1 |
4507645 |
26 |
6.4 |
6 (61%) |
530 |
| 31 |
mitochondrial malate dehydrogenase precursor |
21735621 |
35 |
8.9 |
5 (23%) |
512 |
| 32 |
phosphoglycerate kinase 2 |
31543397 |
45 |
8.7 |
4 (17%) |
506 |
| 33 |
heat shock 70 kDa protein 2 |
13676857 |
70 |
5.5 |
8 (22%) |
423 |
| 34 |
histone cluster 1, H2ae |
10645195 |
14 |
11.0 |
3 (43%) |
392 |
| 35 |
acetyl-Coenzyme A acetyltransferase 1 precursor |
4557237 |
45 |
8.9 |
8 (34%) |
388 |
| 36 |
eukaryotic translation elongation factor 1 alpha 1 |
4503471 |
50 |
9.1 |
7 (30%) |
383 |
| 37 |
ATP synthase, H + transporting, mitochondrial F1 complex, alpha subunit precursor |
4757810 |
59 |
9.1 |
8 (22%) |
378 |
| 38 |
outer dense fiber of sperm tails 1 |
194248060 |
30 |
8.4 |
3 (22%) |
334 |
| 39 |
voltage-dependent anion channel 2 |
42476281 |
32 |
7.4 |
3 (19%) |
332 |
| 40 |
acid phosphatase, prostate short isoform precursor |
6382064 |
44 |
5.8 |
5 (16%) |
326 |
| 41 |
brain creatine kinase |
21536286 |
42 |
5.3 |
7 (29%) |
268 |
| 42 |
saccharopine dehydrogenase (putative) |
55770836 |
47 |
9.2 |
4 (29%) |
265 |
| 43 |
protein disulfide-isomerase A3 precursor |
21361657 |
57 |
5.9 |
2 (9%) |
261 |
| 44 |
eukaryotic translation elongation factor 2 |
4503483 |
96 |
6.4 |
3 (8%) |
261 |
| 45 |
heat shock protein 90 kDa beta, member 1 |
4507677 |
92 |
4.7 |
5 (8%) |
250 |
| 46 |
heat shock protein beta-1 |
4504517 |
22 |
5.9 |
4 (36%) |
191 |
| 47 |
glutamine synthetase |
19923206 |
42 |
6.4 |
3 (14%) |
179 |
| 48 |
valosin-containing protein |
6005942 |
89 |
5.1 |
7 (18%) |
177 |
| 49 |
phospholipase A2, group IIA precursor |
4505849 |
16 |
9.4 |
2 (23%) |
170 |
| 50 |
tyrosine 3/tryptophan 5 -monooxygenase activation protein, zeta polypeptide |
4507953 |
27 |
4.7 |
2 (19%) |
167 |
| 51 |
L-lactate dehydrogenase A isoform 1 |
5031857 |
36 |
8.4 |
2 (7%) |
167 |
| 52 |
fructose-bisphosphate aldolase A |
4557305 |
39 |
8.3 |
3 (16%) |
167 |
| 53 |
chaperonin containing TCP1, subunit 5 (epsilon) |
24307939 |
60 |
5.4 |
3 (9%) |
157 |
| 54 |
voltage-dependent anion channel 3 isoform b |
25188179 |
30 |
8.8 |
4 (26%) |
147 |
| 55 |
peroxiredoxin 6 |
4758638 |
25 |
6.0 |
4 (33%) |
140 |
| 56 |
glutathione peroxidase 4 isoform A precursor |
75709200 |
22 |
8.6 |
5 (43%) |
139 |
| 57 |
transglutaminase 4 (prostate) |
156627577 |
77 |
6.3 |
5 (9%) |
136 |
| 58 |
enolase 1 |
4503571 |
47 |
7.0 |
3 (12%) |
134 |
| 59 |
T-complex protein 1 isoform b |
57863259 |
44 |
7.5 |
5 (30%) |
129 |
| 60 |
CSE1 chromosome segregation 1-like protein |
29029559 |
111 |
5.5 |
2 (3%) |
128 |
| 61 |
fumarate hydratase precursor |
19743875 |
54 |
8.8 |
3 (12%) |
115 |
| 62 |
lectin, mannose-binding 2 precursor |
5803023 |
40 |
6.4 |
2 (13%) |
115 |
| 63 |
clathrin heavy chain 1 |
4758012 |
193 |
5.4 |
6 (7%) |
111 |
| 64 |
chaperonin containing TCP1, subunit 4 (delta) |
38455427 |
58 |
7.9 |
4 (19%) |
107 |
| 65 |
RAB2A, member RAS oncogene family |
4506365 |
23 |
6.0 |
3 (18%) |
106 |
| 66 | chromosome 20 open reading frame 3 | 24308201 | 46 | 5.8 | 3 (19%) | 105 |
Out of the total 74 proteins identified, 47 were overexpressed and 27 underexpressed in the ROS+ group; 15 proteins were significantly (2-fold) differentially expressed in the ROS+ samples compared to the ROS- samples.
Of the differentially expressed proteins, 15 proteins were overexpressed in the ROS+ samples while 5 were underexpressed in comparison to ROS- samples (Table 3). The overexpressed proteins included the histone cluster1; H2ba (HIST1H2BA); mitochondrial malate dehydrogenase precursor (MDH2); heat shock protein 90 kDa beta, member 1 (HSP90B1); heat shock 70 kDa protein 5 (HSPA5); glutamine synthetase (GLUL); transglutaminase 4 (prostate) (TGM4); glutathione peroxidase 4 isoform A precursor (GPX4); sperm acrosomal membrane protein 14 (SPACA4); olfactomedin 4 precursor (OLFM4) and chromosome 20 open reading frame 3 (C20orf3). Proteins that were found to be under expressed were: semenogelin II precursor (SEMG2); peroxiredoxin 6 (PRDX6); clathrin heavy chain 1 (CLTC); eukaryotic translation elongation factor 2 (EEF2) and enolase 1 (ENO1).
Table 3.
