SUMMARY
T-bet is a critical transcription factor for T helper-1 (Th1) cell differentiation. To study the regulation and functions of T-bet, we developed a T-bet-ZsGreen reporter mouse strain. We determined that interleukin-12 (IL-12) and interferon-γ (IFN-γ) were redundant in inducing T-bet in mice infected with Toxoplasma gondii and that T-bet did not contribute to its own expression when induced by IL-12 and IFN-γ. By contrast, T-bet and the transcription factor Stat4 were critical for IFN-γ production whereas IFN-γ signaling was dispensable for inducing IFN-γ. Loss of T-bet resulted in activation of an endogenous program driving Th2 cell differentiation in cells expressing T-bet-ZsGreen. Genome-wide analyses indicated that T-bet directly induced many Th1 cell-related genes but indirectly suppressed Th2 cell-related genes. Our study revealed redundancy and synergy among several Th1 cell-inducing pathways in regulating the expression of T-bet and IFN-γ, and a critical role of T-bet in suppressing an endogenous Th2 cell-associated program.
INTRODUCTION
CD4+ T helper (Th) cells play critical roles in orchestrating adaptive immune responses to pathogens, mainly through cytokine production (Murphy and Reiner, 2002; Zhu et al., 2010). They are also involved in pathological responses to self-antigens and to non-harmful allergens, resulting in autoimmune and allergic diseases, respectively. Activated CD4+ T cells are classified based on their capacity to produce unique cytokines, on their distinct homing properties and on their particular functions during various immune responses. They develop from naïve CD4+ T cells when antigen-specific T cell receptors (TCRs) as well as cytokine receptors on these cells are activated.
In vitro, interleukin-12 (IL-12)- and interferon (IFN)-γ-mediated signaling, through the activation of the transcription factors Stat4 and Stat1 respectively, together with TCR activation, are important for T helper type 1 (Th1) cell differentiation. IL-2 and IL-4 are critical for T helper type 2 (Th2) cell differentiation (Ansel et al., 2006; Oestreich and Weinmann, 2012; Zhu et al., 2010).
T-bet is a critical transcription factor of Th1 cells. It plays an important role in regulating IFN-γ production (Szabo et al., 2000; Szabo et al., 2002). Naïve CD4+ T cells do not express T-bet. IFN-γ has been shown to induce T-bet expression, which results in a potential positive feedback loop during Th1 cell differentiation (Afkarian et al., 2002; Lighvani et al., 2001). IL-12 also induces T-bet in an IFN-γ-Stat1-independent manner in CD8+ T cells (Yang et al., 2007). The relative importance of IL-12 and IFN-γ in T-bet induction as well as IFN-γ expression especially during in vivo Th1 responses has not been established. Furthermore, retroviral T-bet has been reported to induce endogenous T-bet expression (Mullen et al., 2001); however, whether endogenous T-bet directly regulates its own expression or does so indirectly through its up-regulation of IFN-γ and/or IL-12Rβ2 is still unclear.
Similarly, the mechanism of Th2 cell differentiation in vivo has been an enigma since the earliest demonstration of the Th2 cell differentiation process. IL-4 plays an essential role for Th2 cell differentiation in vitro. Assuming a comparable role for IL-4 in vivo, the source of such IL-4 remains controversial, particularly since professional antigen presenting cells (APCs), such as dendritic cells, do not produce IL-4. Recently, it has been proposed that basophils serve as Th2 cell-promoting APCs and that they produce IL-4 and/or thymic stromal lymphpoietin (TSLP) for such a purpose (Perrigoue et al., 2009; Sokol et al., 2008; Sokol et al., 2009; Yoshimoto et al., 2009). However, the role of basophils in antigen presentation remains controversial (Kim et al., 2010). Furthermore, Th2 cell differentiation in several, but not all, parasite infection models occurs in IL-4-, IL-4R or Stat6-deficient mice indicating that IL-4-mediated signaling is not essential for initiating Th2 cell differentiation in vivo (Finkelman et al., 2000; Jankovic et al., 2000; Min et al., 2004; Voehringer et al., 2004). It has been proposed that Th2 cell differentiation may occur through a default pathway; however, CD4+ T cells in IL-12-deficient mice fail to default to Th2 cells in response to intracellular pathogens (Jankovic et al., 2002).
