Abstract
The application of novel and modern techniques in genetic engineering and genomics has resulted in information explosion in genomics. Three major genome databases under International Nucleotide Sequence Database collaboration NCBI, DDBJ and EMBL have been providing a convenient platform for submission of sequences which they share among themselves. Many institutes in India under Indian Council of Agricultural Research have scientists working on biotechnology and bioinformatics research. The various studies conducted by them, generate massive data related to biological information of plants, animals, insects, microbes and fisheries. These scientists are dependent on NCBI, EMBL, DDBJ and other portals for their sequence submissions, analysis and other data mining tasks. Due to various limitations imposed on these sites and the poor connectivity problem prevents them to conduct their studies on these open domain databases. The valued information generated by them needs to be shared by the scientific communities to eliminate the duplication of efforts and expedite their knowledge extended towards new findings. A secured common submission portal system with user-friendly interfaces, integrated help and error checking facilities has been developed in such a way that the database at the backend consists of a union of the items available on the above mentioned databases. Standard database management concepts have been employed for their systematic storage management. Extensive hardware resources in the form of high performance computing facility are being installed for deployment of this portal.
Availability
Background
Genomic sequences provide understanding of the structure, function and evolution of genetically diverse organisms. The recent genomic era has seen a massive explosion in the amount of biological information and data arising from the rapid research and unprecedented progress in molecular biology and genome sequencing [1]. The field of bioinformatics has been intermingled with traditional computational biology and biostatistics. It is not only concerned with handling the information, but also to extract biological meaning from it [2]. The applications of novel and modern techniques are carried out on the biological information base to extract the knowledge [3]. This knowledge has profound impacts on different fields, such as human health, agriculture, environment, energy and biotechnology [2]. Therefore, this biological information needs to be systematically stored and managed using appropriate tools and standard database management practices. It also requires advanced hardware resources and parallel computing facilities for high speed information processing for knowledge extraction. Sharing genomic resources and bio-computing processes is desirable for enhancement in the growth of the biotechnological research and also useful for avoiding redundancy and duplication of efforts. This will not only reduce cost and time for development of biotechnological product but also help in sharing knowledge for the social benefits.
International Nucleotide Sequence Database Collaboration constitutes three major genome databases in the world; (i) National Center for Biotechnology Information (NCBI), (ii) DNA Data Bank of Japan (DDBJ) and (iii) European Molecular Biology Laboratory (EMBL). The sequence submission process of these databases is governed by international collaborative agreement. Sequences submitted to any one of the three databases are automatically added in the other two databases within a few days of their release to the public [4, 5]. To maintain data accuracy and integrity, well-defined procedures exist for submitting and changing entries in these databases [6]. These databases comprise of feature tables giving shared rules to allow information exchange among these databases, qualifiers for explicit referencing of specific sequences and the country qualifier for providing geographical location. These organizations have been providing a convenient and common platform for submission of sequences which they share among themselves [7].
Agricultural research scientists from various organizations of India is also significantly harnessing these resources and contributing to these international genomics and proteomics databases. These scientists are using NCBI, EMBL, DDBJ and other biological resources for their sequence submissions, analysis and other data mining tasks. The Indian Council of Agricultural Research (ICAR) is the apex body for coordinating, guiding and managing research and education in agriculture including horticulture, fisheries and animal sciences in India. ICAR is conducting research at 4 Deemed Universities, 49 Research Institutes, 19 National Research Centres, 6 National Bureaus, 27 Project Directorates and 8 Zonal Project Directorates placed all over India. These institutes have scientists conducting research on different aspects of biotechnology and bioinformatics. The various studies conducted by them, generate massive biological data related to plants, animals, insects, microbes, fisheries etc. These valued information needs to be validated, curated and shared to the scientific communities to eliminate the duplication of efforts and expedite the research related to agriculture. The National Agricultural Biotechnology Information Center (NABIC) developed a Web based relational database for agricultural plants with biotechnology information [8].
