Abstract
Background
Identification of cancer stem cells (CSCs) and their behaviors will provide insightful information for the future control of human cancers. This study investigated CD44 and CD24 cell surface markers as breast cancer CSC markers in vitro and in vivo.
Methods
Flow cytometry with CD44 and CD24 markers was used to sort breast cancer MCF7 cells for scanning electron microscopy (SEM), tumor cell invasion assay, and nude mouse xenograft assay.
Results
Flow cytometry assay using CD44 and CD24 markers sorted MCF7 cells into four subsets, i.e., CD44+/CD24-/low, CD44-/CD24+, CD44+/CD24+, and CD44-/CD24-. The SEM data showed that there were many protrusions on the surface of CD44+/CD24-/low cells. CD44+/CD24-/low cells had many microvilli and pseudopodia. The CD44+/CD24-/low cells had a higher migration and invasion abilities than that of the other three subsets of the cells. The in vivo tumor formation assay revealed that CD44+/CD24- cells had the highest tumorigenic capacity compared to the other three subsets.
Conclusion
CD44 and CD24 could be useful markers for identification of breast CSCs because CD44+/CD24-/low cells had unique surface ultrastructures and the highest tumorigenicity and invasive abilities.
Keywords: Breast cancer stem cells, Ultrastructure, Tumorigenicity, Invasive ability
Background
Cancer stem cells (CSCs) were originally described in hematologic malignancies and have the exclusive ability to self-renew and differentiate into heterogeneous lineages of tumor cells [1,2]. The concept of CSC theory may change our perspective with regard to cancer development, progression, and treatment. CSCs are defined as a small number of tumor cells that have the ability to generate daughter tumor cells, self-renew, and maintain tumor phenotypes. In addition, CSCs are responsible for tumor resistance to chemo- and radiation therapy, recurrence, and metastasis [3-6]. Currently, searching for CSC markers is a very hot topic and some interesting data have been generated. For example, markers for CSCs of the breast, prostate, colon, and pancreas have been reported [7-9], but more work needs to be done. Furthermore, it is evident that the distinguished ultrastructure of cancer cells is linked to the tumorigenic potential for making tumor diagnoses and predicting tumor behaviors (such as recurrence, metastasis, and so on) [10-12]; however, it is unknown whether CSCs possess their own particular ultrastructure partners. Therefore, identification of CSCs and understanding of their behaviors in human cancer will provide us useful information for defining the molecular mechanisms of cancer development and progression and developing novel treatment strategies.
In human breast cancer, it has been shown that breast CSCs have a CD44+/CD24- phenotype [2]. Consistent with this finding, knockdown of CD44 expression has been found to induce breast CSCs to differentiate into regular tumor cells without the capacity to self-renew (defined as non-CSCs) [13]. Nevertheless, in certain breast cancer patients, CD44+/CD24- cells are still sensitive to radiotherapy, suggesting that not all breast CSCs are radioresistant [14] and that not all CD44+/CD24- cells are CSCs. Indeed, clinical data have indicated that there is no significant correlation between CD44+/CD24-/low tumor cell prevalence and tumor progression. The prevalence of a CD44+/CD24-/low tumor cell did not correlate with the event-free and overall survival and did not affect the tumor response to different treatment modalities [15,16]. Thus, further investigation is needed to clarify these two markers for breast CSCs. In this study, we first sorted breast cancer cells with these two markers and then analyzed the ultrastructure of the breast CSC surface in CD44+/CD24-/low genotypes. Furthermore, we systemically analyzed the ultrasturcture, invasion capacity, and tumor formation in breast cancer MCF7 cells with different CD44/CD24 genotypes. Our findings indicated that CD44+/CD24-/low MCF7 cells with high numbers of microvilli and pseudopodia have greater tumorigenicity rates and invasive ability.
Materials and methods
Cell line and culture
The human breast cancer cell line MCF7 was obtained from the Shanghai Cell Bank (Shanghai, China) and maintained in Dulbecco’s modified Eagle’s medium (DMEM) with high glucose and supplemented with 10% fetal calf serum (FCS), 100 units/ml penicillin, and 100 μg/ml streptomycin at 37°C in 5% CO2.