GATA3 is the critical transcription factor for Th2 cell differentiation both in vitro and in vivo (Pai et al., 2004; Zhang et al., 1997; Zheng and Flavell, 1997; Zhu et al., 2004). GATA3 is also indispensable for CD4+ T cell development in the thymus at multiple stages and thus, unlike T-bet which is not expressed in naïve CD4+ T cells, GATA3 is detected in naïve CD4+ T cells (Ho et al., 2009). IL-4-mediated Stat6 activation is the main inducing signal for GATA3 up-regulation in vitro. T cell receptor activation, particularly with low dose antigen, also up-regulates GATA3 expression (Yamane et al., 2005). On the other hand, GATA3 is down-regulated during Th1 cell differentiation. Th2 cells may develop with low amounts of GATA3 expression but even in such cases GATA3 is essential (Zhu et al., 2003); thus, constitutive expression of GATA3 at certain amounts in naïve CD4+ T cells is consistent with an endogenous Th2 program. The failure of CD4+ T cells to default to Th2 cells in an IL-12-deficient setting suggests either this endogenous Th2 cell-inducing program does not exist or there are other Th1 cell-inducing signals, such as IFN-γ, that are responsible for suppressing this program.
To study the CD4+ T cell differentiation process and cross-regulation both in vitro and in vivo, we developed a T-bet reporter mouse strain. Using this research tool, we observed that IL-12 and IFN-γ were redundant in inducing T-bet during in vitro culture and in vivo immune responses to T. gondii. IFN-γ signaling was dispensable for generating IFN-γ-producing T cells under the conditions tested. Furthermore, T-bet collaborated with Stat4 in inducing IFN-γ production but was not required for its own expression in the presence of IL-12 and IFN-γ. When T-bet was absent, T. gondii infection elicited expression of the T-bet reporter while inducing a Th2 cell differentiation program, including IL-4 production and GATA3 up-regulation, in the same cells. Our result suggests that an endogenous program for in vivo Th2 cell differentiation exists and that it is normally repressed by T-bet during Th1 cell differentiation. In the absence of T-bet, this program can be activated in the absence of Th2 cell-stimulating cytokines.
RESULTS
T-bet-ZsGreen reporter faithfully reflects the expression of endogenous T-bet
To study the regulation of T-bet expression and the functions of T-bet-expressing cells both in vitro and in vivo, we generated a bacterial artificial chromosome (BAC) transgenic T-bet reporter mouse strain. The coding region of ZsGreen (ZsG), an improved version of green fluorescent protein (GFP), was inserted into the T-bet translational start site in the BAC clone RP23-237M14. Based on such design, ZsG but not T-bet protein will be expressed from the transgenic BAC; T-bet will continue to be expressed from the endogenous Tbx21 gene. The reporter strain was designated the T-bet green reporter (TBGR).
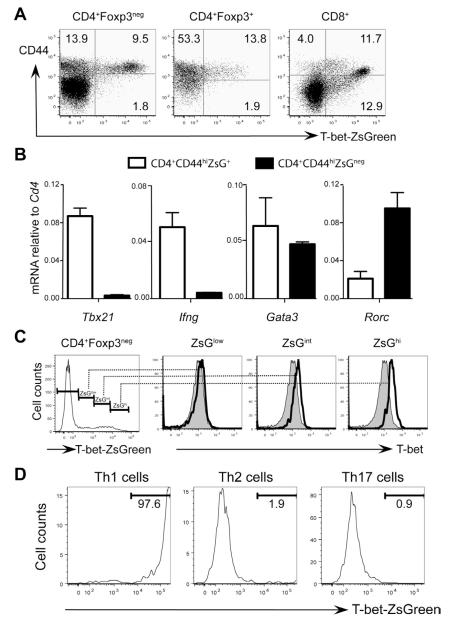
ZsG expression was detected in subsets of splenocytes and lymph node cells from unimmunized TBGR mice (Figures 1A and S1). ~40% of memory phenotype splenic CD4+ T cells (CD4+Foxp3negCD44hi cells) were ZsG positive (ZsG+). All the CD4+CD44low cells were ZsG negative (ZsGneg). A proportion of regulatory T cells (CD4+Foxp3+) cells (~15%) were found to express intermediate amounts of ZsG. In the CD8+ T cell compartment, all the cells that expressed high amounts of ZsG were CD44hi. Some cells that expressed intermediate amounts of ZsG and CD44 may represent cells at a transition stage from naïve to memory-like CD8+ T cells.
Figure 1. T-bet-ZsGreen reporter faithfully reflects the expression of endogenous T-bet.
(A) Splenocytes of naïve TBGR mice were stained for CD4, CD8, CD44 and Foxp3. Dot plots were gated on different populations as indicated.
(B) CD4+CD44hi cells from naïve TBGR mice were separated into ZsG+ and ZsGneg population by cell sorting. The relative expression amounts of different genes normalized to Cd4 were assessed by quantitative PCR after reverse transcription.
(C) Splenocytes from naïve TBGR mice were stained for CD4, Foxp3 and T-bet. Histograms were gated on different populations based on ZsG intensity (low, intermediate and high) as indicated. Shaded histograms represent T-bet staining in CD4+Foxp3negZsGneg cells.