Differentially expressed proteins in ROS- and ROS+ groups along with the spectral count and the spectral count ratio for the two groups
| Protein | NCBI # | SC ROS- | NSC-ROS- | SC ROS+ | NSC-ROS+ | NSC ratio in ROS+/ROS- |
|---|---|---|---|---|---|---|
| acetyl-Coenzyme A acetyltransferase 1 precursor (ACAT1) |
4557237 |
20 |
0.006 |
26.000 |
0.008 |
1.4 |
| acid phosphatase, prostate short isoform precursor (ACPP) |
6382064 |
17 |
0.005 |
27.000 |
0.009 |
1.7 |
| actin, alpha 1, skeletal muscle (ACTA1) |
4501881 |
0 |
0.000 |
8.000 |
0.003 |
ROS + only |
| A-kinase anchor protein 3 (AKAP3) |
21493041 |
76 |
0.022 |
70.000 |
0.022 |
1.0 |
| A-kinase anchor protein 4 isoform 2 (AKAP4) |
21493039 |
497 |
0.145 |
467.000 |
0.147 |
1.0 |
| angiotensin I converting enzyme 1 isoform 1 precursor (ACE) |
4503273 |
5 |
0.001 |
5.000 |
0.002 |
1.1 |
| ATP synthase, H + transporting, mitochondrial F1 complex, alpha subunit precursor (ATP5A1) |
4757810 |
28 |
0.008 |
30.000 |
0.009 |
1.2 |
| beta actin (ACTB) |
4501885 |
53 |
0.015 |
36.000 |
0.011 |
0.7 |
| brain creatine kinase (CKB) |
21536286 |
19 |
0.006 |
21.000 |
0.007 |
1.2 |
| chaperonin containing TCP1, subunit 4 (delta) (CCT4) |
38455427 |
4 |
0.001 |
6.000 |
0.002 |
1.6 |
| chaperonin containing TCP1, subunit 5 (epsilon) (CCT8) |
24307939 |
4 |
0.001 |
7.000 |
0.002 |
1.9 |
| chaperonin containing TCP1, subunit 8 (theta) |
48762932 |
5 |
0.001 |
4.000 |
0.001 |
0.9 |
| chromosome 20 open reading frame 3 (C20orf3) |
24308201 |
2 |
0.001 |
4.000 |
0.001 |
2.2 |
| clathrin heavy chain 1 (CLTC) |
4758012 |
14 |
0.004 |
6.000 |
0.002 |
0.5 |
| clusterin preproprotein (CLU) |
355594753 |
56 |
0.016 |
55.000 |
0.017 |
1.1 |
| CSE1 chromosome segregation 1-like protein (CSE1L) |
29029559 |
3 |
0.001 |
0.000 |
0.000 |
ROS- only |
| enolase 1 (ENO1) |
4503571 |
7 |
0.002 |
2.000 |
0.001 |
0.3 |
| eukaryotic translation elongation factor 1 alpha 1 (EEF1A1) |
4503471 |
28 |
0.008 |
33.000 |
0.010 |
1.3 |
| eukaryotic translation elongation factor 2 (EEF2) |
4503483 |
12 |
0.003 |
5.000 |
0.002 |
0.5 |
| fatty acid synthase (FASN) |
41872631 |
40 |
0.012 |
35.000 |
0.011 |
1.0 |
| fibronectin 1 isoform 3 preproprotein (FN1) |
16933542 |
41 |
0.012 |
21.000 |
0.007 |
0.6 |
| fructose-bisphosphate aldolase A (ALDOA) |
4557305 |
15 |
0.004 |
17.000 |
0.005 |
1.2 |
| fumarate hydratase precursor (FH) |
19743875 |
5 |
0.001 |
7.000 |
0.002 |
1.5 |
| glutamine synthetase (GLUL) |
19923206 |
4 |
0.001 |
12.000 |
0.004 |
3.2 |
| glutathione peroxidase 4 isoform A precursor (GPX4) |
75709200 |
8 |
0.002 |
21.000 |
0.007 |
2.8 |
| glutathione S-transferase mu 3 (GSTM3) |
23065552 |
50 |
0.015 |
65.000 |
0.020 |
1.4 |
| glyceraldehyde-3-phosphate dehydrogenase (GAPDH) |
7669492 |
14 |
0.004 |
22.000 |
0.007 |
1.7 |
| glyceraldehyde-3-phosphate dehydrogenase, spermatogenic (GAPDHS) |
7657116 |
210 |
0.061 |
185.000 |
0.058 |
1.0 |
| heat shock 70 kDa protein 2 (HSPA2) |
13676857 |
26 |
0.008 |
22.000 |
0.007 |
0.9 |
| heat shock 70 kDa protein 5 (HSPA5) |
16507237 |
5 |
0.001 |
11.000 |
0.003 |
2.4 |
| heat shock 90 kDa protein 1, alpha isoform 1 (HSP90AA1) |
153792590 |
39 |
0.011 |
26.000 |
0.008 |
0.7 |
| heat shock protein 90 kDa beta, member 1 (HSP90B1) |
4507677 |
11 |
0.003 |
22.000 |
0.007 |
2.2 |
| heat shock protein beta-1 (HSPB1) |
4504517 |
5 |
0.001 |
4.000 |
0.001 |
0.9 |
| histone cluster 1, H2aa (HIST1H2AA) |
25092737 |
47 |
0.014 |
56.000 |
0.018 |
1.3 |
| histone cluster 1, H2ae (HISTH1AE) |
10645195 |
14 |
0.004 |
13.000 |
0.004 |
1.0 |
| histone cluster 1, H2ba (HIST1H2BA) |
24586679 |
22 |
0.006 |
49.000 |
0.015 |
2.4 |
| lactotransferrin precursor (LTF) |
54607120 |
246 |
0.072 |
292.000 |
0.092 |
1.3 |
| lectin, mannose-binding 2 precursor |
5803023 |
3 |
0.001 |
5.000 |
0.002 |
1.8 |
| L-lactate dehydrogenase A isoform 1 (LDHA) |
5031857 |
10 |
0.003 |
9.000 |
0.003 |
1.0 |
| L-lactate dehydrogenase C (LDHC) |
4504973 |
17 |
0.005 |
12.000 |
0.004 |
0.8 |
| mitochondrial ATP synthase beta subunit precursor (ATP5B) |
32189394 |
50 |
0.015 |
70.000 |
0.022 |
1.5 |
| mitochondrial malate dehydrogenase precursor (MDH2) |
21735621 |
22 |
0.006 |
44.000 |
0.014 |
2.2 |
| olfactomedin 4 precursor (OLFM4) |
32313593 |
1 |
0.000 |
4.000 |
0.001 |
4.3 |
| outer dense fiber of sperm tails 1 (ODF1) |
194248060 |
15 |
0.004 |
14.000 |
0.004 |
1.0 |
| outer dense fiber of sperm tails 2 isoform 2 (ODF2) |
24430183 |
90 |
0.026 |
91.000 |
0.029 |
1.1 |
| peptidylprolyl isomerase A (PPIA) |
10863927 |
4 |
0.001 |
2.000 |
0.001 |
0.5 |
| peroxiredoxin 6 (PRDX6) |
4758638 |
4 |
0.001 |
1.000 |
0.0003 |
0.3 |
| phosphoglycerate dehydrogenase (PHGDH) |
23308577 |
3 |
0.001 |
5.000 |
0.002 |
1.8 |
| phosphoglycerate kinase 2 (PGK2) |
31543397 |
17 |
0.005 |
9.000 |
0.003 |
0.6 |
| phospholipase A2, group IIA precursor (PLA2G2A) |
4505849 |
6 |
0.002 |
10.000 |
0.003 |
1.8 |
| prolactin-induced protein (PIP) |
4505821 |
118 |
0.034 |
101.000 |
0.032 |
0.9 |
| prostate specific antigen isoform 1 preproprotein (KLK3) |
4502173 |
3 |
0.001 |
2.000 |
0.001 |
0.7 |
| protein disulfide-isomerase A3 precursor (PDIA3) |
21361657 |
7 |
0.002 |
7.000 |
0.002 |
1.1 |
| pyruvate kinase, muscle isoform M2 (PKM) |
33286418 |
49 |
0.014 |
31.000 |
0.010 |
0.7 |
| RAB2A, member RAS oncogene family (RAB2A) |
4506365 |
3 |
0.001 |
3.000 |
0.001 |
1.1 |
| ropporin (ROPN1) |
21359920 |
20 |
0.006 |
31.000 |
0.010 |
1.7 |
| ropporin, rhophilin associated protein 1B (ROPN1B) |
59891409 |
73 |
0.021 |
73.000 |
0.023 |
1.1 |
| saccharopine dehydrogenase (putative) (SCCPDH) |
55770836 |
13 |
0.004 |
16.000 |
0.005 |
1.3 |
| semenogelin I isoform a preproprotein (SEMG1) |
4506883 |
171 |
0.050 |
101.000 |
0.032 |
0.6 |
| semenogelin II precursor (SEMG2) |
4506885 |
513 |
0.149 |
226.000 |
0.071 |
0.5 |
| sorbitol dehydrogenase (SORD) |
156627571 |
36 |
0.010 |
27.000 |
0.009 |
0.8 |
| sperm acrosomal membrane protein 14 |
19424138 |
1 |
0.000 |
2.000 |
0.001 |
2.2 |
| sperm autoantigenic protein 17 (SPA17) |
8394343 |
15 |
0.004 |
20.000 |
0.006 |
1.4 |
| sperm protein associated with the nucleus, X chromosome, family member C (SPANX) |
13435137 |
54 |
0.016 |
80.000 |
0.025 |
1.6 |
| transglutaminase 4 (prostate) (TGM4) |
156627577 |
4 |
0.001 |
9.000 |
0.003 |
2.4 |
| triosephosphate isomerase 1 isoform 1 (TPI1) |
4507645 |
28 |
0.008 |
31.000 |
0.010 |
1.2 |
| tubulin alpha 6 (TUBA1C) |
14389309 |
9 |
0.003 |
11.000 |
0.003 |
1.3 |
| tubulin, alpha 3c (TUBA3C) |
17921993 |
117 |
0.034 |
134.000 |
0.042 |
1.2 |
| tubulin, beta 4 (TUBB4A) |
21361322 |
36 |
0.010 |
30.000 |
0.009 |
0.9 |
| tubulin, beta, 2 (TUBB4B) |
5174735 |
232 |
0.068 |
235.000 |
0.074 |
1.1 |
| tyrosine 3/tryptophan 5 -monooxygenase activation protein, zeta polypeptide (YWHAZ) |
4507953 |
4 |
0.001 |
3.000 |
0.001 |
0.8 |
| valosin-containing protein (VCP) |
6005942 |
16 |
0.005 |
9.000 |
0.003 |
0.6 |
| voltage-dependent anion channel 2 (VDAC2) |
42476281 |
11 |
0.003 |
11.000 |
0.003 |
1.1 |
| voltage-dependent anion channel 3 isoform b | 25188179 | 7 | 0.002 | 12.000 | 0.004 | 1.9 |
A cut-off of 2-fold difference in NSC ratio was considered as significantly differentially expressed in ROS +. Overexpressed and underexpressed proteins are shown in bold.