ICAR has recently initiated a World Bank funded project named as National Agricultural Innovation Project (NAIP) under which it is proposed to establish a National Agricultural Bioinformatics Grid (NABG) with lead center at Indian Agricultural Statistics Research Institute (IASRI), New Delhi for providing a biological computing platform to the researchers in agricultural bioinformatics and computational biology [9, 10]. This platform will not only help in development of biological databases/ data warehouses for storage and analysis of indigenous biological information but also provide information in the international biological resources at local server of national super-computing facility for its computational analysis. This consolidated effort would enable accelerated growth in biotechnological research in the country. A portal handling biological databases and derived databases would finally be installed on high-end computational hardware with parallelized processing/computing facilities. A secured common submission portal system for biological data with user-friendly interfaces along with integrated help and error checking facilities would facilitate wider accessibility and acceptability of the stored information. Therefore, a portal with above features is required to be developed. Taking a step towards the development of this, a biological sequence submission portal for genomic sequences has been taken up on priority basis. This portal would help to build and strengthen genomic database in India. The backend database of this genomic portal has been developed using MySQL employing the concepts of Relational Database Management Systems (RDBMS). This database has been designed keeping in view the information contents of NCBI, EMBL and DDBJ databases. This portal can be run on any java enabled internet browser. The sequence and other necessary details can be submitted to the portal by any user after the profile creation. These details include information about the sequence/ reference authors, release date, organism name, organelle/ location, source modifiers, molecular types, genomic completeness, topology, features, qualifiers and so on. The information entered by the user is updated in the database every time a user clicks on “Next” button which enables proper handling of incomplete submissions made by the user.
Data sources
The greatest challenge being faced by molecular biology community today is to make sense of the wealth of data that has been produced by the genome sequencing projects [2]. A large number of laboratories in India and abroad are generating genomic data through their laboratory experiments. The data from these sources can be an important source for populating the database on this portal. In (Figure 1), it shows different data sources which will be used to populate genomic sequences in this portal. The portal is installed on the central server located at Indian Agricultural Statistics Research Institute, New Delhi. The five domain institutions of NABG are National Bureau of Plant Genetic Resources (NBPGR), New Delhi, National Bureau of Fish Genetic Resources (NBFGR), Lucknow, National Bureau of Animal Genetic Resources (NBAGR), Karnal, National Bureau of Agriculturally Important Microbes (NBAIM), Mau and National Bureau of Agriculturally Important Insects (NBAII), Bengaluru. These five domain institutions, associated with this development and implementation work, will be responsible for ensuring data quality in their respective fields. In these institutions, domain expertise is available along with the scientists from the field of computational biology. In addition to these domain institutions, other institutions of National Agricultural Research System (NARS) as well as institutions working in the field of agricultural and allied field would be making use of this portal. However, access to this portal is not limited to any specific user or institute, but can be used by any user, any organization across the globe, as it has been made available in public domain. In (Figure 1), it shows the data flow architecture of the portal.
Figure 1.
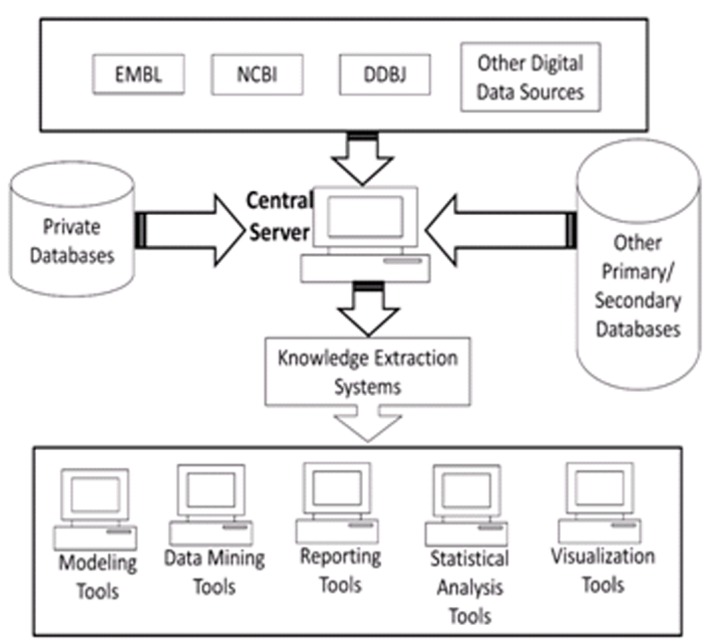
Data flow architecture of sequence submission portal.