(D) Sorted naïve CD62LhiCD44loCD25negZsGnegCD4+ T cells from TBGR mice were activated under Th1, Th2 or Th17 cell-polarizing conditions for 3 days. The expression of T-bet-ZsGreen was assessed by flow cytometric analysis.
Numbers indicate % of cells in each quadrant or gate. Error bars represent means+SD. Data are representative of at least two independent experiments.
As expected, Tbet mRNA (Tbx21) was only detected in sorted CD4+CD44hiZsG+ cells (Figure 1B). Only the ZsG+ cells expressed Ifng mRNA. Rorc mRNA was enriched in ZsGneg cells, suggesting the ZsGneg fraction contains Th17 cells. We have previously reported that Th17 cells express lower GATA3 than Th1 cells do (Wei et al., 2011). The fact that Gata3 mRNA was not differentially expressed in the ZsG+ and ZsGneg cells suggests that the ZsGneg fraction contains some Th2 cells (Figure 1B). A comparison of splenic CD4+Foxp3neg T cells showed an excellent correlation between expression of ZsG and T-bet as assessed by flow cytometric analysis (Figure 1C). These results indicate that the ZsG reporter in TBGR mice faithfully reflects T-bet-expressing cells developed in vivo and that ZsG is much more powerful than anti-T-bet in detecting and separating T-bet-expressing cells from T-bet-non-expressing cells.
To evaluate T-bet-ZsGreen expression under conditions of Th cell polarization, naïve CD4+ T cells (CD4+CD62LhiCD44lowCD25negZsGneg) were stimulated with soluble anti-CD3 plus anti-CD28 under Th1, Th2 or Th17 (IL-17-producing T helper cells) cell-polarizing conditions for 3 days in vitro. Almost all cells in the Th1 cell-polarizing culture (>95%) expressed high amounts of ZsG, whereas, under Th2 or Th17 cell-polarizing conditions, most if not all (>98%) the cells remained ZsG negative (Figure 1D). These results confirm that the ZsG reporter faithfully reflects T-bet-expressing Th1 cells.
IL-12 and IFN-γ are redundant in inducing T-bet expression
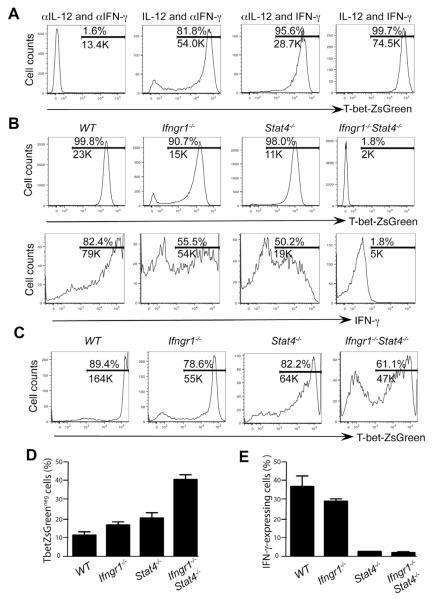
Both IL-12 and IFN-γ have been reported to up-regulate T-bet expression (Oestreich and Weinmann, 2012); however, their relative contribution in inducing T-bet is unknown. Using CD4+ T cells from TBGR mice, we studied T-bet induction by IL-12 and IFN-γ in cells stimulated with anti-CD3 and anti-CD28. When IL-12 and IFN-γ were both neutralized during in vitro priming, no ZsG induction was detected, whereas virtually all the TBGR CD4+ T cells were ZsG+ when both IL-12 and IFN-γ were added to the culture (Figure 2A). Addition of either IL-12 or IFN-γ alone, while neutralizing the alternative cytokine, induced ZsG expression in the majority of the cells although the mean fluorescence intensity (MFI) was somewhat lower.
Figure 2. IL-12 and IFN-γ are redundant in inducing T-bet expression.
(A) Naïve CD4+ T cells were cultured under modified Th1 cell-polarizing conditions as indicated (with anti-IL-4 included) for 3 days.
(B) Naïve CD4+ T cells from various strains were cultured under Th1 cell-polarizing conditions (IL-12 and anti-IL-4) in the presence of exogenous IFN-γ for 3 days.
(C-E) Various mouse strains were infected with T. gondii for 7 days. Cells harvested from spleens were characterized.
The expression of T-bet-ZsGreen was assessed by flow cytomteric analysis. IFN-γ production was assessed by intracellular staining after stimulation with PMA plus ionomycin (B) or plate-bound anti-CD3 plus anti-CD28 (E) for 4 hours. Histogram plots were gated on CD4+CD44hi cells. Numbers indicate % and the MFI of the positive cells in each gate. Error bars represent means+SD. Data are representative of three (A, B) and two (C-E) independent experiments.