Gene ontology annotations of the proteins identified in ROS+ and ROS- samples
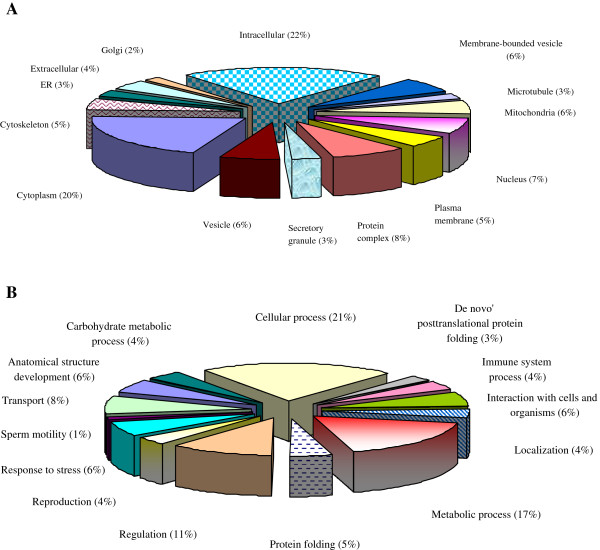
The cellular distributions of the 74 proteins present in the ROS+ and ROS- samples showed that there was a greater distribution of proteins in the intracellular region compared to the extracellular region (Figure 1A). A significant number of proteins were also distributed in the cytoplasmic region. The proteins involved in the biological processes showed a greater distribution in cellular processes (21%) followed by metabolic processes (17%) (Figure 1B).
Figure 1.
Functional annotations from consolidated publicly available software tools A: Cellular distribution showing the maximum distribution was intracellular (22%) followed by cytoplasmic distribution; B: Biological processes distribution of proteins identified in spermatozoa from ROS+ and ROS- group showing the highest distribution was in the cellular processes (21%) followed by metabolic processes.
Gene ontology annotation comparisons of the differentially expressed proteins in ROS+ and ROS- samples
Based on the Gene Ontology classifications, the majority of the differentially expressed proteins that were over or underexpressed in ROS+ group compared to the ROS- group were localized to the cytoplasmic and intracellular regions (Figure 2). Specifically, the cellular compartments of intracellular, organelle, macromolecular complex region and mitochondria were predominantly occupied by the overexpressed proteins. The cytosolic, cytoplasm, extracellular, plasma membrane, protein complex and the vesicular region showed an abundance of underexpressed proteins when compared to the overexpressed ones. Strikingly, the endosome, lipid particle, membrane-bound organelles and the microtubules showed only the presence of overexpressed proteins in the ROS+ group while the proteinaceous extracellular matrix was restricted only to the underexpressed proteins.
Figure 2.
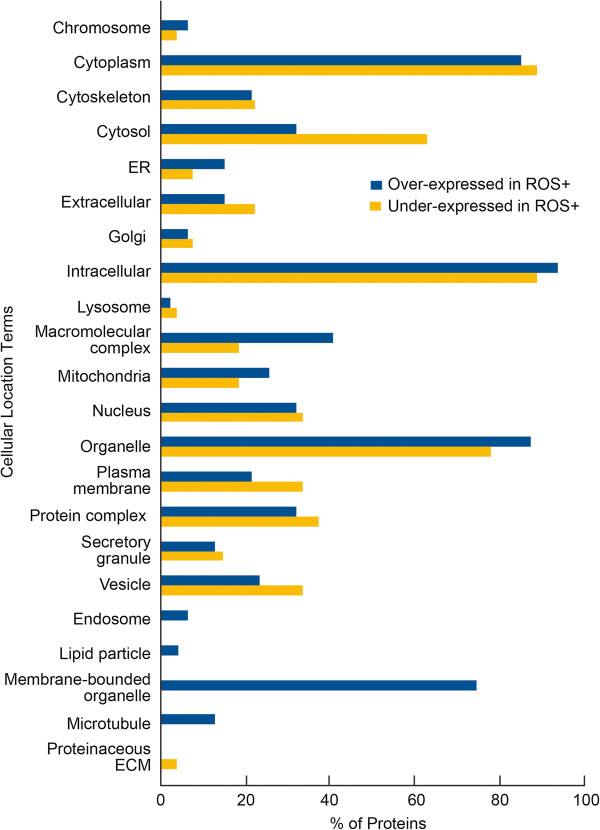
Cellular distribution showing the proteins that were significantly overexpressed in ROS + group were located in the cytoplasm, intracellular, organelle and membrane-bound organelle and underexpressed in ROS + group were located in the cytoplasm, cytosol, intracellular and organellar.
GO analysis also revealed that the majority of over and under expressed proteins in the ROS+ group was found to be involved in cellular processes (Figure 3). The proteins involved in developmental processes, cellular process, interactions with cells and organisms, localization, metabolic processes and transport were common to both under and overexpressed proteins. Overexpressed proteins were abundantly expressed for these functions. Many of the processes such as cellular amino acid metabolic processes, cellular component biogenesis, chromosome organization, cytoskeleton organization, embryo development, gluconeogenesis and homeostatic processes were found to be restricted only to the overexpressed proteins. Similarly, the processes such as carbohydrate catabolic processes, cellular component movement, glycolysis, and response to unfolded protein were found to be restricted to the underexpressed proteins.
Figure 3.
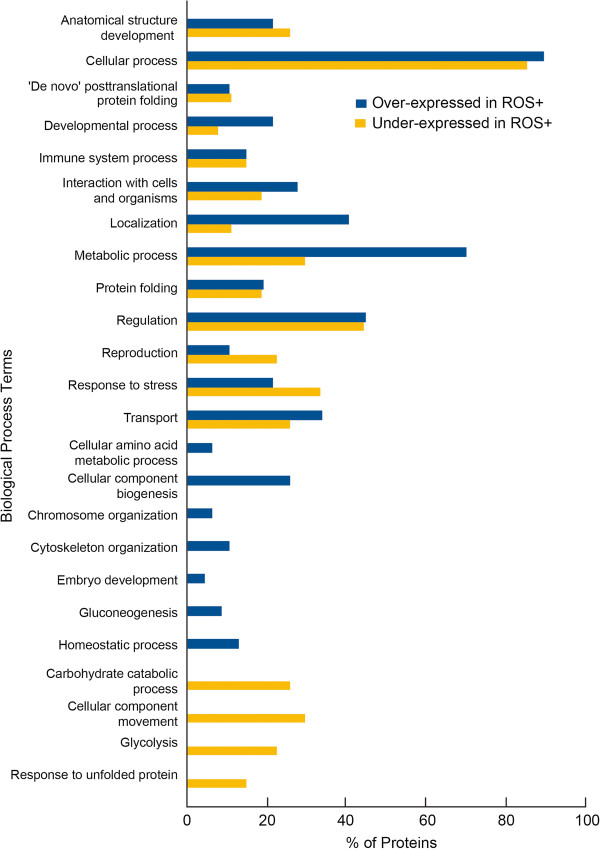
Biological process distribution of proteins of overexpressed were cellular processes, metabolic processes, localization, regulation and transport in spermatozoa from ROS + compared to cellular preocesses, regulation, response to stress, cellular movement and glycolysis in ROS- group.
