Abstract
Polymeric immunoglobulin receptor (pIgR) expression is downregulated in lung cancer, but its implications in lung tumourigenesis remain unknown. We hypothesised that loss of pIgR expression occurs early, and is associated with cell proliferation and poor prognosis.
pIgR expression was evaluated by immunohistochemistry in airways of patients with normal mucosa, pre-invasive lesions and invasive lesions, and correlated with clinical outcomes. 16-HBE and A549 cells stably transfected with pIgR were tested for proliferation, apoptosis and cell cycle progression.
Immunostaining was strong in normal epithelium, but severely reduced in pre-invasive lesions and most lung cancers. Persistent expression was associated with younger age and adenocarcinoma subtype but not survival. pIgR overexpression significantly reduced A549 and 16-HBE proliferation. Growth inhibition was not due to cell cycle arrest, increased apoptosis or endoplasmic reticulum stress, but we observed altered expression of genes encoding for membrane proteins, including NOTCH3. Interestingly, NOTCH3 expression was inversely correlated with pIgR expression in cell lines and tissues.
pIgR expression was lost in most lung cancers and pre-invasive bronchial lesions, suggesting that pIgR downregulation is an early event in lung tumourigenesis. pIgR overexpression in A549 and 16-HBE cells inhibited proliferation. Future investigations are required to determine the mechanisms by which pIgR contributes to cell proliferation.
Keywords: Differentiation, lung adenocarcinoma, lung pre-invasive lesions, polymeric immunoglobulin receptor, proliferation
Transmembrane receptors are critical in cellular signalling and regulate proliferation, host defence mechanisms and cellular homeostasis. Polymeric immunoglobulin (Ig) receptor (pIgR) is a transmembrane glycoprotein expressed in mucosal surfaces that mediates polymeric IgA (pIgA) transcytosis from the basolateral pole to the apical surface of epithelial cells [1, 2]. At the cell’s luminal surface, pIgR is cleaved and its extracellular ligand-binding region, called the secretory component (SC), is released free or bound to pIgA in a complex called secretory IgA (sIgA). sIgA is the first line of immunological defence against environmental antigens to which mucosal surfaces are exposed [3, 4].
Under normal conditions, pIgR is highly expressed in epithelial cells of the upper respiratory tract (mainly serous, but also mucous and ciliated cells), while expression is absent in alveolar cells [5]. pIgR expression has been reported to be downregulated in various lung diseases, including chronic obstructive pulmonary disease [5], fibrosis [6], sarcoidosis [6] and cancer [7–13]. Several studies have shown that pIgR expression is decreased in lung adenocarcinomas (ADCs), with lower expression in poorly differentiated tumours, and absence of expression in other lung cancer types [7–13]. Decreased pIgR expression has also been described in other cancers [14–18]. In the colorectum, pIgR downregulation was observed in cancerous and pre-invasive lesions, with lower expression in poorly differentiated lesions [16, 18]. Moreover, pIgR expression in colorectal cancer was associated with outcome and proposed as a prognostic biomarker [14, 16–18].
Mechanisms leading to pIgR downregulation in lung cancer are poorly understood. pIgR mRNA levels have been correlated with the expression of the constitutive transcription factors upstream regulatory factor 1/2 and activator protein 2α [19].
The temporality of pIgR downregulation in lung tumourigenesis is unknown and we do not know whether pIgR downregulation carries prognostic value. Moreover, biological implications of pIgR downregulation in lung cancer development/progression have not been reported. We therefore formulated the following three hypotheses: pIgR downregulation 1) is an early event in lung tumourigenesis, 2) is associated with poor prognosis and 3) has biological implications in lung cancer development/progression. To test these hypotheses, we determined pIgR expression by immunohistochemistry (IHC) in lung pre-invasive and invasive tumours, correlated pIgR staining to clinical outcomes, and evaluated the effects of pIgR overexpression on proliferation, cell cycle progression and apoptosis. Addressing these hypotheses may improve our understanding of the molecular steps leading to lung cancer and the role of a critical cellular receptor involved in cell trafficking and differentiation regulation.
MATERIAL AND METHODS
Tissues and cell lines
Nonsmall-cell lung cancer (NSCLC) samples from 175 patients without prior medical treatment were assembled into tissue microarrays (TMAs) prepared from paraffin blocks obtained from the archives of the Pathology Department at Vanderbilt University Medical Center and Nashville VA Medical Center (Nashville, TN, USA). Lung cancer diagnosis was confirmed by a lung cancer pathologist (A.L. Gonzalez) and samples were annotated with clinical data elements.
The study was approved by institutional review boards. Snapfrozen tissues from 11 patients with lung ADC and six with squamous cell carcinoma (SCC) were obtained through the Vanderbilt Specialized Program of Research Excellence in Lung (Nashville). The following cell lines were purchased from the American Type Culture Collection (Manassas, VA, USA): BEAS-2B, NCI-H1299, NCI-H460, NCI-H226, NCI-H520, NCI-A549, Calu-3, NCI-H23, NCIH2009, NCI-H1650, NCI-H1819 and NCI-H1993. 16-HBE was a kind gift from D.C. Gruenert (Children’s Hospital, Oakland Research Institute, Oakland, CA, USA). H157, HCC15, HCC95, H549 and HCC78 were kind gifts from J.D. Minna (University of Texas Southwestern Medical Center, Dallas, TX, USA). All cells were cultured under recommended conditions.
Immunohistochemistry
TMAs were stained following a previously reported protocol [5]. Briefly, slides were incubated overnight at 4°C with anti-human pIgR/SC polyclonal goat IgG (1/500 dilution; goat 606 prepared in the laboratory of Y. Sibille, Cliniques Universitaires Mont-Godinne, Université Catholique de Louvain, Yvoir, Belgium). The secondary antibody, biotinylated rabbit anti-goat IgG from the VECTASTAIN Elite ABC system (1/50 dilution; Vector Laboratories, Burlingame, CA, USA), was applied at room temperature for 30 min. Staining intensity was evaluated by two independent observers (A.L. Gonzalez and S. Ocak) as 0 (no staining), 1 (weak staining), 2 (moderate staining) or 3 (strong staining). Staining intensity was multiplied by the percentage of stained tumour cells to obtain the final staining score (scale 0–300) used for the association with clinical outcomes.
Western blotting
Lysates were obtained from cells/tissues following a standard protocol [20]. Blots were probed with antibodies against human pIgR/SC (1 µg•mL−1; polyclonal rabbit IgG 877, prepared in the laboratory of Y. Sibille), calreticulin (1/1,000 dilution; Cell Signaling, Danvers, MA, USA), BIP (1/1,000; Santa Cruz Biotechnology, Santa Cruz, CA, USA), NOTCH3 (1/1,000 dilution; Orbigen, San Diego, CA, USA) or actin (1/5,000; Sigma, St Louis, MO, USA).
Stable transfection with pIgR or empty pcDNA3.1(−) vector
A549 or 16-HBE cells were seeded in 6-cm dishes at 2×105 and 12×105 cells per dish, respectively. The next day, transfection of human pIgR cDNA (kind gift from C.S. Kaetzel, University of Kentucky, Lexington, KY, USA) [21] in pcDNA3.1(−) (Invitrogen, Carlsbad, CA, USA) or empty pcDNA3.1(−) was performed using PolyFect Transfection Reagent (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. 2 days later, cells were split and the medium was supplemented with geneticin (600 mg·mL−1 for A549 and 100 mg·mL−1 for 16-HBE; Gibco, Carlsbad, CA, USA). After 2 weeks, geneticin-resistant clones were picked and tested for pIgR expression.