To rule out the possibility that IL-12 or IFN-γ had been incompletely neutralized, we examined TBGR mice that were singly-(Ifngr1−/− or Stat4−/−) or doubly-(Ifngr1−/−Stat4−/−) deficient for IFN-γR1 and Stat4. Naïve cells from these mice were primed with soluble anti-CD3 and anti-CD28 in the presence of IL-12, IFN-γ and anti-IL-4. Virtually all the wild type (WT) cells were ZsG+, but no ZsG+ cells were detected in Ifngr1−/−Stat4−/− cells in which both IL-12 and IFN-γ signaling are defective (Figure 2B, upper panel). Nonetheless, most of Ifngr1−/− or Stat4−/− TBGR cells expressed ZsG albeit at lower amounts (~50% reduction in MFI) compared to their wild type counterparts. >80% of in vitro-primed wild type cells expressed IFN-γ; ~50% of Ifngr1−/− and Stat4−/− cells were capable of expressing IFN-γ (Figure 2B, lower panel). The MFI of IFN-γ was substantially reduced in the IFN-γ+ cells from Stat4−/− donors while IFN-γ+ cells from Ifngr1−/− mice were almost as bright as those from wild-type donors; no IFN-γ production by Ifngr1−/−Stat4−/− cells was detected. Therefore, either IL-12 or IFN-γ alone can induce T-bet expression in most cells; similarly, these cells are still capable of producing IFN-γ in vitro.
To study T-bet induction by IL-12 and IFN-γ in vivo, we infected TBGR mice with T. gondii, a pathogen that elicits strong Th1 responses (Jankovic et al., 2007), and assessed T-bet and IFN-γ expression one week after infection. Similar to the findings obtained in vitro, deficiency in either IFN-γR1 or Stat4 resulted in a modest reduction in ZsG expression, both in percentage of positive cells and in MFI of ZsG in these cells (Figure 2C). By contrast, some CD4+ T cells from Ifngr1−/−Stat4−/− TBGR mice up-regulated T-bet although the proportion that did so was less than that of singly-deficient cells (Figure 2D). These results indicate that under in vivo conditions T-bet can be induced in an IL-12- and IFN-γ-independent manner and that such inducing signal(s) are absent in our in vitro Th1 cell-polarizing conditions. Addition of type I IFNs or IL-27 to the culture of in vitro differentiating Ifngr1−/−Stat4−/− cells resulted in ZsG expression (Figure S2) suggesting these cytokines may be responsible for IL-12- and IFN-independent T-bet induction in vivo.
In contrast to requirements for in vivo T-bet induction, Stat4 deficiency dramatically diminished the capacity of these cells to produce IFN-γ (Figure 2E). Deficiency in IFN-γR1 had a very small effect on IFN-γ production, despite reduced T-bet expression consistent with a previous report that Stat1 is not required for the development of IFN-γ-producing CD4+ T cells (Lieberman et al., 2004). Since Stat4−/− TBGR cells expressed similar amounts of ZsG as Ifngr1−/− TBGR cells did (Figure 2C), the differential IFN-γ production by these cells implies that Stat4 has an effect on IFN-γ production in addition to its role in inducing T-bet and that T-bet without STAT4 does not induce optimal IFN-γ production.
T-bet and Stat4 synergize in inducing IFN-γ production
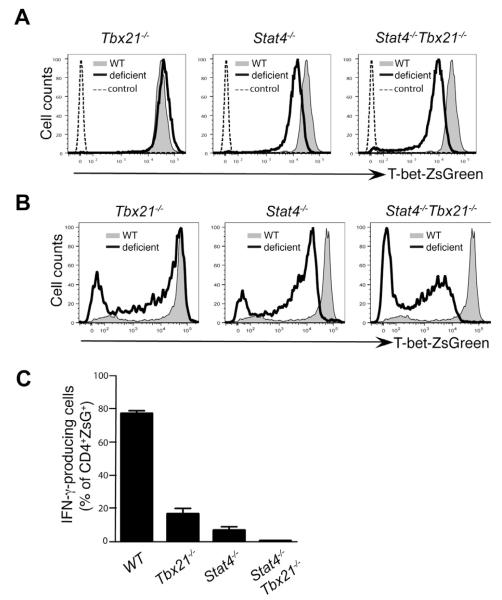
T-bet is regarded as a critical regulator of Th1 cells and directly regulates IFN-γ production by binding to several conserved sites at the Ifng locus (Hatton et al., 2006). However, Stat4 also appears to directly regulate IFN-γ production as shown by the dramatic reduction of IFN-γ production with only a modest decrease of ZsG expression in Stat4−/− TBGR mice. To investigate the relative importance of T-bet and Stat4 in regulating T-bet and IFN-γ production, we generated T-bet singly-deficient (Tbx21−/−) and Stat4-T-bet doubly deficient (Stat4−/−Tbx21−/−) TBGR mice. Our unique model of the T-bet-ZsGreen reporter crossed to Tbx21−/− background also allows us to address the role of endogenous T-bet in controlling its own expression.