Flow cytometry
To sort tumor cells with CD44 and CD24 markers, MCF7 cells were seeded and grown for several days, then washed with phosphate-buffered saline (PBS), and finally harvested with 0.05% trypsin/0.025% EDTA. The cells were then detached and washed with PBS containing 2% FCS before being subjected to antibody binding, i.e., combinations of fluorochrome-conjugated monoclonal antibodies against human CD44 (FITC) and CD24 (PE) or their respective isotype controls were added to the cell suspensions at a concentration recommended by the manufacturer and incubated on ice in the dark for 20 min. After that, cells were washed twice with PBS/2% FCS and resuspended in 0.5 ml (per million cells) of PBS/2% FBS for flow cytometric analysis using a FACSVantage instrument. Anti-human CD44 FITC (Clone: IM7; 11-0441-82) and anti-human CD24 PE (Clone: eBioSN3; 12-0247-42) antibodies were obtained from Affymetrix eBioscience (http://www.affymetrix.com).
Scanning electron microscopy (SEM)
To detect the ultrastructure of CD44+/CD24-/low breast CSCs, we performed SEM experiments by directly using flow cytometry-sorted cells. Briefly, flow cytometry-sorted cells were seeded onto glass cover slips and fixed in situ with 2.5% paraformaldehyde in PBS for 2 h at 4°C. Then, the cells were rinsed in PBS and postfixed with 1% osmium tetroxide in PBS for 1 h, dehydrated through a graded ethanol series, stained en bloc during dehydration with a saturated solution of uranyl acetate in 70% ethanol, and then embedded in Araldite for the ultrastructural study of cells using SEM.
Tumor cell invasion assay
Breast cancer MCF7 cells were cultured in complete cell medium and then stained with anti-CD44-FITC and anti-CD24-PE antibodies (Miltenyi Biotec, Germany). After that, the cells were washed twice with cold PBS and then stained with DAPI before being washed twice with PBS. Next, these cells were first cultured in serum-free DMEM overnight. The next day, the cells were detached with trypsin, counted, and added into the top chambers of Transwell inserts with an 8-μm pore size filter coated with Matrigel in 24-well plates. In the bottom of the chambers, DMEM containing 20% FCS was added, and the cells were then cultured for 24 h. At the end of the experiments, the cells on the top surface of the filter were removed by using a cotton-swab, and the cells on the bottom of the filter were fixed with methyl alcohol and then reviewed and quantified by a laser scanning confocal microscope (FV-1000; Olympus Japan).
Animal experiments
Flow cytometry-sorted cells were collected, washed in PBS, and then injected into the mammary fat pad of 5-week-old severe combined immunodeficient (SCID) mice. Mice were maintained in laminar flow rooms under constant temperature and humidity and received estradiol supplementation (0.4 mg/kg) every week after cell injection. Mice were inspected for tumor appearance daily by observation and palpation for 12 weeks after cell injection. At the end of the experiments, all mice were sacrificed by cervical dislocation, and the presence of each tumor nodule was confirmed by necropsy. Experimental protocols were approved by the Ethics Committee for Animal Experimentation of the institute.
Statistical analyses
All the in vitro experiments were repeated six times, and had similar results. Statistical analysis was performed using SPSS software version 17.0. Statistical significance was tested by using a two-tailed Student’s t-test. P<0.05 was considered statistically significant.