Pathways and network analysis of differentially expressed in ROS+ proteins using Ingenuity Pathway Analysis (IPA) and Metacore™ software packages
A list of significantly enriched or topmost pathways and/or process networks associated with the over and underexpressed proteins in the ROS+ group is shown in Table 3. Cellular processes were comprised of cell cycle, cell adhesion, cellular morphology, cellular movement and cell death and cell survival. Protein modifications included protein folding and degradation; general metabolic pathways were: carbohydrate metabolism (such as glycolysis and gluconeogenesis), amino acid metabolism, energy metabolism and oxidative stress regulation. With respect to the pathways related to carbohydrate metabolism, enzymes operating in gluconeogenesis and glycolysis including phosphoglycerate kinase 2 (PGK2) and glyceraldehyde phosphate dehydrogenase-S (GAPD-S) were found to be underexpressed in the ROS+ group for both gluconeogenesis and glycolysis while glyceraldehyde-3-phosphate dehydrogenase (GAPDH), fructose-biphosphate aldolase A(ALDOA) and mitochondrial malate dehydrogenase precursor (MDH2) were overexpressed. The overexpressed proteins, especially in the glycolytic pathway, were triosephosphate isomerase 1 (isoform 1(TPI1), glyceraldehyde-3 phosphate dehydrogenase (GAPDH) and fructose-biphosphate aldolase A (ALDOA). Two enzymes involved in the fermentation of pyruvate to lactate – L-lactate dehydrogenase C (LDHC) and L-lactate dehydrogenase A isoform 1(LDHA) were underexpressed. In addition, two enzymes functional in sucrose degradation pathways - triosephosphate isomerase 1 (TPI1) and fructose biphosphate aldolase A (ALDO A), were overexpressed.
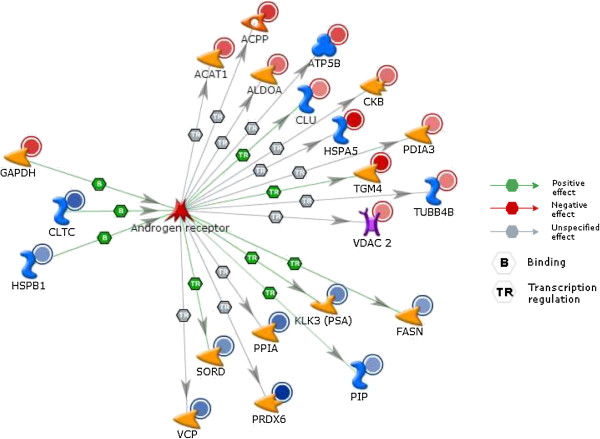
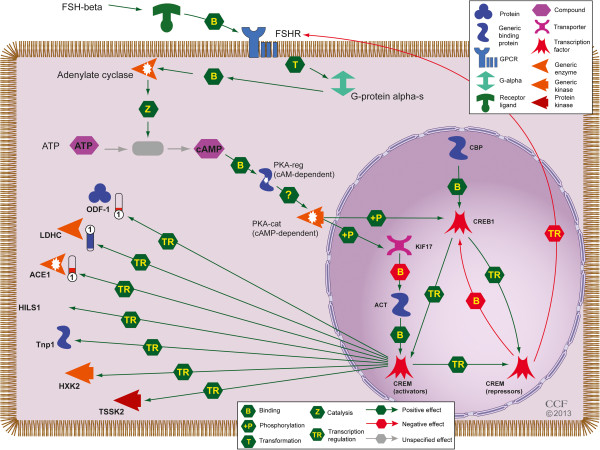
Transcriptional regulatory network analysis of the differentially expressed proteins, using Metacore™, showed that the androgen receptor was one of the topmost regulators with 21 differentially expressed proteins in the ROS+ group interacting with the receptor (Figure 4). As a result of the established connection between germ cell development and the levels of androgens, it was important to elucidate the potential interaction of these over- and underexpressed proteins with the androgen receptor. One of the pathways that may be relevant to spermatogenesis is cAMP responsive element modulator (CREM) signaling in the testis (Figure 5). In this pathway, 3 proteins were found to be differentially expressed. Angiotensin I converting isoform 1 precursor (ACE). The outer dense fiber of sperm tails 1 (ODF1) was found to be underexpressed whereas the L- lactate dehydrogenase C (LDHC) was overexpressed in ROS+ sample compared with ROS- sample.
Figure 4.
Transcriptional regulatory network showing interactions between differentially expressed ROS + proteins and androgen receptor. Proteins with red or blue circles around them are over-expressed (HSPA5 and TGM4) or underexpressed (PRDX6) in spermatozoa from the ROS + (relative to ROS-) group. The levels of expression values are reflected in the intensity of red or blue colors. Green arrows with a hexagon indicate positive effect. TR = Transcription regulation; PGK2 = phosphoglycerate kinase 2; GAPD-S = glyceraldehyde phosphate dehydrogenase-S; GAPDH = glyceraldehyde-3-phosphate dehydrogenase; ALDOA = fructose-biphosphate aldolase A; and MDH2 = mitochondrial malate dehydrogenase precursor.
Figure 5.
CREM signaling in the testis. Proteins in red or blue are the ones over or underexpressed in spermatozoa from the ROS + compared to ROS- group. The color levels in the tubes reflect their expression levels (red-overexpressed and blue underexpressed). B = Binding; Z = catalysis; T = transformation and TR = transcription regulation. The figure shows the generic enxzymes (Adenylate cyclase, LDHC, ACE1, HXK2); Protein kinases (PKA-cat (cAMP –dependent and TSSK2); CREM activators and repressors; generic binding proteins (ACT, CPB and Tnp1 and PKA-reg (cAMP –dependent) and proteins (HILS1 and ODF1) involved in the CREM signaling in the testis. CREM = cAMP responsive element modulator; ACE = Angiotensin I converting isoform 1 precursor; ODF1 = outer dense fiber of sperm tails 1; LDHC = L- lactate dehydrogenase C.
Discussion
Oxidative stress plays a key role in the etiology of male infertility, possibly by altering protein expression levels in spermatozoa, causing molecular and genetic defects. Therefore, understanding the function of each protein involved and the post-translational modifications that occur during sperm-maturation is important and may lead to a useful biomarker for male infertility. In earlier studies, we demonstrated that ROS are independent of the semen parameters [6]. Therefore, in the current study, we compared the sperm proteome from a group of men with high levels of ROS (ROS+) with that from men with low or physiological levels of ROS (ROS-). The ROS grouping was done irrespective of the patient’s clinical diagnoses and semen parameters: the only criteria was the presence or absence of ROS.
We have identified 74 proteins, 15 of which had a >2-fold difference in their expression levels in the ROS+ group when compared to the ROS- group. Based on the spectral counts of the 74 proteins, we identified 47 were overexpressed and 27 that were underexpressed in the ROS+ group compared to the ROS- group. Of the 74 proteins, 17 were determined to be either in their precursor or pre-protein form. The presence of these incompletely processed proteins is indicative of post-translational processing problems as reported by some investigators [31,32]. Accumulation of precursor forms of protein may result in deregulation of the downstream functions of the mature protein [31]. Oxidative stress may result in the accumulation of precursors or preproteins. The cellular distribution of all the proteins as ascertained by GO analysis (Figure 1A) suggested that the majority of the proteins (68 proteins) were intracellular or organellar (62 proteins) in origin, whereas a few were localized in the extracellular (13 proteins) region. The presence of extracellular proteins may be attributed to the presence of round cells in the ejaculate from both the donors and patients. We did not remove these from subsequent processing for proteomic analysis.