Tbx21−/− TBGR cells primed in vitro in the presence of IL-12 and IFN-γ expressed amounts of ZsG similar to comparably treated wild type cells indicating that T-bet is not required for its own expression in the presence of IL-12 and IFN-γ (Figure 3A). A deficiency in Stat4 resulted in a modest reduction of ZsG expression similar to the results shown in Figure 2. T-bet deficiency in the setting of Stat4 deficiency did not result in a further reduction of ZsG expression when the cells were stimulated in the presence of exogenous IL-12 and IFN-γ indicating that T-bet does not participate in IFN-γ-mediated T-bet up-regulation in vitro. Of course, T-bet could still affect its own expression indirectly through its regulation of IFN-γ production. Indeed, at a steady state, Tbx21−/− TBGR cells express lower amounts of ZsG compared to WT TBGR cells (data not shown).
Figure 3. T-bet and Stat4 synergize in inducing IFN-γ production but T-bet does not regulate its own expression in the presence of IL-12 and IFN-γ.
(A) Naïve CD4+ T cells from various strains were cultured under Th1 cell-polarizing conditions (IL-12 and anti-IL-4) in the presence of exogenous IFN-γ for 3 days.
(B-C) Various mouse strains were infected with T. gondii for 7 days. Cells harvested from spleens were characterized.
The expression of T-bet-ZsGreen was assessed by flow cytometric analysis. IFN-γ production was assessed by intracellular staining after stimulation with plate bound anti-CD3 plus anti-CD28 for 4 hours. Histogram plots were gated on CD4+CD44hi cells. Error bars represent means+SD. Data are representative of two independent experiments.
In response to T. gondii infection, Tbx21−/− TBGR cells expressed close to normal amounts of ZsG implying that T-bet is dispensable for its own expression in vivo as well as in vitro (Figure 3B). However, a combination of Stat4 and T-bet deficiency resulted in dramatic reduction in the amounts of ZsG expression in CD4+ T cells from T. gondii-infected mice in the ZsG+ cells. Such a reduction was not observed in in vitro-cultured Stat4−/−Tbx21−/− TBGR cells that had received exogenous IFN-γ. Double deficiency in Stat4 and IFN-γRI in T. gondii-infected mice only resulted in reduction in the frequency of ZsG+ cells but not the mean fluorescence intensity (MFI) of ZsG in ZsG+ cells compared to Stat4 single deficiency (Figure 2C). These results imply that T-bet may participate in its own expression in instances in which the third, yet unknown, signaling pathway is responsible for T-bet expression. Nonetheless, T-bet or Stat4 deficiency alone caused substantial reduction in the frequency of IFN-γ-producing cells and the combination of T-bet and Stat4 deficiency completely abolished IFN-γ production (Figure 3C).
T-bet suppresses IL-4 and GATA3 expression during T. gondii infection
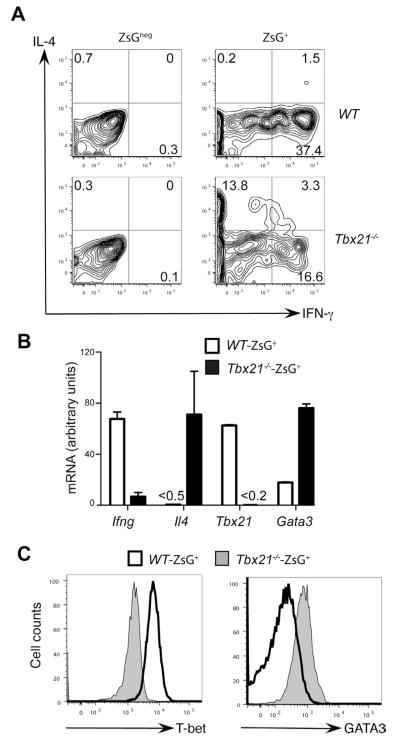
T-bet has been reported to suppress Th2 differentiation when it is over-expressed (Szabo et al., 2000; Usui et al., 2006). When Tbx21−/− mice are infected with Leishmania major, IFN-γ production is dramatically reduced while IL-4 production is induced (Szabo et al., 2002). Since L. major elicits Th1 responses in C57BL/6 mice and Th2 responses in BALB/c mice, it is likely that, in the absence of T-bet, the environment for CD4+ T cell differentiation may have been switched from favoring Th1 to favoring Th2 cell differentiation. On the other hand, T. gondii infection elicits strong Th1 responses both in C57BL/6 and BALB/c mice. The WT and Tbx21−/− TBGR mice allow us to examine the role of T-bet in inducing Th1 and suppressing Th2 responses in the strongly Th1 cell-inducing environment of T. gondii infection; TBGR expression can be used as a readout of CD4+ T cells differentiating towards Th1 direction both in WT and Tbx21−/− TBGR mice.