Proliferation assay
Stably transfected A549 and 16-HBE cells were seeded in 96-well plates at 0.5×103 and 5×103 cells per well, respectively. They were cultured in medium containing 10% fetal bovine serum (FBS) for 6 days. Every day, WST-1 reagent (Roche, Mannheim, Germany) was used for spectrophotometric quantification of cell proliferation according to the manufacturer’s instructions.
Apoptosis assay
Stably transfected A549 cells were seeded in 6-cm dishes at 2×105 cells per dish. 2 days later, cells were stained with 100 µL annexin V binding buffer containing 1 µL AlexaFluor®-488-conjugated annexin V (Invitrogen) and 1 µL propidium iodide (PI) (100 µg·mL−1; Sigma). Staining was quantified by fluorescence-activated cell sorting (FACS) analysis. Data acquisition and analysis were performed with CellQuest Pro (BD Biosciences, San Jose, CA, USA).
Cell cycle analysis
Stably transfected A549 cells were seeded in 6-cm dishes at 2×105 cells per dish. The next day, the 10% FBS medium was replaced with serum-free medium (synchronisation). Cells were pelleted and fixed with ethanol at different time-points (0, 4, 8, 12, 20, 22, 24 and 26 h). Cells were incubated with 1 mL PI (100 µg邷mL−1) and RNase A (100 µg·mL−1; Qiagen). A total of 10,000 stained nuclei were subjected to flow cytometry. Data were collected on a Becton Dickinson FACSCalibur flow cytometer using CellQuest Pro. Cell cycle analysis was performed with ModFit LT software (Verity Software House, Topsham, ME, USA).
Gene expression microarray analysis
Total RNA was isolated from stably transfected A549 after 3 days of culture using TRIzol® (Invitrogen), following the manufacturer’s protocol. RNA was hybridised to a GeneChip® Human Genome U133 Plus 2.0 Array (Affymetrix, Santa Clara, CA, USA). Image analysis was performed by the Vanderbilt University DNA Microarray Core Facility (Nashville, TN, USA). Data were analysed with Expression Console (Affymetrix). Experiments were performed three times.
Statistical analyses
Immunohistochemistry
Wilcoxon rank-sum or Kruskal–Wallis tests were used to correlate average pIgR staining scores with categorical clinical/pathological outcomes, and Spearman’s rank test was used for continuous outcomes. Overall survival was calculated from date of diagnosis to date of death or last date of contact for those alive at the time of analysis. Recurrence/progression-free survival was calculated from date of diagnosis to date of recurrence/progression or last date of contact for those without recurrence/progression at the time of analysis. Score test was used for survival analyses. Kaplan–Meier survival curves were calculated according to pIgR expression (staining score 0 versus >0 and < 20 versus ≥20) and compared using the log-rank test. Descriptive statistics, including median and interquartile ranges for continuous, and percentages or frequencies for categorical variables, were reported. Multivariable analyses were performed using the Cox proportional-hazards model to adjust for age, pack-yrs, lung cancer histology and stage. All tests were two-sided, t-tests were unpaired and p-values <0.05 were considered statistically significant. Analyses were performed using R 2.9.2 (University of Vienna, Vienna, Austria).
Gene expression microarray
Data were normalised using the Robust Multichip Average method. Candidate genes were selected based on a two-fold change of gene expression in A549 cells overexpressing pIgR and unpaired t-test p-values <0.05.
RESULTS
pIgR expression is downregulated early in lung tumourigenesis
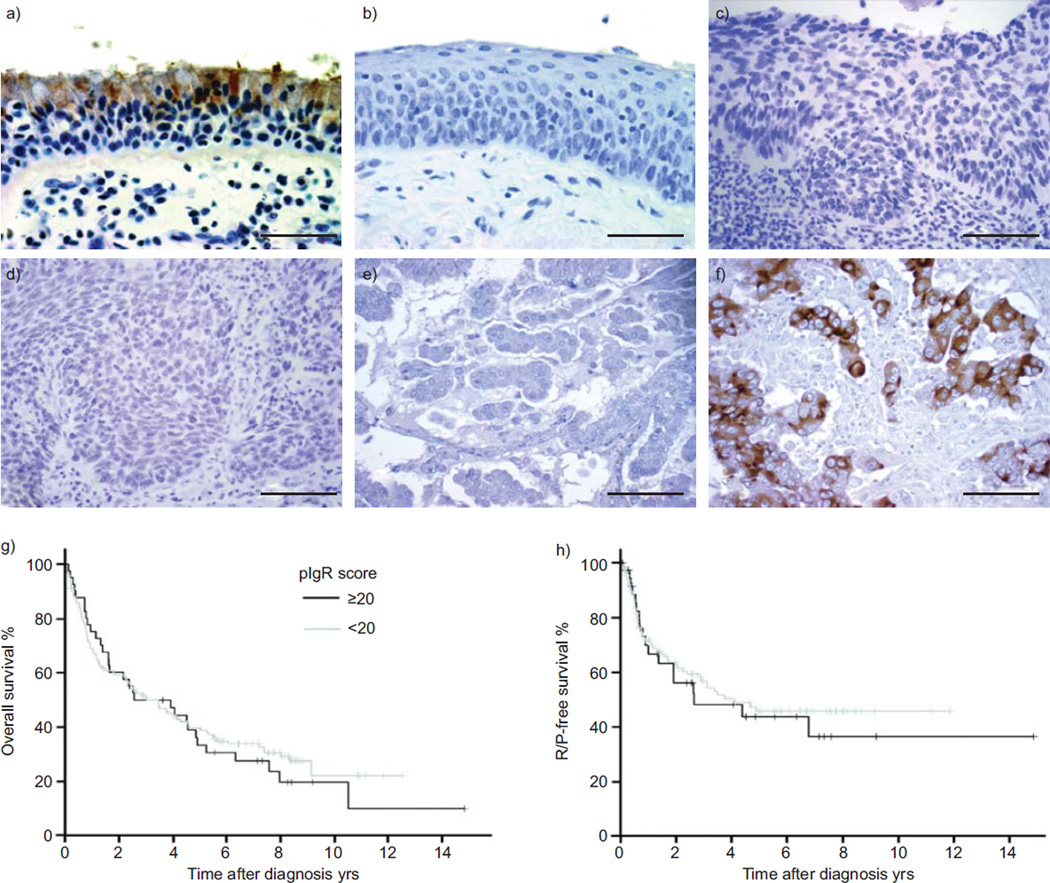
pIgR expression was evaluated by IHC in the airways during bronchial tumour development. The results are summarised in table 1 and representative images are shown in figure 1a–f. pIgR expression was cytosolic and membranous, in accordance with its role as a transcytosed membrane receptor. pIgR was expressed in 10 out of 11 normal bronchial and alveolar tissues, with strong staining intensity in mucous and serous cells of the surface and glandular epithelium, weak intensity in ciliated and an expected absence of staining in alveolar cells. In all tested lung pre-invasive lesions (four low and six high grade), we observed an absence of pIgR expression, suggesting that pIgR downregulation is an early event during lung tumourigenesis. Staining of lung cancer TMAs, including samples from 175 patients, demonstrated an absence of pIgR expression in all undifferentiated NSCLCs, 60 out of 67 SCCs and 47 out of 98 ADCs.