Design of database for sequence submission portal:
In order to achieve scalability and consistency of an integrated genome database, relational database management system (RDBMS) concepts were applied. MySQL RDBMS software has been used to store the submitted data in the form of associated tables. The data consistency and non-redundancy were maintained through the principles of database normalization. The database tables have been created and relationships were established to ensure querying and information extraction. However, few database tables have been kept de-normalized for faster information retrieval. Tables of this database consist of registration details of users, submission/ accession number, features, qualifiers with their data formats, organelles, reference details, molecular types, source modifiers, third party annotation details and so on [11, 12]. The names of entities along with their descriptions can be found in the supplementary Table 1 (see supplementary material). The database tables and their fields along with their descriptions as implemented in the portal database have been shown in Table 2 (see supplementary material) available online. In order to provide easy and interactive features to the user while submitting genomic sequences, the relationships have been established among database tables. The Entity Relationship diagram has been shown in (Figure 2).
Figure 2.
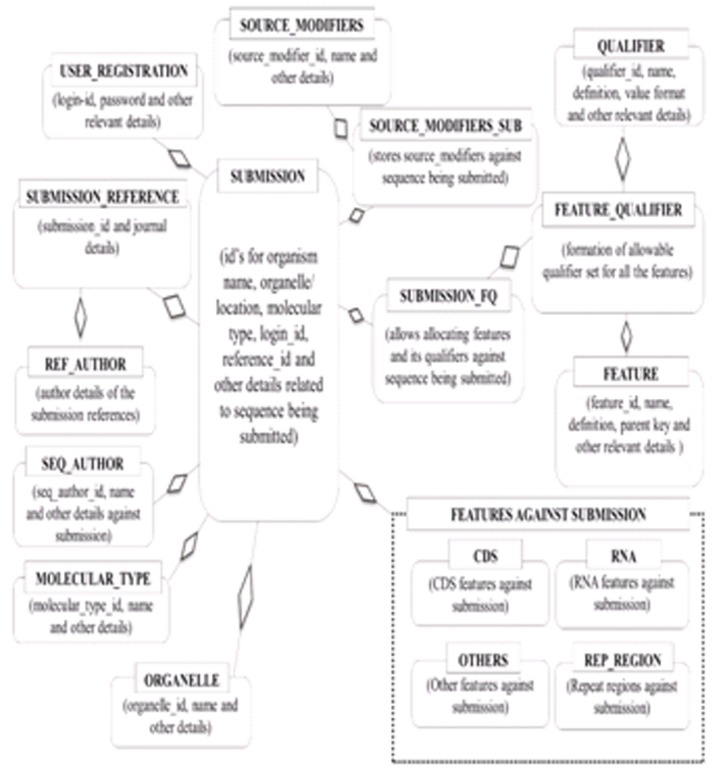
Schematic entity-relationship diagram.
Features of genomic sequence submission database:
An extensive study on the features available with NCBI, EMBL and DDBJ sequence databases have been carried out and it was observed that most of the attributes are same/ similar in all three databases except few attributes. This is otherwise also expected as these databases exchange their biological information among them. Therefore, an attempt was made to develop the database taking superset of all these features. Table 3 (see supplementary material) in the supplementary material provides detailed comparison of the features/ fields available in the databases of NCBI, EMBL and DDBJ with this database. Sequence submission portal available on NCBI, DDBJ and EMBL contains information on locus, definition, accession number, version, keywords, source, organism, reference, authors, title, journal, PubMed, comment, features, base count and origin of the submitted sequence. In case of EMBL, the information about sequence version number and data class under locus description were found, which is not available on other two databases.
Network architecture
The portal has been presently installed on a Windows server machine (central server) which uses WAMP bundled package [13]. After establishment of High Performance Computing system this is likely to be reconfigured to this parallel computing environment. WAMP is a Windows based package of independently-created programs that uses Apache web server, MySQL open-source database, and scripting language PHP, Perl or Python. However, for development of this portal PHP was used [13]. WAMP can manipulate information stored in a database and generate Web pages dynamically for every hit by a browser. The central server communicates with MySQL server, whenever, a user accesses it, for any operation. This portal can be accessed from LAN for internal access and from outside LAN on the web address http://cabindb.iasri.res.in:8080/sequence_portal/ or http://nabg.iasri.res.in through a firewall for secured access. The architecture for user access of this portal has been shown in (Figure 3). The sequence submission is implemented in three layers - user registration, sequence submission and storage as shown in (Figure 4).
Figure 3.