Similar comparative studies have been performed previously by other groups in an effort to understand the sperm proteome [31,33,34]. Most notable amongst these are two studies where the expression profile of proteins was compared in asthenozoospermic and normozoospermic donors [31,33]. In these studies, 10 proteins and 17 spots were identified using 2-DE suggesting their role in motility-related male infertility. Xu et al conducted a comparative study between infertile patients and normozoospermic group and identified 24 differentially expressed proteins that could potentially serve as diagnostic markers in identifying the underlying pathology of male infertility [35]. Our findings in this study are novel, as we have compared and identified the proteins that are differentially expressed in spermatozoa obtained from seminal ejaculates with high levels of ROS (ROS+) to those normal levels of ROS (ROS-).
On examining the association of the differentially expressed proteins with the biological processes (Figure 1B), we concluded that the majority of the proteins were involved in cellular, metabolic, and regulatory processes. A smaller distribution of proteins (3-5%) was also found in biological processes such as de novo post translational (8 proteins) and protein folding (14 proteins), and these were identified as ‘enriched processes’ for our list of proteins, by the GO Term Finder.
Even though the spermatozoa become transcriptionally inactive after spermatogenesis, we found the presence of the translational proteins and this may be indicative of the presence of some leftovers from inefficient spermatogenesis. They may also have a role in the normal physiology of the spermatozoa or affect fertilization and embryo development. Further validation through the pathway and process network analysis using IPA and Metacore™, suggested that the topmost function of the differentially expressed proteins was carbohydrate metabolism and included pathways of gluconeogenesis, glycolysis, fermentation of pyruvate to lactate and sucrose degradation. There is a great controversy pertaining to the major pathway of energy metabolism during sperm motility [36-39]. In this context, our findings showed that while oxidative phosphorylation is important for sperm function, the major pathway for energy production for sperm motility was via the glycolytic pathway. To support this view, various respiratory enzymes involved in the glycolytic pathway including phosphoglycerate kinase 2 (PGK2), glyceraldehyde-3phosphate dehydrogenase (GAPDH), fructose- biphosphate aldolase A (ALDOA), glyceraldehyde-3 phosphate dehydrogenase-S (GAPDHS), triosephosphate isomerase 1 isoform (TPI1) were identified in our study. Furthermore, GAPDH-S protein was overexpressed in the ROS+ sample. Oxidation of GAPDH-S as a result of H2O2 generation has been reported to decrease sperm motility and inhibit GAPDH-S activity [40].
Various isoforms of histone proteins were also identified and these included histone cluster 1, H2aa (HIST1H2AA), histone cluster1 H2ae (HISTH1AE) and histone cluster1 and H2ba (HIST1H2BA). Histones are a group of proteins that are replaced by the protamines during sperm maturation in the epididymal region [41]. Their presence in the ejaculated spermatozoa is indicative of improper packaging of sperm chromatin and subsequent DNA damage. These findings have been attributed to oxidative stress [5,41].
We found 17 precursor proteins to be dysregulated; of these, 10 were over-expressed and 5 were underexpressed with a 2-fold difference between the ROS+ and ROS- groups. The differentially expressed proteins were mainly intracellular, cytoplasmic or organellar in distribution. This is contrary to the findings of Xu et al, who reported a higher distribution of differentially expressed proteins in the extracellular region in the infertile groups with normal semen parameters [35]. This difference may be attributed to the import/export of epididymal and seminal vesicle proteins. Furthermore, the distribution of overexpressed and underexpressed proteins in the ROS+ group revealed their involvement in various processes. We noted that many proteins showing significant changes in expression between the ROS+ and ROS- groups had roles in sexual reproduction, metabolic processes and stress response (Figures 2 and 3).
Our data suggests that semenogelin II precursor (SEMG2), which is involved in sexual reproduction, was underexpressed in the ROS+ group. Overexpression of semenogelin I and II precursor has been reported in previous comparative studies [31,35]. Xu et al reported an increase in both isoforms of semenogelin in the spermatozoa of infertile patients compared to the normozoospermic men [35]. Semenogelin -1 and semenogelin -2 have been reported in men with varicocele only as well as in men with varicocele with a history of moderate as well as heavy smoking. Semenogelin is known to cause a decrease in sperm motility, but it was within the normal range in the ROS+ samples. Semenogelin (SEMG) is the major protein of seminal fluid proteins (20-40%) and is secreted from seminal vesicles, epididymis, and prostate [42,43]. It is an androgen dependent protein that exists in two forms I (52 kDa) and II (71-76 kDa). SEMG helps in the formation of the semen coagulum that is degraded later by prostate-specific antigen. SEMG counteracts oxidative stress by reducing the generation of free radicals through several mechanisms: slow sperm motility of the entrapped sperm, reduced energy consumption, free radical generation and inhibition of superoxide radical generation through direct interference with sperm NADH oxidase [44].
A great number of proteins are involved in the energy production required for the spermatozoon tail movement. We identified several proteins that are involved in energy producing processes such as the carbohydrate metabolic pathways of glycolysis and gluconeogenesis. Enolase I (ENO1) was found to be significantly underexpressed while respiratory enzyme mitochondrial malate dehydrogenase precursor (MDH2) was found to be overexpressed. MDH2 is an ROS producing enzyme localized in the sperm mitochondria [45]. It catalyzes the last step of the Krebs cycle when malate is converted into oxaloacetate using NAD+/NADH as intermediates. Several of the chaperone proteins such as heat shock protein 90 kDa beta, member 1 (HSP90B1) and heat shock 70 kDa protein 5 (HSPA5) were also seen to be overexpressed in response to stress. The association of these proteins with various processes related to cellular stressors such as heat, glucose deprivation, free radical attack, and infection indicates that ROS induces a stress response in the spermatozoa [46-48].
The GO annotations further revealed the involvement of overexpressed proteins in cellular amino acid metabolic processes. Examples of proteins that were found to be overexpressed included transglutaminases (TGM4) and glutamine synthetases. Transglutaminases are a family of calcium-dependent enzymes that catalyze the post-translational modification of the selected glutamine residues on proteins by cross-linking with the peptide-bound lysine residues or incorporating polyamines [49,50]. Transglutaminase that is secreted from the prostate is known as TGM4 prostate. A recent study showed increased levels of transglutaminase in response to oxidative stress in the prostate [51]. Excessive production of ROS has been reported to induce cell senescence, oxidative protein modifications, and cell membrane damage, increase intracellular calcium and activate certain cytokines such as TGFb [51]. Oxidative stress impacts the aggregation, increased molecular weight, unfolding, denaturation and proteolysis of cellular proteins. These effects may result in the initiation of cellular hyperamonemia, which probably triggers the expression of glutamate synthetase to redirect the production of the released ammonium into glutamate [52].
Glutathione peroxidase 4 isoform A precursor (GPX4) and peroxiredoxin 6 (PRDX6) function to reduce oxidative stress. The former was overexpressed and the latter was underexpressed in the ROS+ group. GPX4 is a well-known antioxidant enzyme and its increased expression in sperm in the ROS+ samples is reflective of its influence on genes synthesizing this enzyme. PRDX6 also belongs to a family of antioxidant enzyme known as peroxiredoxins. They detoxify H2O2 similar to catalase or glutathione peroxidases (GPXs) [53,54]. Peroxiredoxin 6 is localized in the sperm head (postacrosomal region and equatorial segment) and sperm tail [55]. Reduced levels of peroxiredoxins (PRDX1 and PRDX6) have been reported in infertile men with oxidative stress. Reduced levels of PRDX6 have also been reported in the seminal plasma from men with varicocele. Differential amounts of peroxiredoxin 6 have been reported (41% in men with varicocele and 65% in men with idiopathic infertility) [56]. Human spermatozoa are reported to have high sensitivity to ROS and may be a result of reduced recycling of peroxiredoxins after exposure to oxidative stress [55].