TABLE 1.
Polymeric immunoglobulin receptor (pIgR) expression in normal, pre-invasive and lung cancer tissues
| Cytology/histology | Patients | pIgR-positive tissues | Average pIgR staining score |
|
|---|---|---|---|---|
| All tissues | pIgR-positive tissues | |||
| Normal | ||||
| Mucous cells | 11 | 10 (91) | 227 ± 85 | 250 ± 41 |
| Serous cells | 11 | 10 (91) | 227 ± 85 | 250 ± 41 |
| Ciliated cells | 11 | 10 (91) | 45 ± 25 | 50 ± 20 |
| Alveolar cells | 11 | 0 | 0 | 0 |
| Pre-invasive | ||||
| Low grade | 4 | 0 | 0 | 0 |
| High grade | 6 | 0 | 0 | 0 |
| Cancer | ||||
| ADC | 98 | 47 (48) | 26 ± 43 | 55 ± 47 |
| SCC | 67 | 7 (10) | 7 ± 27 | 62 ± 62 |
| Undifferentiated NSCLC | 10 | 0 | 0 | 0 |
Data are presented as n, n (%) or mean±SD. ADC: adenocarcinoma; SCC: squamous cell carcinoma; NSCLC: nonsmall-cell lung cancer.
FIGURE 1.
Polymeric immunoglobulin receptor (pIgR) expression in normal bronchial epithelium, pre-invasive lung lesions and lung cancers evaluated by immunohistochemistry (IHC). a) IHC on normal lung tissues revealed a strong staining for pIgR in mucous and serous cells in surface and glandular epithelium, a weak staining in ciliated cells, and an expected absence of staining in alveolar cells. b, c) There was no pIgR expression in pre-invasive lung lesions: b) squamous metaplasia; c) severe dysplasia. pIgR expression was also absent in d) 90% of squamous cell carcinomas and e, f) 52% of adenocarcinomas (ADCs): e) ADC with absence of pIgR staining and f) ADC with strong pIgR staining. Scale bars=20 µm. Kaplan–Meier analysis of g) overall survival (p=0.66) and h) recurrence (R)- or progression (P)-free survival (p=0.67) did not show a survival difference between lung cancer patients with or without pIgR expression.
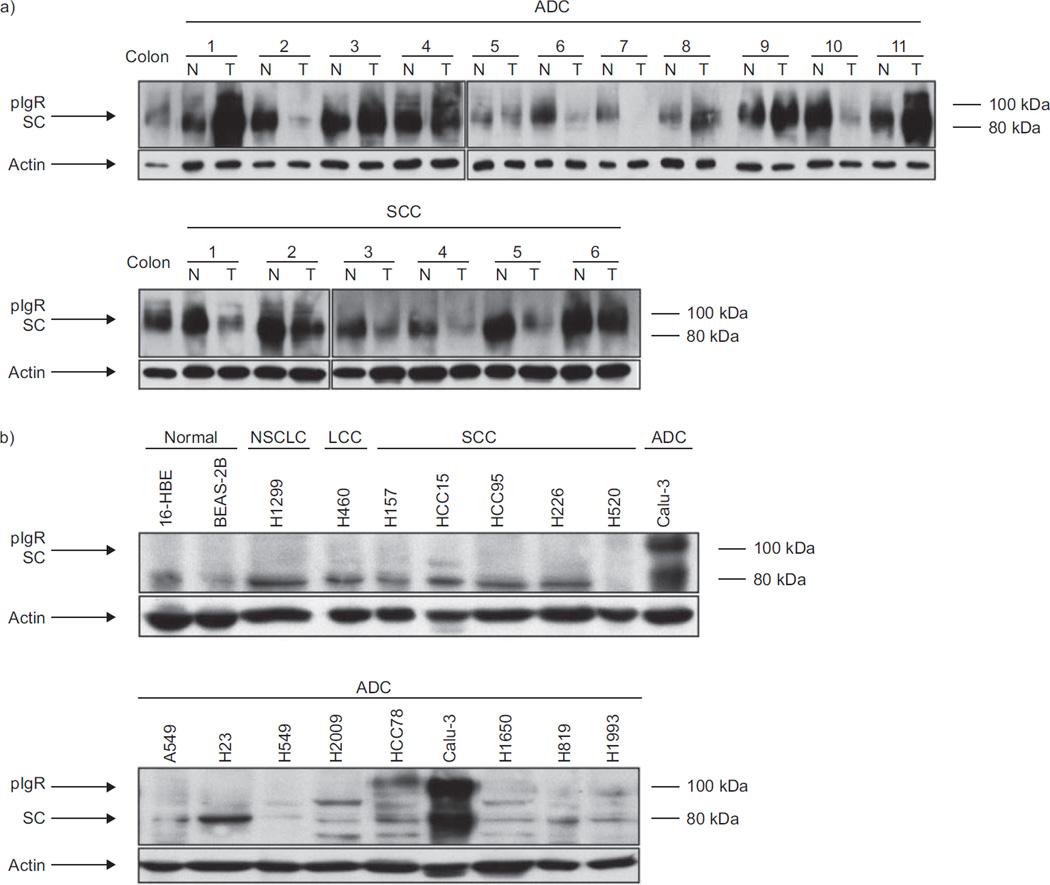
pIgR expression was then evaluated by immunoblotting in lung cancer and normal lung tissues from the same patients (fig. 2a). pIgR expression was downregulated in six out of six SCCs. In ADCs, downregulation was observed in five out of 11, while there was no expression difference in two out of 11 and overexpression in four out of 11 tissues. These results were consistent with those observed by IHC. pIgR expression was low in two out of two immortalised normal bronchioepithelial, one out of one NSCLC, one out of one large cell carcinoma, five out of five SCC and seven out of nine ADC cell lines. Expression was moderate in HCC78 and strong in Calu-3 cells, both lines being of the ADC subtype (fig. 2b).
FIGURE 2.
Polymeric immunoglobulin receptor (pIgR) expression in lung tissues and cell lines evaluated by immunoblotting. Whole tissue or cell lysates were resolved by sodium dodecylsulfate–polyacrylamide gel electrophoresis and blots were incubated with anti-pIgR antibody. pIgR expression levels were normalised to actin. a) Lung cancer tissues (T) were compared to normal lung tissues (N) from the same patients. The results show downregulation of pIgR expression in six out of six squamous cell carcinomas (SCCs) and in five out of 11 adenocarcinomas (ADCs), while pIgR was overexpressed in four out of 11 ADCs and no expression difference was noted in two out of 11 ADCs. b) pIgR expression was low in two out of two transformed normal bronchial epithelial cell lines, one out of one undifferentiated nonsmall cell lung cancer (NSCLC) cell line, one out of one large cell carcinoma (LCC) cell line, five out of five SCC cell lines and seven out of nine ADC cell lines. pIgR expression was moderate to strong in two out of nine ADC cell lines. SC: secretory component.
pIgR expression is associated with age and histology
pIgR staining scores were correlated with clinical and pathological variables. Patient characteristics and the statistical significance of the correlations between pIgR staining and these clinicopathological variables are summarised in table 2. Staining scores treated as continuous variables were significantly higher in younger patients (p=0.041) and those with ADC (p<0.0001). Staining scores were also treated as categorical variables (0 versus >0 and >20 versus ≥20), and this revealed similar results (data not shown). Staining scores were not significantly associated with other variables, especially disease stage, and overall (fig. 1g) and recurrence/progression-free survivals (fig. 1h). These observations were also true in the ADC subgroup.