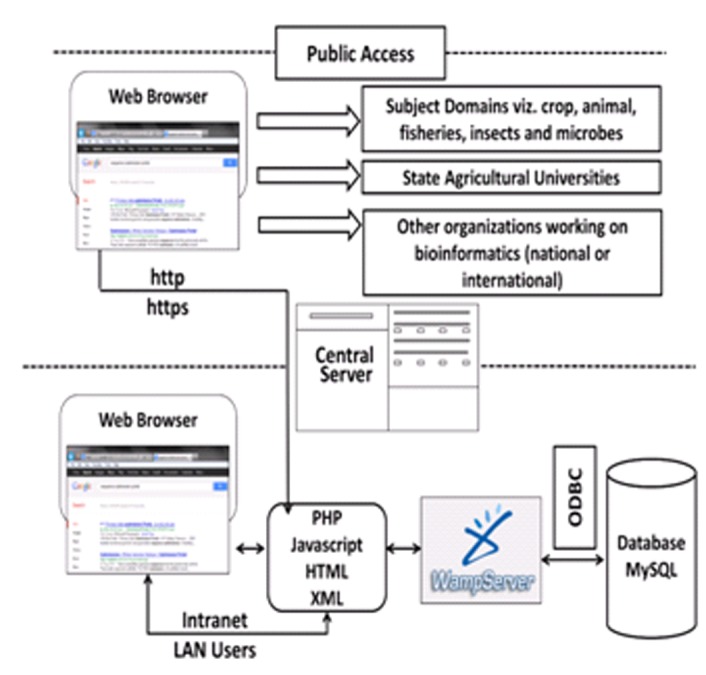
Network architecture of portal.
Figure 4.
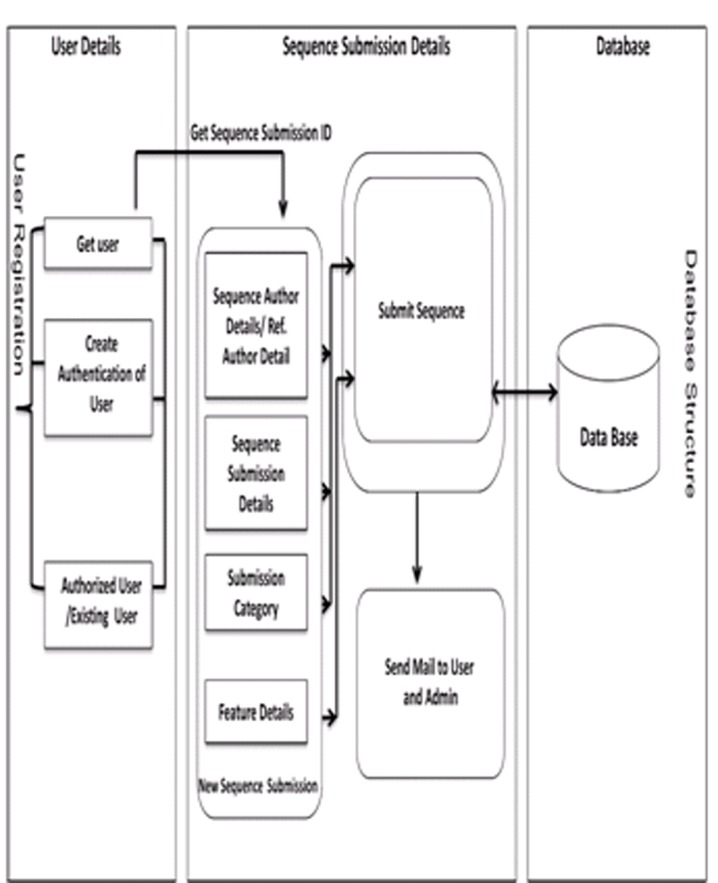
Sequence submission process.
First section includes user authentication and profile creation, which is a mandatory requirement for any user. After profile creation, a user can login, by entering login-id and the password. A user can submit as many sequences as required. This portal maintains the history of sequence submissions for each user in the database. For submitting a new sequence or viewing already submitted sequence, the portal extracts information from sequence submission section, where all details related to sequence are stored. Each sequence submitted on the portal is linked with a unique submission/accession number. This submission number is generated immediately after a user chooses to submit a new sequence. The submission number is generated through date, time and a counter as [YYYYMMDD][hhmmss][n] where YYYY is year, MM is month, DD is date, hh, mm and ss are time in hour, minute and second. The last character in the submission number is a counter, shown as n. The details about the sequence and reference author, submission category, features and qualifiers are entered by the user in a set of wizard like pages. The third section stores all the details entered by the user pertaining to a particular submission number.