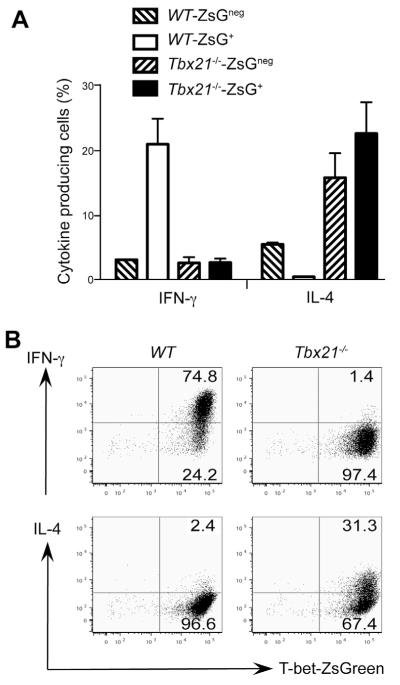
ZsG+ cells from wild type TBGR mice expressed IFN-γ but no IL-4 after T. gondii infection; however, IL-4 production was detected in ~20% of ZsG+ cells from infected Tbx21−/− TBGR mice (Figure 4A). Interestingly, some of these IL-4-producing cells also produced low amounts of IFN-γ. On the other hand, ZsGneg cells from infected T-bet deficient TBGR mice did not produce IL-4. These results suggest that IL-4-producing T-bet-deficient cells developed in the presence of signals typically resulting in Th1 cell differentiation. While Ifng mRNA was strikingly reduced in the ZsG+ cells from T-bet deficient TBGR mice, Il4 mRNA was dramatically induced in these Tbx21−/−ZsG+ “Th1” cells (Figure 4B). Gata3 mRNA was also induced in the absence of T-bet suggesting the signals involved in up-regulating T-bet expression, are not sufficient to directly suppress GATA3 in the absence of T-bet. Intracellular staining for GATA3 confirmed that GATA3 expression was much higher in Tbx21−/−ZsG+ “Th1” cells than in wild type Th1 cells (Figure 4C). These data show that in the absence of T-bet, Tbx21−/− mice are still capable of generating T-bet-inducing signals after T. gondii infection, presumably from innate cells. Indeed, serum concentration of IL-12 was comparable between WT and Tbx21−/− mice that were infected with T. gondii (data not shown).
Figure 4. T-bet suppresses IL-4 and GATA3 expression during T. gondii infection.
Wild type (WT) or T-bet deficient (Tbx21−/−) TBGR mice were infected with T. gondii for 7 days. Cells harvested from spleens were characterized.
(A) IFN-γ and IL-4 production was assessed by intracellular staining after stimulation with plate-bound anti-CD3 plus anti-CD28 for 4 hours. Histogram plots were gated on CD4+ZsGneg and CD4+ZsG+ cells. Numbers indicate % of cells in each quadrant.
(B) CD4+ZsG+ cells from were sorted. The relative expression amounts of different genes were assessed by quantitative PCR after reverse transcription. Error bars represent means+SD.
(C) T-bet and GATA3 expression were assessed by intracellular staining. Histogram plots were gated on CD4+Foxp3negZsG+ cells.
Data are representative of two independent experiments.
Suppression of Th2 cell-specific gene expression by T-bet is CD4+ T cell intrinsic
To determine if the effect of T-bet in suppressing IL-4 is CD4+ T cell intrinsic, we transferred naïve CD4+ T cells from WT or Tbx21−/− TBGR mice into WT recipients prior to infecting the recipients with T. gondii. Unfortunately, the number of transferred cells recovered in this type of experiment was quite low, probably due to massive Th1 response of the endogenous CD4+ T cells. Therefore, we transferred naïve CD4+ T cells from WT and Tbx21−/− TBGR mice into Tcra−/− mice, in which homeostatic proliferation of the transferred cells is expected. Interestingly, 3 weeks after transfer, about half of the transferred cells had become ZsG+ (data not shown). Re-stimulation of transferred cells resulted in IFN-γ production mainly by the ZsG+ cells and modest amounts of IL-4 from the ZsGneg cells from the wild type donors (Figure 5A). In contrast, among cells from the Tbx21−/− donors, 15-20% of both the ZsG+ and ZsGneg cells produced IL-4 but only 2-3% produced IFN-γ. This result indicates that T-bet plays a critical cell intrinsic positive role in Th1 cell differentiation and negative role in Th2 cell differentiation in homeostatic proliferation just as it does in T. gondii infection.