TABLE 2.
Patient characteristics and statistical significance of the association with polymeric immunoglobulin receptor (pIgR) staining intensity
| Characteristic | Result | p-value |
|---|---|---|
| Sex | 0.192 | |
| Male | 113 (65) | |
| Female | 62 (35) | |
| Race | 0.066 | |
| Caucasian | 162 (93) | |
| African-American | 13 (7) | |
| Age at diagnosis yrs | 66 (58–72) | 0.041 |
| ≤60 | 61 (35) | 0.031 |
| >60 | 114 (65) | |
| Smoking status | ||
| Current smoker | 59 (34) | 0.61 |
| Ex-smoker | 104 (60) | |
| Never-smoker | 10 (6) | |
| Smoking history pack-yrs | 50 (30–80) | 0.086 |
| Dl,co % pred | 76 (57–96) | 0.976 |
| FEV1 % pred | 68 (54–85) | 0.703 |
| Lung cancer histology | <0.0001 | |
| ADC | 98 (56) | |
| SCC | 67 (38) | |
| Undifferentiated NSCLC | 10 (6) | |
| Pathological stage I | 102 (58) | 0.151 |
| II | 19 (11) | |
| III | 35 (20) | |
| IV | 19 (11) | |
| Pathological tumour stage | 0.460 | |
| T1 | 68 (40) | |
| T2 | 63 (37) | |
| T3 | 18 (11) | |
| T4 | 22 (13) | |
| Pathological node stage | 0.095 | |
| N0 | 132 (78) | |
| N1 | 12 (7) | |
| N2 | 26 (15) | |
| Pathological metastasis stage | 0.332 | |
| M0 | 156 (89) | |
| M1 | 19 (11) | |
| R/P-free survival yrs | ||
| Median (95% CI) | 4 (2.6–6.8) | 0.411 |
| R/P-free at 3 yrs | 109 (63) | 0.580 |
| Overall survival yrs | ||
| Median (95% CI) | 3 (2.4–4.5) | 0.484 |
| Alive at 3 yrs | 90 (51) | 0.978 |
| Average pIgR staining score | ||
| 0 | 121 (69) | |
| ≥20 | 41 (23) | |
| ≥50 | 27 (15) | |
| ≥100 | 12 (7) |
Data are presented as n (%) or median (interquartile range), unless otherwise stated. Dl,co: diffusing capacity of the lung for carbon monoxide; % pred: % predicted; FEV1: forced expiratory volume in 1 s; ADC: adenocarcinoma; SCC: squamous cell carcinoma; NSCLC: nonsmall cell lung cancer; R recurrence; P: progression. Bold indicates statistically significant p-values.
pIgR overexpression inhibits lung cancer cell proliferation
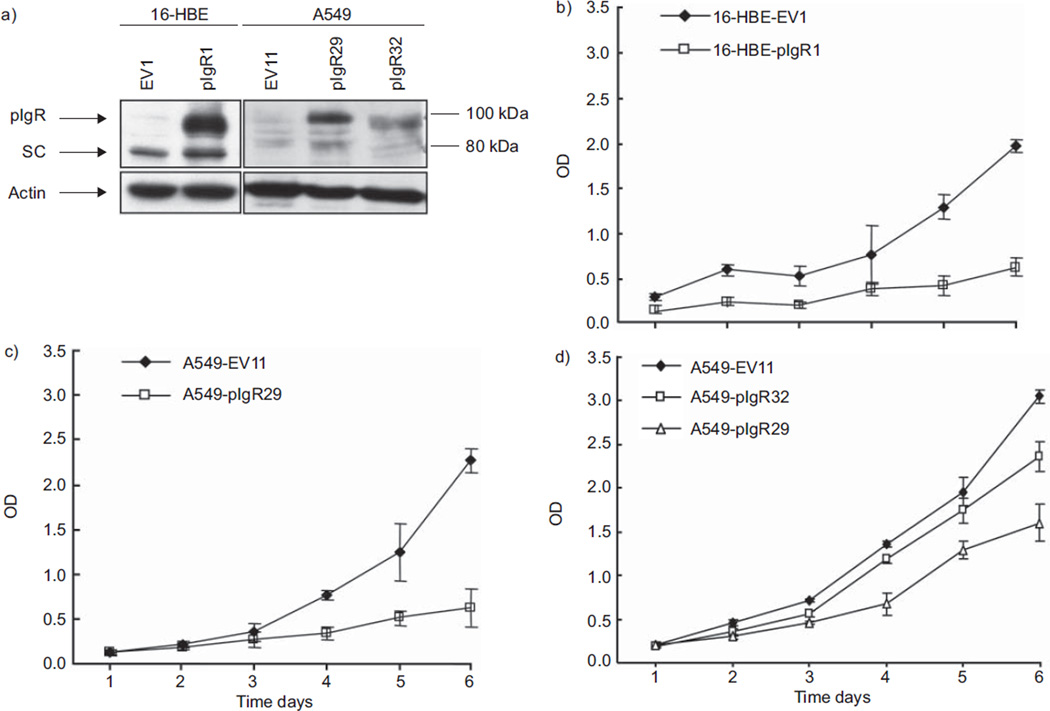
Although pIgR is downregulated in lung cancer, its implication in tumourigenesis remains unknown. Therefore, we tested whether pIgR contributes to cellular functions important for cancer development/progression. We stably transfected pIgR cDNA into two cell lines with low basal pIgR expression: the immortalised normal bronchioepithelial cell line 16-HBE and the lung ADC cell line A549 (fig. 3a). For further functional studies, we selected the clones with the strongest stable pIgR expression (16-HBE-pIgR1 and A549-pIgR29) and also one clone with moderate stable pIgR expression (A549-pIgR32). All the clones transfected with empty vector displayed low basal pIgR expression; therefore, we randomly selected control clones (16-HBE-EV1 and A549-EV11). Then, we evaluated pIgR effects on cell growth over 6 days. Inhibition of cell proliferation was observed by WST-1 assay in both 16-HBE-pIgR1 (fig. 3b) and A549-pIgR29 (fig. 3c) (p<0.001). Moreover, a dose-dependent effect of pIgR on cell growth was observed when WST-1 assay was performed with A549-EV11 (low basal pIgR expression), A549-pIgR32 (moderate pIgR expression) and A549-pIgR29 (strong pIgR expression) cells (fig. 3d).
FIGURE 3.