Plan of submitting sequences
The portal would be deployed on HPC system at IASRI under NABG. The HPC system, which is presently in the process of implementation, will have capacity of around 70 teraflop with 256 nodes computational power controlled by 2 masters in linux operating system environment with a total of 500 terra byte of storage. Sequence file submitted at the central server will populate the database automatically using developed programs. This portal accepts the FASTA format sequence for submission. The submitter details are also stored in the database to keep track of the sequences submitted by a particular researcher. During the process of submitting sequences, various parameters such as author details, registration details, molecular type, source name, source modifier, journal name, references, organism name, third party annotation details etc. are stored in the database. The sequence data will then be made available for public access. The sequence submission plan has been shown in (Figure 5).
Figure 5.
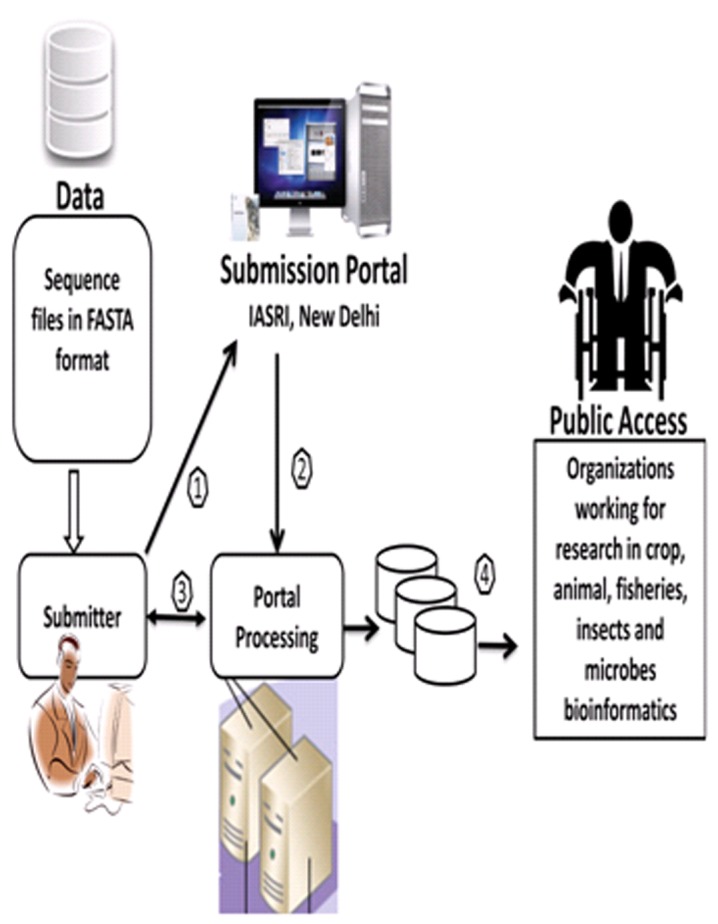
Architectural plan of sequence submission.
Portal description
In order to achieve scalability and consistency of an integrated genome database, relational database management system (RDBMS) concepts were applied. MySQL RDBMS software has been used to store the submitted data in the form of associated tables. The data consistency and non-redundancy were maintained through the principles of database normalization. The database tables have been created and relationships were established to ensure querying and information extraction. However, few database tables have been kept de-normalized for faster information retrieval. Tables of this database consist of registration details of users, submission/ accession number, features, qualifiers with their data formats, organelles, reference details, molecular types, source modifiers, third party annotation details and so on [11, 12]. The names of entities along with their descriptions have been given in the (Table 1).
The database tables and their fields along with their descriptions as implemented in the portal database have been shown in (Table 2). In order to provide easy and interactive features to the user while submitting genomic sequences, the relationships have been established among database tables. The Entity Relationship diagram has been shown in (Figure 2).