Optimal levels of expression and function of androgens are central to the process of spermatogenesis. To understand the dependency of the differentially expressed proteins in the ROS+ groups, a network was derived to identify proteins regulated by androgen receptors. Spermatogenesis is dependent on androgen action and androgens act by stimulating the receptors present on the Sertoli cells [57,58]. Transcriptional network analysis (Figure 4) showed that 21 proteins were regulated by the androgen receptor. In our study, several proteins appeared to be transcriptionally activated by the androgen receptor but the effect of the androgen receptor on several other proteins is still unknown (Figure 5). Stanton et al reported that any loss to the androgen stimulus during meiosis induces changes in the proteins that are associated with various molecular processes such as apoptosis, cell signaling, oxidative stress and RNA processing [58]. These investigators further validated this observation by immunostaining for oxidized DNA adducts – they showed that spermatocytes that undergo oxidative stress induced DNA damage during androgen suppression. The completion of meiosis requires androgen action via the Sertoli cell [59]. Low levels of testicular androgens support meiosis while a high level is required for spermiogenesis.
Testicular testosterone suppression has been shown to adversely influence antioxidant activity [60]. The present study utilizing Network analysis revealed heat shock protein beta-1 (HSPB1) and clathrin heavy chain 1 (CLTC), which are known to bind and activate androgen receptor were actually underexpressed in the ROS+ group. We also observed that GAPDH levels were overexpressed, and that it was bound with the androgen receptor to impart a stimulatory effect in the ROS+ sample. The mechanism by which the androgen stimulus is transduced by the Sertoli cell to the germ cells is still unclear. In addition, it is well known that in majority of the infertility cases attributed to a male factor, spermatogenic arrest is a common feature. The process of spermatogenesis is dependent on the cAMP responsive element modulator (CREM) signaling pathway in the testis which operates in coordination with the CREM modulators (repressors and activators) [61]. CREM is essential for male fertility and the absence of CREM-dependent transcription in post-meiotic germ cells is known to result in an arrest of spermatid differentiation and apoptosis [62,63]. CREM repressors are expressed during pre-meiotic germ cells while CREM activators are expressed in the post-meiotic germ cells. In order to understand the impact of oxidative stress on impaired spermatogenesis, we examined the CREM signaling in testis. Our findings revealed that three of the identified proteins that were differentially expressed in the ROS+ group were modulated by CREM activators. This included ODF1 and ACE1 proteins that were underexpressed as well LDHC, which was overexpressed. These findings suggest that oxidative stress may affect the switching of genes at the spermatogenic level. Comparative studies on CREM signaling in testis have been conducted on normo- and oligozoospermic men and men with round spermatids vs. round spermatid maturation arrest [64]. These findings further support the fact that a lack of a switch in the expression of CREM gene isoforms may have an adverse impact on spermatogenesis in humans.
To summarize, in this study, we have identified various proteins that are implicated in a multitude of functions associated with response and management of oxidative stress. The increased expression of histone cluster 1H2ba (HIST1H2BA), mitochondrial malate dehydrogenase precursor (MDH2), transglutaminase 4 (TGM4), glutathione peroxidase 4 isoform A precursor (GPX4), glutamine synthetase (GLUL), heat shock proteins (HSP90B1 and HSPA5) in the ROS+ sample suggests that these proteins can serve as potential biomarkers of oxidative stress. Thus alterations reported in our study in men with and without oxidative stress may help explain pathways leading to the altered semen phenotype especially in men exhibiting oxidative stress both in infertile men as well as in infertile men with varicocele. However, the interaction between the proteins and the androgen receptors is still unclear and warrants further investigation. It is important to accurately assess the protein levels and verify that the differentially expressed proteins are indeed involved in the process of oxidative stress and are possibly modified as a result of oxidative stress. In future studies, as a follow-up to the present study, quantitative estimation of the differentially (overexpressed and underexpressed) expressed proteins, utilizing western blotting and antibodies is warranted. The findings of our study provide the groundwork for further testing including the proposition that these newly identified sperm proteins play crucial roles in oxidative stress and the pathophysiology of male infertility.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
RK: participated in the study conception/design, review of the data and writing of the manuscript and final approval; SD: acquisition and preparation of samples for analysis; BW, acquisition of samples for analysis; interpretation of the results; SY: acquisition of the samples, interpretation of the data, discussion o results; BG: contributed to bioinformatic analysis, data interpretation and participated in the paper redaction; AH: drafting of the article; GM participated in the review of the data and writing of the manuscript; AA contributed to the study design, and review of the data. All authors read and approved the final manuscript.
Contributor Information
Rakesh Sharma, Email: sharmar@ccf.org.
Ashok Agarwal, Email: Agarwaa@ccf.org.
Gayatri Mohanty, Email: gayatri_mohanty@yahoo.com.
Alaa J Hamada, Email: ala1977h@yahoo.com.
Banu Gopalan, Email: banugopalan@yahoo.com.
Belinda Willard, Email: willarb@ccf.org.
Satya Yadav, Email: yadavs@ccf.org.
Stefan du Plessis, Email: ssdp@sun.ac.za.
Acknowledgements
This study was supported by Cleveland Clinic, Research Program Committee. Authors wish to thank the Cleveland Clinic Andrology laboratory personnel for their help with scheduling of subjects used in this study. This research project was supported in part by funds from the Cleveland Clinic Research Program Committee and the Center for Reproductive Medicine. Visit of GM supported in part by the INSPIRE fellowship from the Department of Science and Technology, New Delhi, India.