Effects of polymeric immunoglobulin receptor (pIgR) overexpression on 16-HBE and A549 proliferation. a) Whole cell lysates were resolved by sodium dodecylsulfate–polyacrylamide gel electrophoresis and blots were incubated with anti-pIgR antibody. pIgR expression levels were normalised to actin. Higher expression of pIgR was confirmed by immunoblotting in immortalised normal bronchial epithelial cell line 16-HBE and lung adenocarcinoma cell line A549 stably transfected with pIgR pcDNA3.1(−) (16-HBE-pIgR1, A549-pIgR29 and A549-pIgR32) as compared with those transfected with the empty vector (16-HBE-EV1 and A549-EV11). pIgR expression was higher in A549-pIgR29 (clone with the strongest pIgR expression) than in A549-pIgR32 (clone with moderate pIgR expression). SC: secretory component. b) Inhibition of cell growth was observed by WST-1 assay in 16-HBE-pIgR1 grown for 6 days as compared with 16-HBE-EV1. Optical density (OD) in the y-axis reflects the proportion of metabolically active cells. c) Inhibition of cell growth was observed by WST-1 assay in A549-pIgR29 grown for 6 days as compared with A549-EV11. d) Dose-dependent effect on cell growth inhibition in A549 cells. Inhibition of cell growth was less pronounced in A549-pIgR32 (clone with moderate expression of pIgR) than in A549-pIgR29 (clone with higher expression of pIgR). Error bars represent standard deviation (n=5). All the graphs represent one of three independent experiments with similar results.
pIgR overexpression effect on cell death
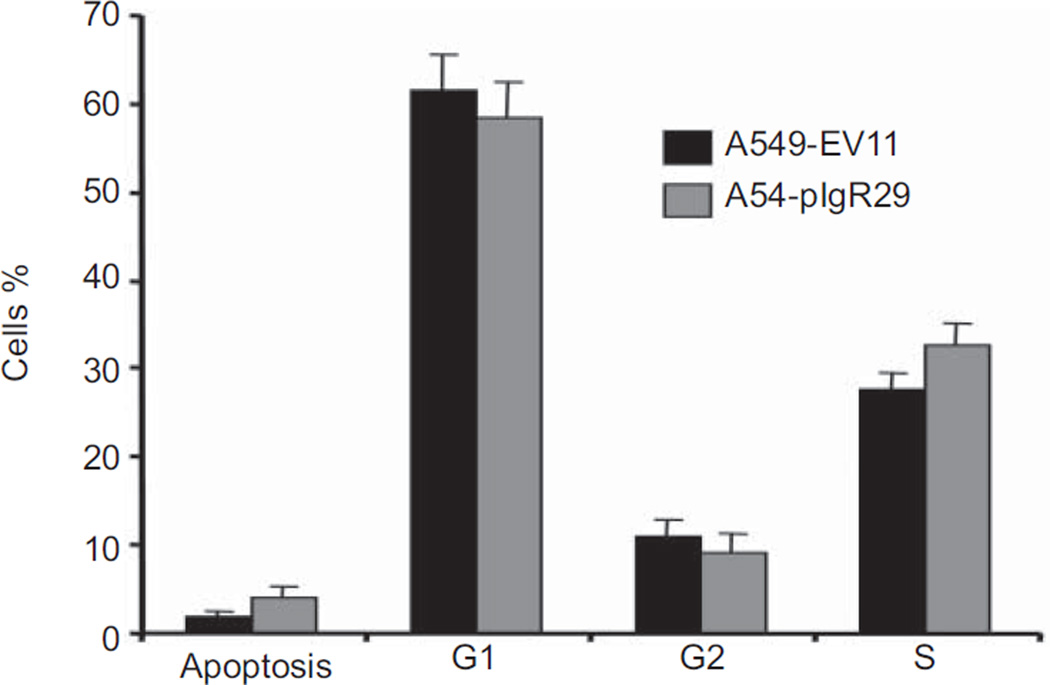
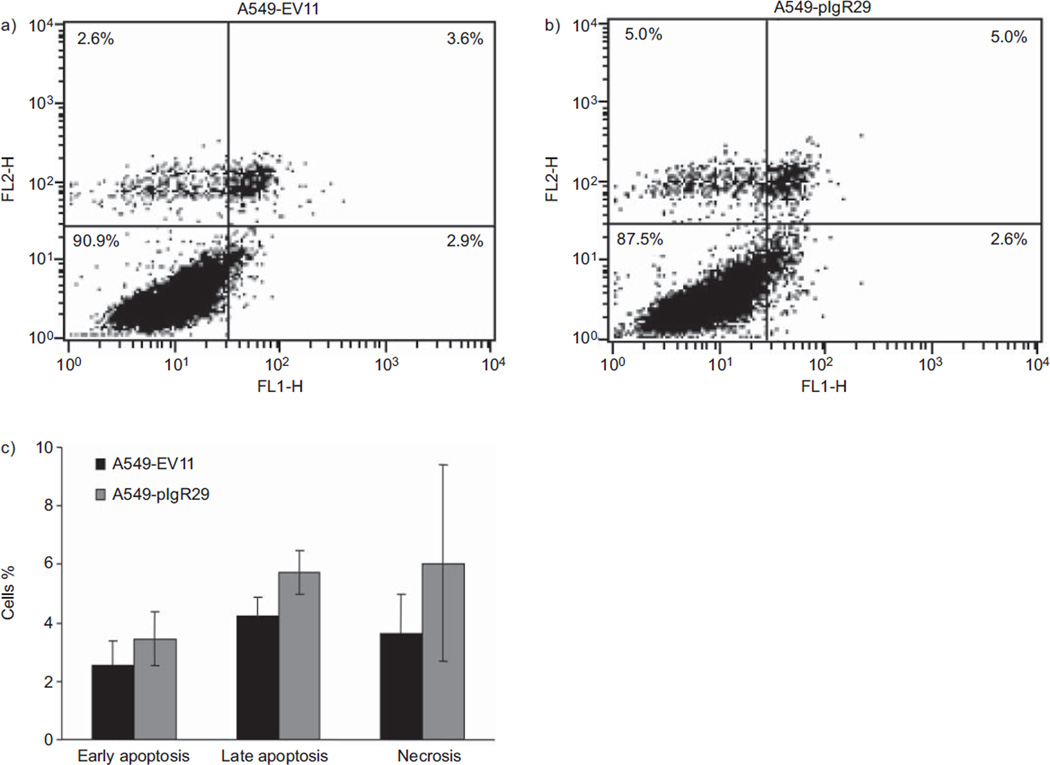
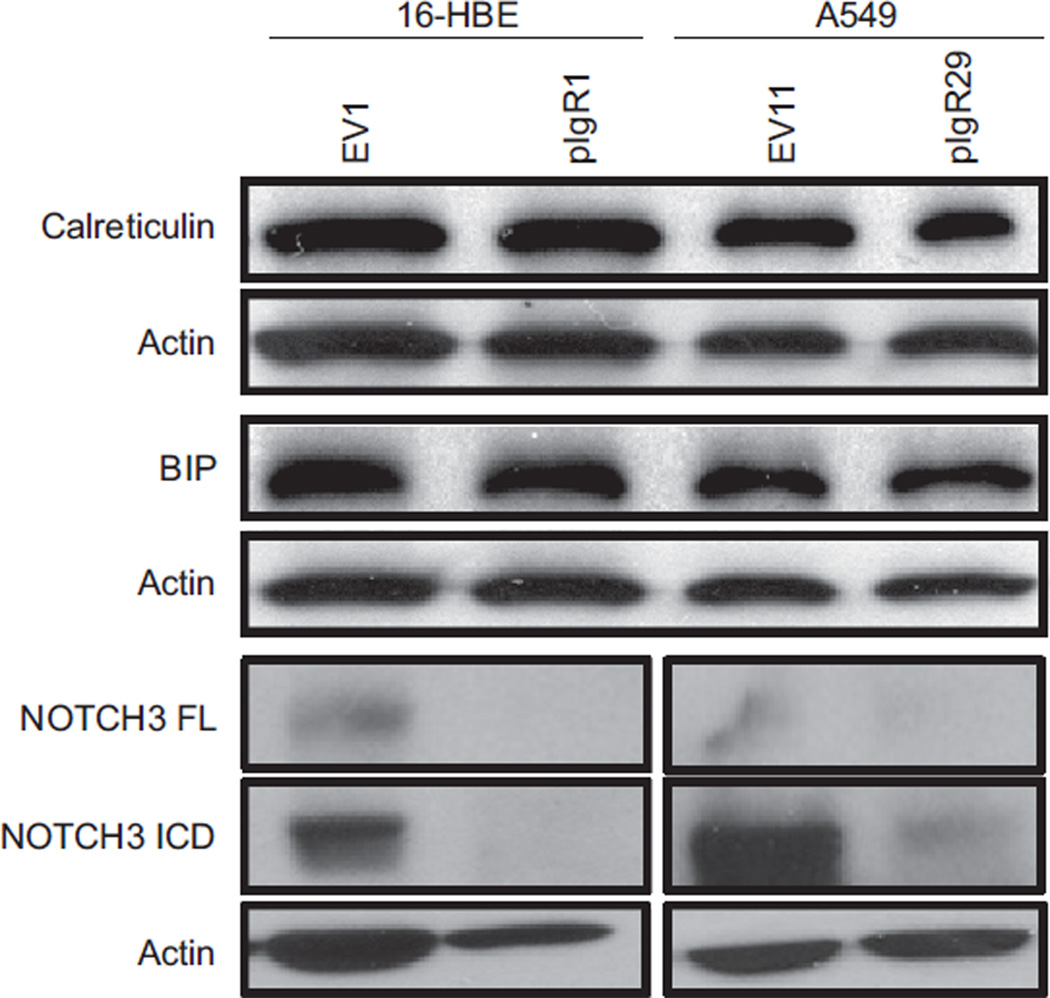
To understand the mechanisms by which pIgR decreased cell proliferation, we questioned whether it induces cell cycle arrest and/or increases apoptosis. In our experiments, pIgR transfection did not impact on cell cycle in A549-pIgR29, independently of serum starvation for ≤26 h (fig. 4). The apoptotic rate of A549-pIgR29 at baseline was not significantly different than that of A549-EV11, but there was a tendency for increased apoptosis and necrosis in A549-pIgR29 (fig. 5), which may contribute to the decreased cell proliferation. Next, we questioned whether growth inhibition was related to endoplasmic reticulum (ER) stress induced by pIgR sequestration in the ER. Expression of calreticulin and BIP, two ER stress markers, was not modified in A549-pIgR29 and 16-HBE-pIgR1 (fig. 6), suggesting the absence of ER stress.
FIGURE 4.
Effects of polymeric immunoglobulin receptor (pIgR) overexpression on A549 cell cycle evaluated by flow cytometry with nuclear propidium iodide staining. The percentage of cells in pre-G1 (apoptosis), G1, G2 and S phases was not statistically different in A549-EV11 and A549-pIgR29. Average of three independent experiments is shown. Error bars represent standard deviation (n=3). The raw data are shown in figure S1.
FIGURE 5.
Effects of polymeric immunoglobulin receptor (pIgR) overexpression on A549 apoptosis evaluated by annexin V apoptosis assay. Stably transfected cells were stained with annexin V and propidium iodide (PI), and the staining was quantified by fluorescence-activated cell sorting analysis. Quadrant dot blot analysis of a) A549-EV11 and b) A549-pIgR29 cells stained for apoptosis using an annexin V detection of phosphatidyl serine expression along the x-axis, counterstained with PI to detect late apoptotic or necrotic cells along the y-axis. In the lower left quadrant, viable nonapoptotic cells are found, and in the lower right quadrant, apoptotic cells binding annexin V. Cells in the upper right are late apoptotic cells binding annexin V and taking up PI, and in the upper left are necrotic cells taking up PI but not binding annexin V. These figures are representative of three independent experiments. c) The spontaneous level of early apoptosis, late apoptosis and necrosis had a tendency to be higher in cells overexpressing pIgR (A549-pIgR29), but the difference was not statistically significant. Average of three independent experiments is shown. Error bars represent standard deviation (n=3).