Results
Sorting breast cancer MCF7 cells using CD44 and CD24 markers
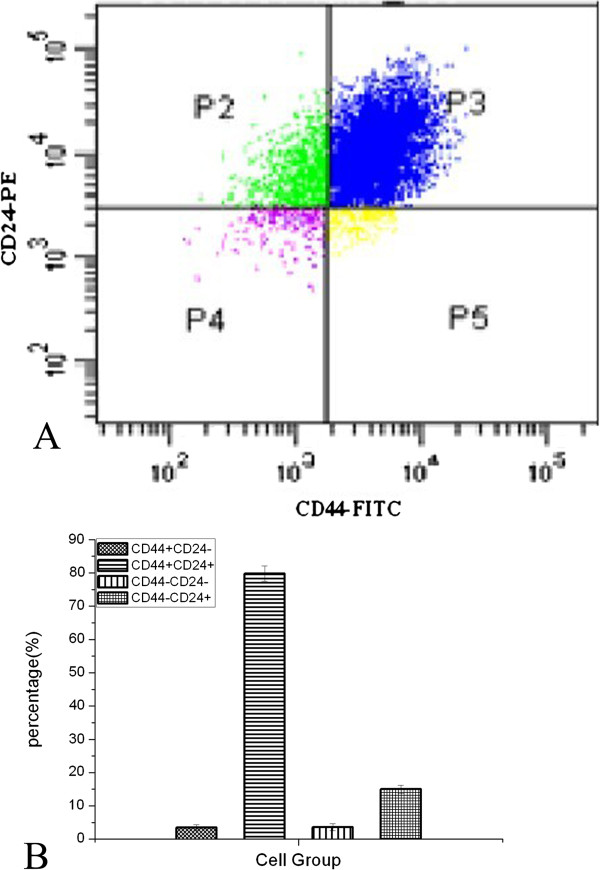
To obtain putative breast CSCs from MCF7 cell line, we performed flow cytometry assay using antibodies against CD44 and CD24 cell surface markers. We obtained four subtypes of tumor cells (i.e., CD44+/CD24-/low, CD44+/CD24+, CD44-/CD24+, and CD44-/CD24-; Figure 1). The percentages of CD44+/CD24-/low, CD44+/CD24+, CD44-/CD24+, and CD44-/CD24- cells were 3.5%, 79.8%, 15.0%, and 3.6%, respectively (Figure 1). According to the literature, the CD44+/CD24-/low cells were breast CSCs (approximately 3.5%; Figure 1). The purity of this sorted cell type was further confirmed using flow cytometry. As shown in Figure 1C, the purity of the CD44+/CD24-/low cell type was more than 90%.
Figure 1.
Flow cytometry sorting of MCF7 cells using CD44 and CD24 markers. A, MCF7 cells were analyzed by fluorescence-activated cell sorting (FACS) using anti-CD44 and anti-CD24 antibodies. B, Quantification of the four subsets of MCF7 cells. C, Confirmation of the purity of the CD44+/CD24-/low cell type using flow cytometry.
Detection of the cell surface ultrastructure of flow cytometry-sorted MCF7 cells
To characterize the ultrastructural features of flow cytometry-sorted MCF7 cells, we performed SEM experiments. Our data showed that there are numerous protrusions on the surface of CD44+/CD24-/low cells (Figure 2A). These cells also showed many microvilli and pseudopodia compared to the other three cell subsets (Figure 2). In contrast, CD44+/CD24+ cells showed a rough surface with several but less abundant protrusions and fewer pseudopodia on the surface compared to CD44+/CD24-/low cells (Figure 2B). Again, CD44-/CD24+ cells showed a smooth surface with fewer protrusions and pseudopodia (Figure 2C), while CD44-/CD24- cells showed a smooth surface with fewer protrusions and no pseudopodia (Figure 2D). These data indicate that the cell surface ultrastructure is quite different among these four subtypes of MCF7 cells.
Figure 2.
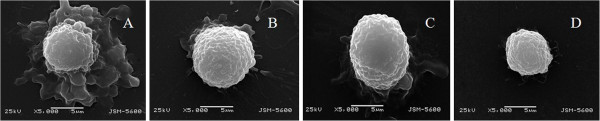
Cell surface ultrastructure of the four different subsets of MCF7 cells. A, CD44+/CD24-/low; B, CD44+/CD24+; C, CD44-/CD24+; and D, CD44-/CD24-.