References
- Sharma R, Aaron T, Kothari S, Agarwal A. In: Free Radical Biomedicine. Pantopoulos K, Schipper H, editor. Hauppauge NY: Nova Science Publishers Inc; 2012. A comprehensive overview - Oxidative stress in male reproduction; pp. 305–328. Chapter 13. [Google Scholar]
- Agarwal A, Cocuzza M, Abdelrazik H, Sharma RK. In: Handbook of Chemiluminescenct Methods in Oxidative Stress Assessment. Popov I, Lewin G, editor. Kerala, India: Transworld Research Network; 2009. Oxidative stress measurement in patients with male or female factor infertility; pp. 195–218. [Google Scholar]
- Mahfouz R, Sharma R, Thiyagarajan A, Kale V, Gupta S, Sabanegh E, Agarwal A. Semen characteristics and sperm DNA fragmentation in infertile men with low and high levels of seminal reactive oxygen species. Fertil Steril. 2003;94:2141–2146. doi: 10.1016/j.fertnstert.2009.12.030. [DOI] [PubMed] [Google Scholar]
- Pasqualotto FF, Sharma RK, Nelson DR, Thomas AJ Jr, Agarwal A. Relationship between oxidative stress, semen characteristics, and clinical diagnosis in men undergoing fertility investigation. Fertil Steril. 2000;73:459–464. doi: 10.1016/S0015-0282(99)00567-1. [DOI] [PubMed] [Google Scholar]
- Agarwal A, Makker K, Sharma R. Clinical relevance of oxidative stress in male factor infertility: an update. Am J Reprod Immunol. 2008;59:2–11. doi: 10.1111/j.1600-0897.2007.00559.x. [DOI] [PubMed] [Google Scholar]
- Agarwal A, Sharma RK, Nallella KP, Thomas AJ Jr, Alvarez JG, Sikka SC. Reactive oxygen species as an independent marker of male factor infertility. Fertil Steril. 2006;86:878–885. doi: 10.1016/j.fertnstert.2006.02.111. [DOI] [PubMed] [Google Scholar]
- Sharma RK, Pasqualotto FF, Nelson DR, Thomas AJ Jr, Agarwal A. The reactive oxygen species-total antioxidant capacity score is a new measure of oxidative stress to predict male infertility. Hum Reprod. 1999;14:2801–2807. doi: 10.1093/humrep/14.11.2801. [DOI] [PubMed] [Google Scholar]
- Pasqualotto FF, Sharma RK, Potts JM, Nelson DR, Thomas AJ, Agarwal A. Seminal oxidative stress in patients with chronic prostatitis. Urology. 2000;55:881–885. doi: 10.1016/S0090-4295(99)00613-5. [DOI] [PubMed] [Google Scholar]
- Agarwal A, Prabakaran S, Allamaneni SS. Relationship between oxidative stress, varicocele and infertility: a meta-analysis. Reprod Biomed Online. 2006;12:630–633. doi: 10.1016/S1472-6483(10)61190-X. [DOI] [PubMed] [Google Scholar]
- Agarwal A, Sharma RK, Desai NR, Prabakaran S, Tavares A, Sabanegh E. Role of oxidative stress in pathogenesis of varicocele and infertility. Urology. 2009;73:461–469. doi: 10.1016/j.urology.2008.07.053. [DOI] [PubMed] [Google Scholar]
- Pasqualotto FF, Sundaram A, Sharma RK, Borges E Jr, Pasqualotto EB, Agarwal A. Semen quality and oxidative stress scores in fertile and infertile patients with varicocele. Fertil Steril. 2008;89:602–607. doi: 10.1016/j.fertnstert.2007.03.057. [DOI] [PubMed] [Google Scholar]
- Hamada A, Sharma R, du Plessis S, Willard B, Yadav S, Sabanegh E, Agarwal A. Two dimensional differential in-gel electrophoresis-based proteomics of male gametes in relation to oxidative stress. Fertil Steril. 2013;99:1216–1226. doi: 10.1016/j.fertnstert.2012.11.046. [DOI] [PubMed] [Google Scholar]
- Vazquez L, Mónica H. Proteomic analysis and sperm physiopathology: the two-dimensional difference in gel electrophoresis approach. Fertil Steril. 2013;99:1199–1200. doi: 10.1016/j.fertnstert.2012.12.051. [DOI] [PubMed] [Google Scholar]
- de Mateo S, Estanyol JM, Oliva R. Methods for the analysis of the sperm proteome. Methods Mol Biol. 2013;927:411–422. doi: 10.1007/978-1-62703-038-0_35. [DOI] [PubMed] [Google Scholar]
- de Mateo S, Martínez-Heredia J, Estanyol JM, Domínguez-Fandos D, Vidal-Taboada JM, Ballescà JL, Oliva R. Marked correlations in protein expression identified by proteomic analysis of human spermatozoa. Proteomics. 2007;7:4264–4277. doi: 10.1002/pmic.200700521. [DOI] [PubMed] [Google Scholar]
- Oliva R, De Mateo S, Castillo J, Azpiazu R, Oriola J, Ballescà JL. Methodological advances in sperm proteomics. Hum Fertil. 2010;13:263–267. doi: 10.3109/14647273.2010.516877. [DOI] [PubMed] [Google Scholar]
- Baker MA. The ’omics revolution and our understanding of sperm cell biology. Asian J Androl. 2011;13:6–10. doi: 10.1038/aja.2010.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindaraju A, Dogan S, Rodriguez-Osorio N, Grant K, Kaya A, Memili E. Delivering value from sperm proteomics for fertility. Cell Tissue Res. 2012;349:783–793. doi: 10.1007/s00441-012-1452-2. [DOI] [PubMed] [Google Scholar]
- Baker MA, Nixon B, Naumovski N, Aitken RJ. Proteomic insights into the maturation and capacitation of mammalian spermatozoa. Syst Biol Reprod Med. 2012;58:211–217. doi: 10.3109/19396368.2011.639844. [DOI] [PubMed] [Google Scholar]
- Parte PP, Rao P, Redij S, Lobo V, D’Souza SJ, Gajbhiye R, Kulkarni V. Sperm phosphoproteome profiling by ultra performance liquid chromatography followed by data independent analysis (LC-MS(E)) reveals altered proteomic signatures in asthenozoospermia. J Proteomics. 2012;75:5861–5871. doi: 10.1016/j.jprot.2012.07.003. [DOI] [PubMed] [Google Scholar]
- Baker MA, Naumovski N, Hetherington L, Weinberg A, Velkov T, Aitken RJ. Head and flagella subcompartmental proteomic analysis of human spermatozoa. Proteomics. 2013;13:61–74. doi: 10.1002/pmic.201200350. [DOI] [PubMed] [Google Scholar]
- World Health Organization. WHO Laboratory Manual for the Examination and Processing of Human Semen. 5th ed. 5. Cambridge: World Health Organization; 2010. [Google Scholar]
- Benjamin D, Sharma RK, Moazzam A, Agarwal A. In: Studies on Men’s Health and Fertility. Agarwal A, Aitken RJ, Alvarez JG, editor. New York, NY: Springer Science+Business Media; 2012. Methods for the detection of ROS in human sperm samples; pp. 257–273. [Google Scholar]
- Boyle E, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G. GO: TermFinder-open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GO term mapper. http://go.princeton.edu/cgi-bin/GOTermMapper.
- The UniProt Consortium. Reorganizing the protein space at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2011;40:D71–D75. doi: 10.1093/nar/gkr981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia VN, Perlman DH, Costello CE, McComb ME. Software tool for researching annotations of proteins: open-source protein annotation software with data visualization. Anal Chem. 2009;81:9819–9823. doi: 10.1021/ac901335x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge CL, Haase J, Janes J, Huss JW 3rd, Su AI. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingenuity Pathway Analysis (IPA) from Ingenuity® Systems Website Title: http://www.ingenuity.com23813567
- Metacore™ from GeneGo Inc Website: http://www.genego.com23826137
- Martínez-Heredia J, de Mateo S, Vidal-Taboada JM, Ballescà JL, Oliva R. Identification of proteomic differences in asthenozoospermic sperm samples. Hum Reprod. 2008;23:783–791. doi: 10.1093/humrep/den024. [DOI] [PubMed] [Google Scholar]
- de Yebra L, Ballescá JL, Vanrell JA, Corzett M, Balhorn R, Oliva R. Detection of P2 precursors in the sperm cells of infertile patients who have reduced protamine P2 levels. Fertil Steril. 1998;69:755–759. doi: 10.1016/S0015-0282(98)00012-0. [DOI] [PubMed] [Google Scholar]
- Zhao C, Huo R, Wang FQ, Lin M, Zhou ZM, Sha JH. Identification of several proteins involved in regulation of sperm motility by proteomic analysis. Fertil Steril. 2007;87:436–438. doi: 10.1016/j.fertnstert.2006.06.057. [DOI] [PubMed] [Google Scholar]
- Rajeev SK, Reddy KV. Sperm membrane protein profiles of fertile and infertile men: identification and characterization of fertility-associated sperm antigen. Hum Reprod. 2004;19:234–242. doi: 10.1093/humrep/deh066. [DOI] [PubMed] [Google Scholar]