FIGURE 6.
Effects of stable polymeric immunoglobulin receptor (pIgR) overexpression on endoplasmic reticulum (ER) stress markers evaluated by immunoblotting. Whole cell lysates were resolved by sodium dodecylsulfate–polyacrylamide gel electrophoresis, and blots were incubated with anti-pIgR, calreticulin, BIP and NOTCH3 antibodies. Expression levels of these proteins were normalised to actin. pIgR overexpression did not affect the expression of the ER stress markers calreticulin and BIP, but decreased NOTCH3 expression. FL: full length; ICD: intracellular domain.
pIgR overexpression is associated with NOTCH3 downregulation
In search of a mechanism explaining the inhibitory effect of pIgR on cell growth, we investigated whether the expression of downstream target genes was modified. We used an Affymetrix gene expression array, and compared the gene expression profiles of A549-pIgR29 and A549-EV11. Among 16,830 genes represented on the array, 46 were significantly upregulated (more than two-fold) (table S1) and 40 were significantly downregulated (more than two-fold) (table S2) in A549-pIgR29. We determined that differentially expressed genes were encoding for membrane and secreted proteins. While membrane proteins represent only 20–30% of total cell proteins [22], 20 (43%) out of 46 of upregulated and 18 (45%) out of 40 of downregulated genes in A549-pIgR29 encoded membrane proteins. Moreover, five upregulated and 10 downregulated genes encoded secreted proteins. Overall, 25 (54%) out of 46 upregulated and 28 (70%) out of 40 downregulated genes encoded membrane or secreted proteins. Among the upregulated genes, we found pIgR and eight genes involved in induction of differentiation (EGR1, GATA3, IL24, NDN, PLCB4, Rab3B, SPARC and TLE4). There were also six genes implicated in tumour suppression through cell adhesion and cell–cell/cell–matrix interactions (COL13A1, CYR61, FRMD3, PPAP2B, SPARC and TFPI2). Among the downregulated genes, we found genes involved in metabolic reactions contributing to tumour growth (AQP3 and CA12), inhibition of differentiation (NOTCH3), and migration and invasion (CNTN1, CSPG2, FN1 and VAV3).
We observed that NOTCH3 expression was significantly decreased in A549-pIgR29 and 16-HBE-pIgR1 (fig. 6), validating, at the protein level, the observations we made by gene expression array. We also tested NOTCH3 expression in lung cancer and normal tissues previously tested for pIgR expression (fig. 2a). We found that NOTCH3 and pIgR were inversely correlated (pIgR upregulation was associated with NOTCH3 downregulation and pIgR downregulation was associated with NOTCH3 upregulation) in three out of five lung ADCs and three out of five SCCs (fig. S2).
DISCUSSION
The main objectives of this study were to test whether pIgR expression is downregulated in lung pre-invasive lesions, has prognostic implications in patients with lung cancer and plays a role in cellular functions important to cancer development/progression.
To address the first hypothesis, we analysed pIgR expression by IHC in 10 lung pre-invasive lesions. We found that pIgR expression was lost in all tested tissues, suggesting that its downregulation occurs early in lung tumourigenesis. This is a first report of pIgR expression in pre-invasive bronchial lesions. Our results are consistent with observations made in colorectal cancer development [16, 18] and suggest a broader role of this protein in tumourigenesis.