Figure 5. Suppression of Th2 cell-specific gene expression by T-bet is CD4+ T cell intrinsic.
(A) Naive CD4+ T cells from WT and Tbx21−/− TBGR mice were sorted and transferred into Tcra−/− mice. Three weeks later, splenocytes were harvested and cytokine production was assessed by intracellular staining after PMA plus ionomycin stimulation. % of cytokine producing cells was calculated within CD4+ZsGneg and CD4+ZsG+ subsets. Error bars represent means+SD.
(B) Naïve CD4+ T cells were cultured under Th1 cell-polarizing conditions (IL-12 and anti-IL-4) for 3 days. IFN-γ and IL-4 production was assessed by intracellular staining after stimulation with PMA plus ionomycin. Numbers indicate % of cells in each quadrant.
Data are representative of two (A) and three (B) independent experiments.
IL-4 plays a major role in Th2 cell differentiation in vitro. However, many in vivo Th2 cell-associated responses are either independent of the IL-4-Stat6 pathway or only partially dependent upon it. T-bet deficiency leads to up-regulation of Th2 cell signature genes even in the cells expressing the T-bet reporter suggesting that in the absence of T-bet, Th2 cell differentiation proceeds without “conventional” exogenous Th2 cell inducing stimuli. Indeed, culturing naïve Tbx21−/− TBGR CD4+ T cells in vitro under “Th1” cell differentiation conditions (i.e. anti-CD3, anti-CD28, IL-12 and anti-IL-4), although it led to virtually all the responding cells being ZsG+, resulted in a substantial percentage of these cells being able to produce IL-4 in response to phorbol myristate acetate (PMA) and ionomycin (Figure 5B). Thus, under classical Th1 cell-polarizing conditions, in which IL-4 is neutralized, the absence of T-bet allows Th2 cell differentiation to go forward in at least a portion of the cells under study.
T-bet and GATA3 cross-regulation affects many Th1 and Th2 cell-specific genes
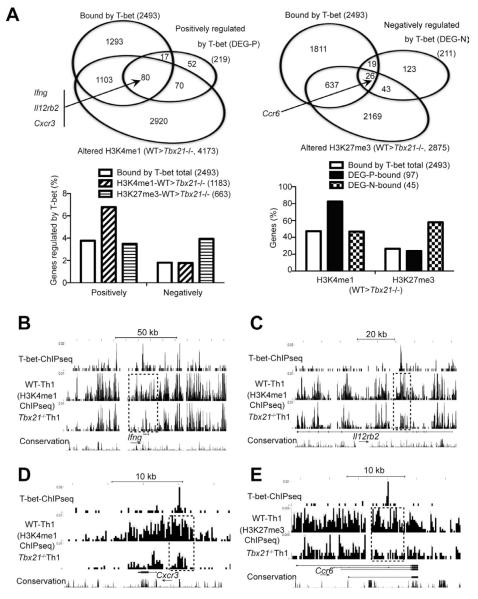
To further explore the mechanisms of T-bet-mediated gene regulation, we performed a T-bet ChIPseq to map T-bet binding sites genome-wide in WT Th1 cells. Modification-specific histone ChIPseq as well as RNAseq were carried out with WT and Tbx21−/− TBGR+ Th1 cells to identify genes regulated by T-bet.
RNAseq results showed that 219 and 211 genes were positively or negatively regulated by T-bet, respectively, with >2 fold change between WT and Tbx21−/− cells (Figure 6A, upper panels). ChIPseq results indicated that T-bet bound to 2493 genes in Th1 cells; in the absence of T-bet, H3K4me1 and H3H27me3 modification peaks within 4173 and 2875 genes, respectively, were reduced (at least one peak per gene). Overall, only ~4% and ~2% of the T-bet bound genes were positively and negatively regulated by T-bet at a transcriptional level, respectively (Figure 6A, lower left). Among the 1183 genes that were bound by T-bet and displayed reduced H3K4me1 modification in the absence of T-bet, the percentage of the genes that were positively regulated by T-bet was increased to ~7%, whereas, the percentage of the genes that were negatively regulated by T-bet remained ~2%. On the other hand, among the 663 genes that were bound by T-bet and displayed reduced H3K27me3 modification in the absence of T-bet, the percentage of the genes that were negatively regulated by T-bet was enriched to ~4%, whereas, the percentage of the genes that are positively regulated by T-bet remained the same as that of the total T-bet bound genes. In the absence of T-bet, 47% and 27% of the total 2493 T-bet bound genes displayed reduced H3K4me1 and H3K27me3, respectively (Figure 6A, lower right). Higher percentages of T-bet bound genes were regulated by T-bet in histones modifications than in gene expression suggesting T-bet may regulate gene expression at an epigenetic level. Among the 97 genes that were bound and positively regulated by T-bet, 82% had reduced H3K4me1 modification in the absence of T-bet whereas among the 45 genes that were bound and negatively regulated by T-bet, 58% displayed reduced H3K27me3 in the absence of T-bet. Therefore, T-bet may promote H3K4me1 or H3K27me3 modifications through which it positively or negatively regulates gene expression.