The resources available with BankIt submission tool have been very helpful for designing web forms for this submission portal [14]. The sequence submission process through this portal consists of the following activities:
• User authentication • Sequence submission details • Reference details • Source information • Molecular types • Submission report
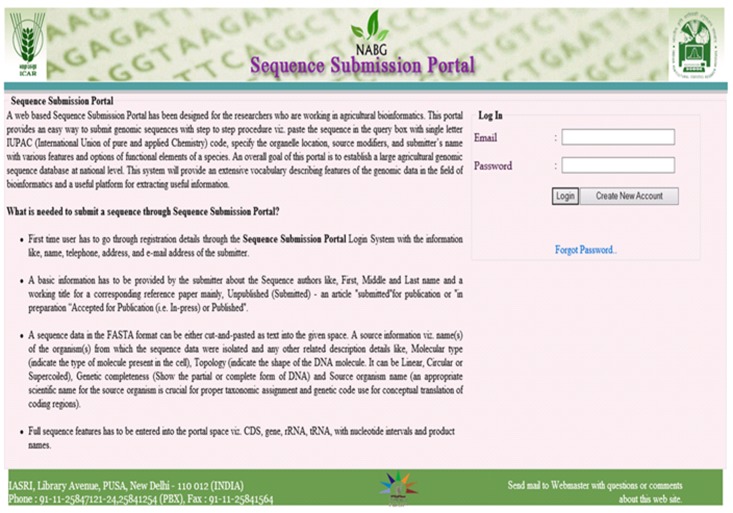
Registration is must for a submitter for submission of sequence. The registration page needs details about the user for creation of user account. The user login is linked with the submitter's email address. Upon successful registration the sequence submission portal page is displayed for immediate use to submit genomic sequence through this portal. The home page of the submission portal containing login screen and a brief description of the portal has been shown in (Figure 6 & Figure 7). shows the signup screen which can be used by a new user to enter his registration details to the portal database.
Figure 6.
Home page of the submission portals
Figure 7.
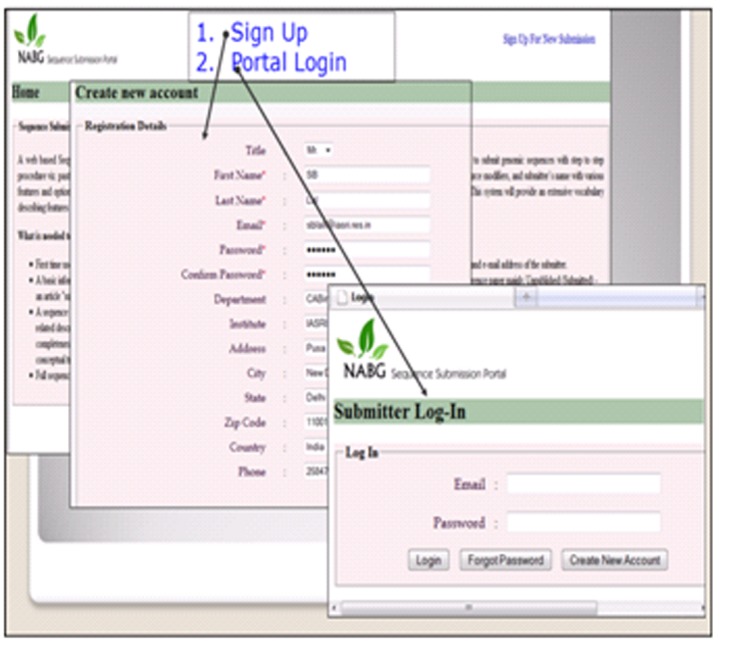
User registration on the portal.
After logging in to the portal, submitter needs to provide descriptive information of the sequence to be submitted. Table 4 (see supplementary material) lists item names along with their possible values either on the text boxes or on the combo boxes of the web pages of the portal. For example - nucleotide information page seeks information about submitter name, reference information, sequence author, submission release date, molecular type, topology, genomic completeness, organism name, sequence and definition line, organelle, source modifiers and submission category. The portal accepts submission of FASTA sequence to be pasted on a text box. On the other pages of the portal requires adding features of the sequence. Every feature being added against the sequence needs some mandatory information to be supplied. Each feature can include adding many optional qualifiers and their values. Supplementary Table 5 (Available with Author) lists all the features along with their allowable qualifiers and value formats. In (Figure 8), it shows the web page where a user can see his previously made submissions and an option for new submission. It also shows a screen where the submitter can add sequence authors. A screen has been shown in (Figure 9), where a user can specify required details about the sequence and paste the sequence.
Figure 8.