To test whether pIgR staining is associated with clinical outcomes, we analysed pIgR expression by IHC in lung cancer tissues from 175 patients, and correlated staining scores with clinical and pathological characteristics. pIgR expression was absent in all undifferentiated NSCLCs, 90% of SCCs and 52% of ADCs. Overall, pIgR expression was significantly higher in lung cancers of ADC subtype, which is consistent with previous observations [7, 9, 13]. pIgR expression was also significantly higher in younger patients. Decreased pIgR expression with ageing was reported in rat liver [23], and mouse kidney and gut [24], but never in human lung. Mechanisms of this age-dependent downregulation are unknown, even though post-transcriptional modifications have been suggested in rat [23]. In our study, pIgR expression was not associated with disease stage or survival. Results were similar whether the analysis was performed with all 175 patients or limited to the 98 patients with lung ADC. This is in contrast with observations made in other cancers. In gastro-oesophageal ADCs, tumours without pIgR expression had more frequent lymph node metastases, suggesting that pIgR downregulation identifies a more aggressive phenotype [25]. In colorectal cancers, several studies proposed pIgR as a prognostic biomarker [14, 16–18, 26]. In three of them, pIgR expression was associated with histological grade [14, 16, 18]. In another study, tumours with low pIgR expression had higher disease stage, shorter recurrence-free survival and increased number of tumour-related deaths [17]. In most of our lung cancer tissues expressing pIgR, staining was heterogeneous, which may explain the absence of association with stage or survival and limit the prognostic value of pIgR.
Finally, to determine whether pIgR plays a role in functions important to cancer development/progression, we tested the effect of pIgR overexpression on proliferation of stably transfected A549 cells. We found that pIgR overexpression significantly reduced their proliferation over time and in a dose-dependent manner. This suggests that loss of pIgR expression contributes to lung tumourigenesis and is, to the best of our knowledge, a first report of an effect of pIgR on cell proliferation.
We further pursued our investigations to understand the mechanisms by which pIgR decreases cell proliferation. Our studies showed no evidence for cell cycle arrest or ER stress induced by pIgR overexpression. However, we observed a tendency for increased apoptosis and necrosis in A549-pIgR29, which may contribute to growth inhibition. We also analysed the gene expression profiles of A549-pIgR29 and A549-EV11 by transcriptome analysis. Among differentially expressed genes, we found a high proportion of genes encoding membrane and secreted proteins, with several upregulated genes involved in induction of differentiation, and a downregulated gene (NOTCH3) involved in inhibition of differentiation in A549-pIgR29. Moreover, several genes upregulated in A549-pIgR29 were involved in cell adhesion and cell–cell/cell–matrix interactions, which regulate cell differentiation. Based on the observation that pIgR is expressed in a highly polarised way in epithelial cells [1] and that disruption of cell polarity disturbs pIgR trafficking [27], it was previously hypothesised that loss of pIgR expression may have a role in tumour progression through disruption of cell architecture or physiology [28]. Taken together, our results also suggest that inhibition of proliferation in A549-pIgR29 may be related to increased cellular differentiation. Disruption of normal differentiation is recognised as a hallmark of human cancers [29], and studies in many cell types have shown that proliferation and differentiation are inversely correlated processes, independent of the cell cycle [30]. Therefore, pIgR may have an indirect role in the inhibition of cell proliferation by maintaining cellular integrity and differentiation.
To test this hypothesis, we evaluated NOTCH3 protein expression in stably transfected A549 and 16-HBE cells. NOTCH3 is a cell surface receptor overexpressed in several epithelial malignancies, including lung cancers [31] and preventing normal terminal differentiation during oncogenesis [32]. We found that pIgR overexpression in A549 and 16-HBE induced NOTCH3 downregulation, validating the observations made by gene expression analysis. However, all these analyses were performed with cell lines grown under nonpolarising conditions, which make it difficult to support the hypothesis that increased differentiation is the mechanism by which pIgR inhibits cell proliferation. We also tested NOTCH3 expression in primary human lung cancers and, interestingly, we found that NOTCH3 and pIgR were inversely correlated in six out of 10 lung cancer tissues. Further studies are warranted to determine the interplay between pIgR and NOTCH3 in cellular differentiation.
In summary, loss of pIgR expression in lung pre-invasive lesions suggests that pIgR downregulation is an early event during lung tumourigenesis that is not associated with survival. pIgR over-expression in the lung ADC cell line A549 induced inhibition of cell proliferation, upregulation of genes involved in cell differentiation and downregulation of a gene involved in inhibition of differentiation, NOTCH3 [32], suggesting that pIgR may play a role in lung tumourigenesis by maintaining cell differentiation.
Supplementary Material
ACKNOWLEDGEMENTS
SUPPORT STATEMENT
This work was partially supported by a Discovery grant from Vanderbilt University (Nashville, TN, USA) and the Vanderbilt Specialized Programs of Research Excellence in Lung Cancer (CA90949). S. Ocak was supported by a grant (Bourse Clinicien-Chercheur) from UniversitéCatholique de Louvain (Brussels, Belgium).
We thank C.S. Kaetzel (University of Kentucky, Lexington, KY, USA) for kindly providing human pIgR cDNA in pcDNA3.1(–). We thank the Vanderbilt University DNA Microarray Core Facility (Nashville, TN, USA) for performing Affymetrix gene expression microarray analysis.
Footnotes
This article has supplementary material available from www.erj.ersjournals.com
STATEMENT OF INTEREST
None declared.