Invasion capacity of breast cancer MCF7 cells with different expressions of CD44 and CD24 markers
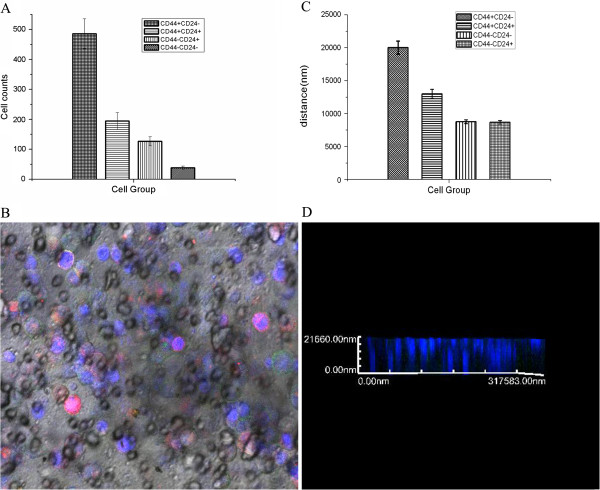
To examine the invasion capacity of these four different subsets of MCF7 cells, we first labeled them with different colors (Green: CD44+/CD24-/low; Red: CD44-/CD24+, Yellow: CD44+/CD24+; and Blue: CD44-/CD24-). A laser confocal microscope randomly selected five fields (×400) to count the number of these four subsets of cells that invaded through the basement membrane (Figure 3). The numbers of CD44+/CD24-/low, CD44+/CD24+, CD44-/CD24+, and CD44-/CD24- cells that invaded through the basement membrane were 485.7 ± 43.628, 194.4 ± 27.778, 126.7 ± 13.333, and 38.0 ± 3.189, respectively (Figure 3A). The number of CD44+/CD24-/low cells that invaded through the basement membrane was significantly higher than that of the other three subsets (Figure 3B). The migration distances of CD44+/CD24-/low, CD44+/CD24+, CD44-/CD24+, and CD44-/CD24- cells were 20011.0 ± 999.476 nm, 12996.3 ± 699.028 nm, 8664.5 ± 157.124 nm, and 8764.6 ± 99.620 nm, respectively (Table 1, Figure 3C, D). Thus, the migration distance of CD44+/CD24-/low was significantly longer than that of the other three subsets (Table 2, Figure 3C, D). These results suggest that CD44+/CD24-/low cells have the highest invasion capacity compared to the other three subsets of MCF7 cells.
Figure 3.
Invasion assay of these four subsets of MCF7 cells. A, Cell number quantification of invasion. B, Confocal images of these four subsets of MCF7 cells (Green: CD44+/CD24-/low; Red: CD44-/CD24+; Yellow: CD44+/CD24+; and Blue: CD44-/CD24-). C, Quantification of tumor cell migration distance. D, Images of cell migration distance.
Table 1.
Invasion capacity of these different MCF7 subtypes
| Cell subtypes | Invasion cell number | n | t | p |
|---|---|---|---|---|
| CD44-/CD24+ |
126.7 ± 13.33 |
6 |
5.77 |
0.004 |
| CD44+/CD24+ |
38.0 ± 3.18 |
6 |
20.69 |
0.000 |
| CD44-/CD24- |
194.4 ± 27.77 |
6 |
4.21 |
0.013 |
| CD44+/CD24- | 485.7 ± 43.62 | 6 | — | — |
Table 2.
Migration distances of these different MCF7 subtypes
| Cell subtypes | Migration distances | n | t | p |
|---|---|---|---|---|
| CD44-/CD24+ |
8664.3 ± 157.12 |
6 |
−19.45 |
0.000 |
| CD44+/CD24+ |
12996.2 ± 699.02 |
6 |
7.25 |
0.002 |
| CD44-/CD24- |
8764.0 ± 99.62 |
6 |
−9.80 |
0.001— |
| CD44+/CD24- | 20011.0 ± 999.47 | 6 | — |
Xenograft formation and growth capacity among these four subtypes of CD44 and CD24-expressing breast cancer cells in nude mice
To compare the tumor-initiating capability of different MCF7 cell subsets, we injected the four flow cytometry-sorted cell subsets at different cell numbers (1×103, 1×104, and 1×105) into the mammary left fat pad of nonobese diabetic (NOD)/SCID mice. All mice were observed for 3 months for tumor formation and growth (Table 3 and Figure 4). Briefly, 13 days after the injection of 1×103 CD44+/CD24-/low cells, a palpable tumor formed in one of six mice. Meanwhile, after the injection of 1×104 CD44+/CD24-/low cells, tumors formed in four of six mice at 11 ± 1.2 days. Furthermore, an injection of 1×105 CD44+/CD24-/low cells formed tumors in all six mice at 9 ± 2.3 days. In contrast, of the other three subsets of MCF7 cells, even with an injection of 1×105 cells, only CD44+/CD24+ cells formed tumors in two of six mice at day 21 and none of the CD44-/CD24- and CD44-/CD24+ cells were able to form tumors. These data suggest that CD44+/CD24- cells have the highest tumorigenic capacity compared to the other three cell subsets.
Table 3.