- Xu W, Hu H, Wang Z, Chen X, Yang F, Zhu Z, Fang P, Dai J, Wang L, Shi H, Li Z, Qiao Z. Proteomic characteristics of spermatozoa in normozoospermic patients with infertility. J Proteomics. 2012;75:5426–5436. doi: 10.1016/j.jprot.2012.06.021. [DOI] [PubMed] [Google Scholar]
- Nakamura N, Mori C, Eddy EM. Molecular complex of three testis-specific isozymes associated with the mouse sperm fibrous sheath: hexokinase 1, phosphofructokinase M, and glutathione S-transferase mu class 5. Biol Reprod. 2010;82:504–515. doi: 10.1095/biolreprod.109.080580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento JM, Shi LZ, Tam J, Chandsawangbhuwana C, Durrant B, Botvinick EL, Berns MW. Comparison of glycolysis and oxidative phosphorylation as energy sources for mammalian sperm motility, using the combination of fluorescence imaging, laser tweezers, and real-time automated tracking and trapping. J Cell Physiol. 2008;217:745–751. doi: 10.1002/jcp.21549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey BT. Mammalian sperm metabolism: oxygen and sugar, friend and foe. Int J Dev Biol. 2008;52:427–437. doi: 10.1387/ijdb.072522bs. [DOI] [PubMed] [Google Scholar]
- Terrell KA, Wildt DE, Anthony NM, Bavister BD, Leibo SP, Penfold LM, Marker LL, Crosier AE. Oxidative phosphorylation is essential for felid sperm function, but is substantially lower in cheetah (Acinonyx jubatus) compared to domestic cat (Felis catus) ejaculate. Biol Reprod. 2011;85:473–481. doi: 10.1095/biolreprod.111.092106. [DOI] [PubMed] [Google Scholar]
- Elkina YL, Atroshchenko MM, Bragina EE, Muronetz VI, Schmalhausen EV. Oxidation of glyceraldehyde-3-phosphate dehydrogenase decreases sperm motility. Biochemistry. 2011;76:268–272. doi: 10.1134/s0006297911020143. [DOI] [PubMed] [Google Scholar]
- Sharma R, Agarwal A. In: Human sperm chromatin: Structure and Function. Sperm Chromatin. Biological and Clinical Applications in Male Infertility and Assisted Reproduction. Zini A, Agarwal A, editor. New York: Springer Science + Business Media; 2012. Spermatogenesis: An overview; pp. 19–44. [Google Scholar]
- Lilja H, Abrahamsson PA, Lundwall A. Semenogelin, the predominant protein in human semen. Primary structure and identification of closely related proteins in the male accessory sex glands and on the spermatozoa. J Biol Chem. 1989;264:1894–1900. [PubMed] [Google Scholar]
- Lilja H, Lundwall A. Molecular cloning of epididymal and seminal vesicular transcripts encoding a semenogelin-related protein. Proc Natl Acad Sci USA. 1992;89:4559–4563. doi: 10.1073/pnas.89.10.4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lamirande E, Yoshida K, Yoshiike TM, Iwamoto T, Gagnon C. Semenogelin, the main protein of semen coagulum, inhibits human sperm capacitation by interfering with the superoxide anion generated during this process. J Androl. 2001;22:672–679. [PubMed] [Google Scholar]
- Auger J, Eustache F, Maceiras P, Broussard C, Chafey P, Lesaffre C, Vaiman D, Camoin L, Auer J. Modified expression of several sperm proteins after chronic exposure to the antiandrogenic compound vinclozolin. Toxicol Sci. 2010;117:475–484. doi: 10.1093/toxsci/kfq199. [DOI] [PubMed] [Google Scholar]
- Feder ME, Hofmann GE. Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu Rev Physiol. 1999;61:243–282. doi: 10.1146/annurev.physiol.61.1.243. [DOI] [PubMed] [Google Scholar]
- Katschinski DM. On heat and cells and proteins. News Physiol Sci. 2004;19:11–15. doi: 10.1152/nips.01403.2002. [DOI] [PubMed] [Google Scholar]
- Sung DY, Guy CL. Physiological and molecular assessment of altered expression of Hsc70-1 in Arabidopsis. Evidence for pleiotropic consequences. Plant Physiol. 2003;132:979–987. doi: 10.1104/pp.102.019398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folk JE. Transglutaminases. Annu Rev Biochem. 1980;49:517–531. doi: 10.1146/annurev.bi.49.070180.002505. [DOI] [PubMed] [Google Scholar]
- Lorand L, Conrad SM. Transglutaminases. Mol Cell Biochem. 1984;58:9–35. doi: 10.1007/BF00240602. [DOI] [PubMed] [Google Scholar]
- Shin DM, Jeon JH, Kim CW, Cho SY, Lee HJ, Jang GY, Jeong EM, Lee DS, Kang JH, Melino G, Park SC, Kim IG. TGF beta mediates activation of transglutaminase 2 in response to oxidative stress that leads to protein aggregation. FASEB J. 2008;22:2498–2507. doi: 10.1096/fj.07-095455. [DOI] [PubMed] [Google Scholar]
- Li Y, Okumura K, Nomura S, Maeda N, Miyasho T, Yokota H. Oxidatively damaged proteins in the early stage of testicular toxicities in male rats by orally administered with a synthetic oestrogen, diethylstilbestrol. Reprod Toxicol. 2011;31:26–34. doi: 10.1016/j.reprotox.2010.09.007. [DOI] [PubMed] [Google Scholar]
- Immenschuh S, Baumgart-Vogt E, Tan M, Iwahara S, Ramadori G, Fahimi HD. Differential cellular and subcellular localization of heme-binding protein 23/peroxiredoxin I and heme oxygenase-1 in rat liver. J Histochem Cytochem. 2003;51:1621–1631. doi: 10.1177/002215540305101206. [DOI] [PubMed] [Google Scholar]
- Wood ZA, Poole LB, Karplus PA. Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science. 2003;300:650–653. doi: 10.1126/science.1080405. [DOI] [PubMed] [Google Scholar]
- O’Flaherty C, de Souza AR. Hydrogen peroxide modifies human sperm peroxiredoxins in a dose-dependent manner. Biol Reprod. 2011;84:238–247. doi: 10.1095/biolreprod.110.085712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Gabriel MC, Zini A, Chan P, O’Flaherty C. Low amounts and high thiol oxidation of peroxiredoxins in spermatozoa from infertile men. J Androl. 2012;33:1342–1351. doi: 10.2164/jandrol.111.016162. [DOI] [PubMed] [Google Scholar]
- Rey RA, Musse M, Venara M, Chemes HE. Ontogeny of the androgen receptor expression in the fetal and postnatal testis: its relevance on Sertoli cell maturation and the onset of adult spermatogenesis. Microsc Res Tech. 2009;72:787–795. doi: 10.1002/jemt.20754. [DOI] [PubMed] [Google Scholar]
- Stanton PG, Sluka P, Foo CF, Stephens AN, Smith AI, McLachlan RI, O’Donnell L. Proteomic changes in rat spermatogenesis in response to in vivo androgen manipulation; impact on meiotic cells. PLoS One. 2012;7:e41718. doi: 10.1371/journal.pone.0041718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RS, Yeh S, Tzeng CR, Chang C. Androgen receptor roles in spermatogenesis and fertility: lessons from testicular cell-specific androgen receptor knockout mice. Endocr Rev. 2009;30:119–132. doi: 10.1210/er.2008-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J, O’Neill C, Handelsman DJ. Induction of spermatogenesis by androgens in gonadotropin-deficient (hpg) mice. Endocrinology. 1995;136:5311–5321. doi: 10.1210/en.136.12.5311. [DOI] [PubMed] [Google Scholar]
- Tamai KT, Monaco L, Nantel F, Zazopoulos E, Sassone-Corsi P. Coupling signalling pathways to transcriptional control: nuclear factors responsive to cAMP. Recent Prog Horm Res. 1997;52:121–139. discussion 139-40. [PubMed] [Google Scholar]
- Hogeveen KN, Sassone-Corsi P. Regulation of gene expression in post-meiotic male germ cells: CREM-signalling pathways and male fertility. Hum Fertil. 2006;9:73–79. doi: 10.1080/14647270500463400. [DOI] [PubMed] [Google Scholar]
- Monaco L, Kotaja N, Fienga G, Hogeveen K, Kolthur US, Kimmins S, Brancorsini S, Macho B, Sassone-Corsi P. Specialized rules of gene transcription in male germ cells: the CREM paradigm. Int J Androl. 2004;27:322–327. doi: 10.1111/j.1365-2605.2004.00494.x. [DOI] [PubMed] [Google Scholar]
- Weinbauer GF, Behr R, Bergmann M, Nieschlag E. Testicular cAMP responsive element modulator (CREM) protein is expressed in round spermatids but is absent or reduced in men with round spermatid maturation arrest. Mol Hum Reprod. 1998;4:9–15. doi: 10.1093/molehr/4.1.9. [DOI] [PubMed] [Google Scholar]