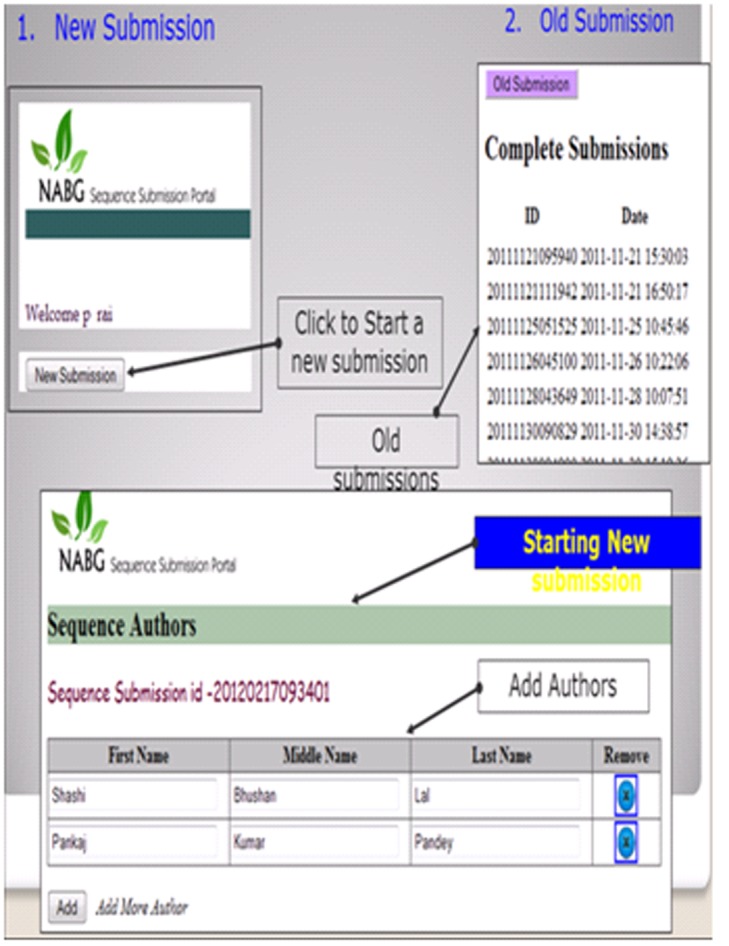
A view of old and new submissions made by the user.
Figure 9.
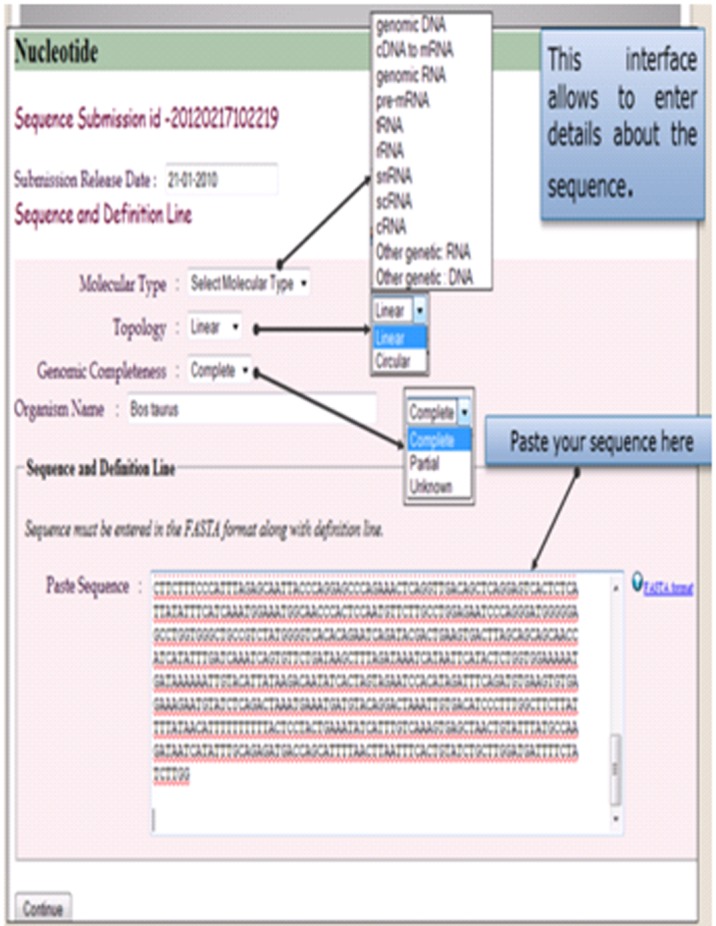
Pasting the sequence and other related information.