In vivo tumor xenograft assay of these four subsets of MCF7 cells
|
MCF7 cell subset |
Tumor formation with the indicated number of cells |
||
|---|---|---|---|
| 1×103 | 1×104 | 1×105 | |
| CD44+/CD24-/low |
1/6 |
4/6 |
6/6 |
| CD44-/CD24+ |
0/6 |
0/6 |
0/6 |
| CD44+/CD24+ |
0/6 |
0/6 |
2/6 |
| CD44-/CD24- | 0/6 | 0/6 | 0/6 |
Figure 4.
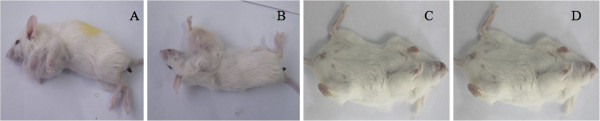
In vivo tumor xenograft assay of these four subsets MCF7 cells. A, CD44+/CD24-/low; B, CD44+/CD24+; C, CD44-/CD24+; and D, CD44-/CD24-.
Discussion
Breast CSCs, like all other CSCs, have been suggested to contribute to tumor development and progression [17,18]. Thus, identification of breast CSCs is very important not only for understanding the mechanism of breast cancer but also providing novel strategies for its successful treatment. Previous studies have used specific molecular markers and ultrastructures to identify and isolate breast CSCs [19-22]. It is well known that the surface ultrastructure of cancer cells is unique and important for cancer development [23,24], since the surface ultrastructure of cells can predict cell-cell and cell-matrix contact, movability, and adhesion ability. However, to date, information regarding the cell surface ultrastructure of breast CSCs is limited. Herein, in this study, we analyzed the surface ultrastructure of different subsets of MCF7 cells after sorting by the known breast CSC markers CD44 and CD24. We found that the CD44+/CD24-/low breast cancer cells showed numerous protrusions on the cell surface and had many microvilli and pseudopodia, indicating such ultrastructures may contribute to higher tumorigenesis and invasion ability. The association between surface marker and surface ultrastructure of CSCs is quite interesting, i.e., CSCs might have a special surface ultrastructure and provide valuable information to identify CSCs and biological significance. Further understanding of these features could help us to develop a novel clinical cancer-targeted therapy. However, at the current stage, due to lack of standard CSC surface ultrastructure data, it is difficult to quantify the CSCs pseudopodia. We will develop a methodology to quantify them in the near future.
The CD44 antigen, encoded by the CD44 gene on chromosome 11, is a cell-surface glycoprotein involved in cell–cell interactions, cell adhesion, and migration [25]. CD44 has been reported as a cell surface marker for some breast and prostate CSCs, but it is associated with an increased survival duration of epithelial ovarian cancer patients [5,26]. Likewise, CD24 is encoded by the CD24 gene and is also a cell adhesion molecule [27]. The expression of CD24 is associated with tumor development and plays a critical role in various cancer metastases [28,29]. However, more recently, researchers in the field of CSCs have used these two proteins as markers to identify breast CSCs with high CD44 expression and negative or low CD24 expression [2,3]. In this study, we also sorted breast cancer MCF7 cells using these two markers to obtain four subpopulations of MCF7 cells. Consistent with previous studies, we found that the CD44+/CD24-/low population had high tumorigenic potential and invasion capacity.
Because in vivo tumor formation is the gold standard for identification of CSCs [30], our current data further confirmed that CD44+/CD24- cells have the highest tumorigenic capacity compared to the other three subsets of MCF7 cells; however, the tumorigenic capacity of CD44+/CD24- cells was not as strong as that found in a previous report [2]. In our current study, only one of six mice formed a breast cancer xenograft with 1×103 cells/injection. However, our current data also showed that CD44+/CD24+ cells formed tumor xenografts in two of six mice after the injection of 1×105 cells. This finding indicates that CD24 is not as good as CD44 as a marker for the identification of breast CSCs. We further analyzed the tumor weight and size in different groups; however, we did not observe any significant differences among them, indicating that CSCs might contribute to cancer initiation but not tumor progression.
Conclusion
In summary, this study systematically investigated the ability of CD44 and CD24 proteins as markers to identify breast CSCs among MCF7 cells using flow cytometry to sort the MCF7 cells with these two proteins into four subsets. Moreoever, CD44+/CD24- cells showed a unique surface ultrastructure by SEM and had the highest tumorigenic potential in tumor cell invasion and nude mouse xenograft assays. Further studies will determine whether CD44 directly contributes to the unique surface ultrastructure and tumorigenic capacity.