REFERENCES
- 1.Kaetzel CS, Mostov K. Immunoglobulin transport and the polymeric immunoglobulin receptor. In: Mestecky J, Bienenstock J, Lamm M, et al., editors. Mucosal Immunology. 3rd Edn. San Diego: Academic Press; 2005. pp. 211–250. [Google Scholar]
- 2.Norderhaug IN, Johansen FE, Schjerven H, et al. Regulation of the formation and external transport of secretory immunoglobulins. Crit Rev Immunol. 1999;19:481–508. [PubMed] [Google Scholar]
- 3.Pilette C, Ouadrhiri Y, Godding V, et al. Lung mucosal immunity: immunoglobulin-A revisited. Eur Respir J. 2001;18:571–588. doi: 10.1183/09031936.01.00228801. [DOI] [PubMed] [Google Scholar]
- 4.Underdown BJ, Schiff JM. Immunoglobulin A: strategic defense initiative at the mucosal surface. Annu Rev Immunol. 1986;4:389–417. doi: 10.1146/annurev.iy.04.040186.002133. [DOI] [PubMed] [Google Scholar]
- 5.Pilette C, Godding V, Kiss R, et al. Reduced epithelial expression of secretory component in small airways correlates with airflow obstruction in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2001;163:185–194. doi: 10.1164/ajrccm.163.1.9912137. [DOI] [PubMed] [Google Scholar]
- 6.Delacroix DL, Marchandise FX, Francis C, et al. Alpha-2-macro-globulin, monomeric and polymeric immunoglobulin A, and immunoglobulin M in bronchoalveolar lavage. Am Rev Respir Dis. 1985;132:829–835. doi: 10.1164/arrd.1985.132.4.829. [DOI] [PubMed] [Google Scholar]
- 7.Brown RW, Clark GM, Tandon AK, et al. Multiple-marker immunohistochemical phenotypes distinguishing malignant pleural mesothelioma from pulmonary adenocarcinoma. Hum Pathol. 1993;24:347–354. doi: 10.1016/0046-8177(93)90080-z. [DOI] [PubMed] [Google Scholar]
- 8.Espinoza CG, Balis JU, Saba SR, et al. Ultrastructural and immunohistochemical studies of bronchiolo-alveolar carcinoma. Cancer. 1984;54:2182–2189. doi: 10.1002/1097-0142(19841115)54:10<2182::aid-cncr2820541019>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 9.Harris JP, South MA. Secretory component: a glandular epithelial cell marker. Am J Pathol. 1981;105:47–53. [PMC free article] [PubMed] [Google Scholar]
- 10.Kawai T, Torikata C, Suzuki M. Immunohistochemical study of pulmonary adenocarcinoma. Am J Clin Pathol. 1988;89:455–462. doi: 10.1093/ajcp/89.4.455. [DOI] [PubMed] [Google Scholar]
- 11.Kondi-Paphitis A, Addis BJ. Secretory component in pulmonary adenocarcinoma and mesothelioma. Histopathology. 1986;10:1279–1287. doi: 10.1111/j.1365-2559.1986.tb02571.x. [DOI] [PubMed] [Google Scholar]
- 12.Loosli H, Hurlimann J. Immunohistological study of malignant diffuse mesotheliomas of the pleura. Histopathology. 1984;8:793–803. doi: 10.1111/j.1365-2559.1984.tb02395.x. [DOI] [PubMed] [Google Scholar]
- 13.Popper H, Wirnsberger G, Hoefler H, et al. Immunohistochemical and histochemical markers of primary lung cancer, lung metastases, and pleural mesotheliomas. Cancer Detect Prev. 1987;10:167–174. [PubMed] [Google Scholar]
- 14.Arends JW, Wiggers T, Thijs CT, et al. The value of secretory component (SC) immunoreactivity in diagnosis and prognosis of colorectal carcinomas. Am J Clin Pathol. 1984;82:267–274. doi: 10.1093/ajcp/82.3.267. [DOI] [PubMed] [Google Scholar]
- 15.Chang Y, Lee TC, Li JC, et al. Differential expression of osteoblast-specific factor 2 and polymeric immunoglobulin receptor genes in nasopharyngeal carcinoma. Head Neck. 2005;27:873–882. doi: 10.1002/hed.20253. [DOI] [PubMed] [Google Scholar]
- 16.Isaacson P. Immunoperoxidase study of the secretory immuno-globulin system in colonic neoplasia. J Clin Pathol. 1982;35:14–25. doi: 10.1136/jcp.35.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koretz K, Schlag P, Quentmeier A, et al. Evaluation of the secretory component as a prognostic variable in colorectal carcinoma. Int J Cancer. 1994;57:365–370. doi: 10.1002/ijc.2910570313. [DOI] [PubMed] [Google Scholar]
- 18.Krajci P, Meling GI, Andersen SN, et al. Secretory component mRNA and protein expression in colorectal adenomas and carcinomas. Br J Cancer. 1996;73:1503–1510. doi: 10.1038/bjc.1996.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khattar NH, Lele SM, Kaetzel CS. Down-regulation of the polymeric immunoglobulin receptor in non-small cell lung carcinoma: correlation with dysregulated expression of the transcription factors USF and AP2. J Biomed Sci. 2005;12:65–77. doi: 10.1007/s11373-004-8185-5. [DOI] [PubMed] [Google Scholar]
- 20.Qian J, Zou Y, Rahman JS, et al. Synergy between phosphatidy-linositol 3-kinase/Akt pathway and Bcl-xL in the control of apoptosis in adenocarcinoma cells of the lung. Mol Cancer Ther. 2009;8:101–109. doi: 10.1158/1535-7163.MCT-08-0973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tamer CM, Lamm ME, Robinson JK, et al. Comparative studies of transcytosis and assembly of secretory IgA in Madin-Darby canine kidney cells expressing human polymeric Ig receptor. J Immunol. 1995;155:707–714. [PubMed] [Google Scholar]
- 22.Stevens TJ, Arkin IT. Do more complex organisms have a greater proportion of membrane proteins in their genomes? Proteins. 2000;39:417–420. doi: 10.1002/(sici)1097-0134(20000601)39:4<417::aid-prot140>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 23.van Bezooijen RL, Wang RK, Lechner MC, et al. Aging effects on hepatic NADPH cytochrome P450 reductase, CYP2B1&2, and polymeric immunoglobulin receptor mRNAs in male Fischer 344 rats. Exp Gerontol. 1994;29:187–195. doi: 10.1016/0531-5565(94)90050-7. [DOI] [PubMed] [Google Scholar]
- 24.Yanagihara T, Kumagai Y, Norose Y, et al. Age-dependent decrease of polymeric Ig receptor expression and IgA elevation in ddY mice: a possible cause of IgA nephropathy. Lab Invest. 2004;84:63–70. doi: 10.1038/labinvest.3700012. [DOI] [PubMed] [Google Scholar]
- 25.Gologan A, Acquafondata M, Dhir R, et al. Polymeric immuno-globulin receptor-negative tumours represent a more aggressive type of adenocarcinomas of distal oesophagus and gastroesophageal junction. Arch Pathol Lab Med. 2008;132:1295–1301. doi: 10.5858/2008-132-1295-PIRTRA. [DOI] [PubMed] [Google Scholar]
- 26.Rognum T, Elgjo K, Brandtzaeg P, et al. Plasma carcinoembryonic antigen concentrations and immunohistochemical patterns of epithelial marker antigens in patients with large bowel carcinoma. J Clin Pathol. 1982;35:922–933. doi: 10.1136/jcp.35.9.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chintalacharuvu KR, Piskurich JF, Lamm ME, et al. Cell polarity regulates the release of secretory component, the epithelial receptor for polymeric immunoglobulins, from the surface of HT-29 colon carcinoma cells. J Cell Physiol. 1991;148:35–47. doi: 10.1002/jcp.1041480105. [DOI] [PubMed] [Google Scholar]
- 28.Traicoff JL, De Marchis L, Ginsburg BL, et al. Characterization of the human polymeric immunoglobulin receptor (PIGR) 39UTR and differential expression of PIGR mRNA during colon tumorigenesis. J Biomed Sci. 2003;10:792–804. doi: 10.1159/000073967. [DOI] [PubMed] [Google Scholar]
- 29.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 30.Brown G, Hughes PJ, Michell RH. Cell differentiation and proliferation – simultaneous but independent? Exp Cell Res. 2003;291:282–288. doi: 10.1016/s0014-4827(03)00393-8. [DOI] [PubMed] [Google Scholar]
- 31.Dang TP, Gazdar AF, Virmani AK, et al. Chromosome 19 translocation, overexpression of Notch3, and human lung cancer. J Natl Cancer Inst. 2000;92:1355–1357. doi: 10.1093/jnci/92.16.1355. [DOI] [PubMed] [Google Scholar]
- 32.Dang TP, Eichenberger S, Gonzalez A, et al. Constitutive activation of Notch3 inhibits terminal epithelial differentiation in lungs of transgenic mice. Oncogene. 2003;22:1988–1997. doi: 10.1038/sj.onc.1206230. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.