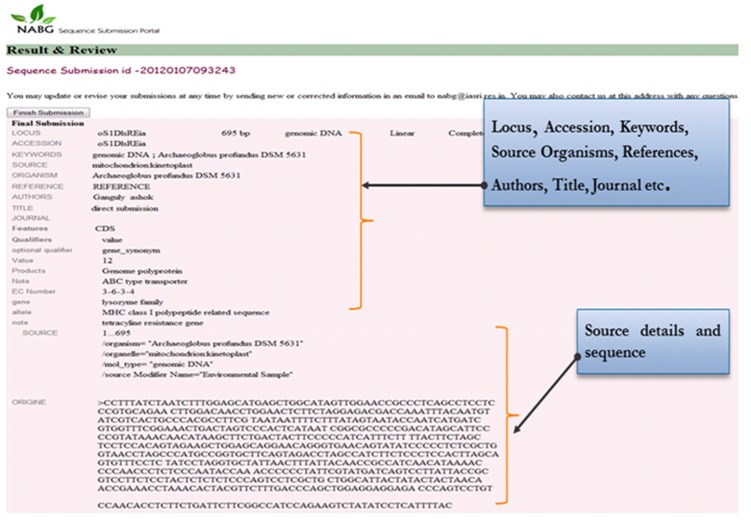
Final output is generated after successful submission of the sequence (in a FASTA format). The portal displays a final output in a customary flat-file with all the information filled at the time of submission. A screen showing the generated submission report is given the (Figure 10).
Figure 10.
Submission report.
Conclusion
Scientific databases are nowadays essential for the progress of science as they provide means for data sharing (compatible and complementary to traditional scientific publications) and longterm preservation (archive) of data to enable further analysis [3]. Currently, the enormous volume of genomic data is generated by agricultural researchers regularly which are inaccessible to the public domain for further research. Therefore, this genomic database has been developed, integrated and deployed for benefit of agricultural as well as other biological researchers. A sequence submission portal provides a distinctive workflow from data acquisition, storage and submission of knowledge enriched genomic sequences. The database resources generated through this portal would be deployed on the high performance computing environment to achieve the speedy access and enable the user in carrying out computational and statistical analysis for important findings.
The integration of genomic data pertaining to various agriculturally important species would be accessible through the implementation of sequence submission portal and provide higher level of data storage for faster access to the user. It would be made available on the public domain and facilitate information exchange through global exchange programmes, national, international consortiums for sharing resources with proper credit/ acknowledgement to the contributor for their findings. It would be feasible to extract meaningful biological information for enhancement of agricultural productivity through development and deployment of the parallel computing tools to enable faster access of the resources available on this portal.
Supplementary material
Footnotes
Citation:Lal et al, Bioinformation 9(11): 588-598 (2013)
References
- 1.Kolatkar PR, et al. Pac Symp Biocomput. 1998:735. [PubMed] [Google Scholar]
- 2.Singh VK, et al. Int J of Bioinf. Res. 2011;3:221. [Google Scholar]
- 3.Chagoyen M, Montano AP. Proc. of the ECAI 2004 Workshop. 2004:8. [Google Scholar]
- 4. http://www.ncbi.nlm.nih.gov/books/NBK21105/
- 5.Benson DA, et al. Nucleic Acids Res. 1996;24:1. doi: 10.1093/nar/24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Givan SA, et al. BMC Bioinformatics. 2007;18:479. doi: 10.1186/1471-2105-8-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Federhen S, et al. Nucleic Acids Res. 2012;40:D136. doi: 10.1093/nar/gkr1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim C, et al. Bioinformation. 2011;6:246. doi: 10.6026/97320630006246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. http://www.igovernment.in/site/icar-set-agriculturalbioinformatics- grid-39254.
- 10. http://www.dnaindia.com/money/report_indianfarmers- to-get-bioinformatics-grid_1505325.
- 11. www.ddbj.nig.ac.jp/FT/FT.pdf.
- 12. www.ehu.es/biofisica/juanma/data_bases/pdf/ft.pdf.
- 13. http://www.wampserver.com/en/
- 14. http://www.ncbi.nlm.nih.gov/books/NBK63590/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.