Abbreviations
CSCs: Cancer stem cells; SEM: Scanning electron microscopy; DMEM: Dulbecco’s modified eagle’s medium; SCID: Severe combined immunodeficient; FCS: Fetal calf serum; PBS: Phosphate-buffered saline.
Competing interest
The authors declare that they have no competing interest.
Authors’ contributions
WH, YC, YY, and TW designed research; WH, YC, HZ, and TW performed research; WH, YC, YY, and HZ analyzed data; WH, and YW wrote the paper. All authors read and approved the final manuscript.
Contributor Information
Wenxing Yan, Email: kubi213@163.com.
Yubing Chen, Email: yb0707@163.com.
Yueliang Yao, Email: yaoyueling@163.com.
Hongmei Zhang, Email: 420293417@qq.com.
Tiejun Wang, Email: chinawtj218@163.com.
References
- Al-Hajj M, Becker MW, Wicha M, Weissman I, Clarke MF. Therapeutic implications of cancer stem cells. Curr Opin Genet Dev. 2004;14:43–47. doi: 10.1016/j.gde.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charafe-Jauffret E, Ginestier C, Iovino F, Wicinski J, Cervera N, Finetti P, Hur MH, Diebel ME, Monville F, Dutcher J, Brown M, Viens P, Xerri L, Bertucci F, Stassi G, Dontu G, Birnbaum D, Wicha MS. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res. 2009;69:1302–1313. doi: 10.1158/0008-5472.CAN-08-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Tiede B, Kang Y, Massagué J. Beyond tumorigenesis: cancer stem cells in metastasis. Cell Res. 2007;17:3–14. doi: 10.1038/sj.cr.7310118. [DOI] [PubMed] [Google Scholar]
- Velasco-Velázquez MA, Popov VM, Lisanti MP, Pestell RG. The role of breast cancer stem cells in metastasis and therapeutic implications. Am J Pathol. 2011;179:2–11. doi: 10.1016/j.ajpath.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5:275–284. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- O'Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- Zhang S, Balch C, Chan MW, Lai HC, Matei D, Schilder JM, Yan PS, Huang TH, Nephew KP. Identification and characterization of ovarian cancer-initiating cells from primary human tumors. Cancer Res. 2008;68:4311–4320. doi: 10.1158/0008-5472.CAN-08-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya S, Li F. Electron microscopic findings for diagnosis of breast lesions. Med Mol Morphol. 2005;38:216–224. doi: 10.1007/s00795-005-0300-9. [DOI] [PubMed] [Google Scholar]
- Zhao YD, Huang Q, Zhang TY, Dong J, Wang AD, Ding F, Lan Q, Gu XS, Qin ZH. [Ultrastructural analysis of glioma stem cells-progenitors] Zhonghua Zhong Liu Za Zhi. 2008;30:663–667. [PubMed] [Google Scholar]
- Paniz Mondolfi AE, Slova D, Fan W, Attiyeh FF, Afthinos J, Reidy J, Pang Y, Theise ND. Mixed adenoneuroendocrine carcinoma (MANEC) of the gallbladder: a possible stem cell tumor? Pathol Int. 2011;61:608–614. doi: 10.1111/j.1440-1827.2011.02709.x. [DOI] [PubMed] [Google Scholar]
- Pham PV, Phan NL, Nguyen NT, Truong NH, Duong TT, Le DV, Truong KD, Phan NK. Differentiation of breast cancer stem cells by knockdown of CD44: promising differentiation therapy. J Transl Med. 2011;9:209. doi: 10.1186/1479-5876-9-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielske SP, Spalding AC, Wicha MS, Lawrence TS. Ablation of breast cancer stem cells with radiation. Transl Oncol. 2011;4:227–233. doi: 10.1593/tlo.10247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipitsin M, Campbell LL, Argani P, Weremowicz S, Bloushtain-Qimron N, Yao J, Nikolskaya T, Serebryiskaya T, Beroukhim R, Hu M, Halushka MK, Sukumar S, Parker LM, Anderson KS, Harris LN, Garber JE, Richardson AL, Schnitt SJ, Nikolsky Y, Gelman RS, Polyak K. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;11:259–273. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- Abraham BK, Fritz P, McClellan M, Hauptvogel P, Athelogou M, Brauch H. Prevalence of CD44+/CD24-/low cells in breast cancer may not be associated with clinical outcome but may favor distant metastasis. Clin Cancer Res. 2005;11:1154–1159. [PubMed] [Google Scholar]
- Wicha MS. Cancer stem cell heterogeneity in hereditary breast cancer. Breast Cancer Res. 2008;10:105. doi: 10.1186/bcr1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Lee HE, Li H, Shipitsin M, Gelman R, Polyak K. Heterogeneity for stem cell-related markers according to tumor subtype and histologic stage in breast cancer. Clin Cancer Res. 2010;16:876–887. doi: 10.1158/1078-0432.CCR-09-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, Jacquemier J, Viens P, Kleer CG, Liu S, Schott A, Hayes D, Birnbaum D, Wicha MS, Dontu G. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GH, Chepko G. Mammary epithelial stem cells. Microsc Res Tech. 2001;52:190–203. doi: 10.1002/1097-0029(20010115)52:2<190::AID-JEMT1005>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Chepko G, Dickson RB. Ultrastructure of the putative stem cell niche in rat mammary epithelium. Tissue Cell. 2003;35:83–93. doi: 10.1016/S0040-8166(02)00107-6. [DOI] [PubMed] [Google Scholar]
- Pierce GB, Nakane PK, Martinez-Hernandez A, Ward JM. Ultrastructural comparison of differentiation of stem cells of murine adenocarcinomas of colon and breast with their normal counterparts. J Natl Cancer Inst. 1977;58:1329–1345. doi: 10.1093/jnci/58.5.1329. [DOI] [PubMed] [Google Scholar]
- Black PH. Shedding from the cell surface of normal and cancer cells. Adv Cancer Res. 1980;32:75–199. doi: 10.1016/s0065-230x(08)60361-9. [DOI] [PubMed] [Google Scholar]
- Nicolson GL, Poste G. The cancer cell: dynamic aspects and modifications in cell-surface organization (first of two parts) N Engl J Med. 1976;295:197–203. doi: 10.1056/NEJM197607222950405. [DOI] [PubMed] [Google Scholar]
- Spring FA, Dalchau R, Daniels GL, Mallinson G, Judson PA, Parsons SF, Fabre JW, Anstee DJ. The Ina and Inb blood group antigens are located on a glycoprotein of 80,000 MW (the CDw44 glycoprotein) whose expression is influenced by the In(Lu) gene. Immunology. 1988;64:37–43. [PMC free article] [PubMed] [Google Scholar]
- Sillanpää S, Anttila MA, Voutilainen K, Tammi RH, Tammi MI, Saarikoski SV, Kosma VM. CD44 expression indicates favorable prognosis in epithelial ovarian cancer. Clin Cancer Res. 2003;9:5318–5324. [PubMed] [Google Scholar]
- Li D, Zheng L, Jin L, Zhou Y, Li H, Fu J, Shi M, Du P, Wang L, Wu H, Chen GY, Zheng P, Liu Y, Wang FS, Wang S. CD24 polymorphisms affect risk and progression of chronic hepatitis B virus infection. Hepatology. 2009;50:735–742. doi: 10.1002/hep.23047. [DOI] [PubMed] [Google Scholar]
- Nestl A, Von Stein OD, Zatloukal K, Thies WG, Herrlich P, Hofmann M, Sleeman JP. Gene expression patterns associated with the metastatic phenotype in rodent and human tumors. Cancer Res. 2001;61:1569–1577. [PubMed] [Google Scholar]
- Aigner S, Ruppert M, Hubbe M, Sammar M, Sthoeger Z, Butcher EC, Vestweber D, Altevogt P. Heat stable antigen (mouse CD24) supports myeloid cell binding to endothelial and platelet P-selectin. Int Immunol. 1995;7:1557–1565. doi: 10.1093/intimm/7.10.1557. [DOI] [PubMed] [Google Scholar]
- Magee JA, Piskounova E, Morrison SJ. Cancer stem cells: impact, heterogeneity, and uncertainty. Cancer Cell. 2012;21:283–296. doi: 10.1016/j.ccr.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]