Figure 6. T-bet directly regulates (either positively or negatively) many key molecules of Th1 and Th17 cells.
Naïve CD4+ T cells from WT and Tbx21−/− TBGR mice were cultured under Th1 cell-polarizing conditions for 3-4 days. After resting in IL-2-containing medium for 2 days, cells were sorted for ZsG+ and further expanded under Th1 cell-polarizing conditions for another 3-4 days. Anti-T-bet and histone ChIPseq as well as RNAseq were carried out.
(A) The Venn diagrams show the overlap of genes that are bound by T-bet, positively (upper left) or negatively (upper right) regulated by T-bet at a transcriptional or epigenetic level in Th1 cells. The association of histone modifications and gene regulation were determined in sub-groups of the genes that are bound by T-bet (lower panels). DEG-P and DEG-N stand for differentially expressed genes (DEG) that are regulated by T-bet positively (P) or negatively (N).
(B-E) UCSC genome browser view of T-bet binding, H3K4me1 or H3K27me3 modifications at the Ifng (B), Il12rb2 (C), Cxcr3 (D) and Ccr6 (E) locus are shown. Dotted boxes indicate differences in epigenetic modifications between WT and Tbx21−/− cells around the T-bet binding sites. Arrows indicate the direction of the genes.
T-bet bound to the Ifng, Il12rb2 and Cxcr3 loci and promoted H3K4me1 modification around the binding sites of the genes (Figure 6B-D). At the Ifng locus, T-bet bound to many sites including CNS-6 (6 kb upstream of transcription start site) at which H3K4me1 modification was dramatically reduced in the absence of T-bet (Figure 6B). Similarly, T-bet bound to an intron of the Il12rb2 and the 5′ enhancer of the Cxcr3; without T-bet, H3K4me1 modification was reduced around these binding sites (Figure 6C and 6D). On the other hand, T-bet directly suppressed the expression of CCR6, a Th17 cell-specific chemokine receptor; T-bet bound to an intron of the Ccr6 gene and promoted H3K27me3 around this binding site. Previously, T-bet has been reported to suppress Runx1-mediated RORγt up-regulation (Lazarevic et al., 2011). While we did not detect increased expression of RORγt in T-bet-deficient “Th1” cells, our data indicate T-bet also binds to the Rorc gene and promotes H3K27me3 around its binding site (Figure S3).
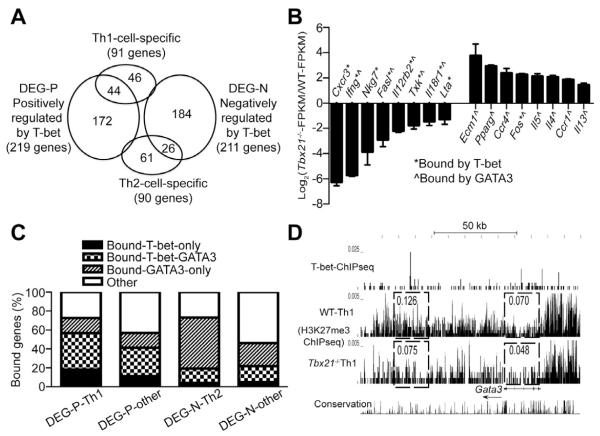
Since we noticed that T-bet deficient “Th1” cells produce IL-4, which is a signature Th2 cell cytokine, we analyzed gene regulation of Th1 and Th2 cell-specific genes by T-bet at a genome-wide level. We have previously identified ~90 Th1 and Th2 cell-specific genes (Wei et al., 2011). ~50% of the Th1 cell-specific genes were positively regulated by T-bet (Figure 7A). Interestingly, 26 out of 90 Th2-specific genes were negatively regulated by T-bet. Genes positively regulated by T-bet included many well-known Th1 cell-specific genes such as Cxcr3, Ifng, Nkg7, Fasl, Il12rb2, Txk, Il18r1 and Lta (Figure 7B, left side). Genes that were negatively regulated by T-bet included dozens of Th2 cell-specific genes such as Ecm1, Pparg, Ccr4, Fos, Il5, Il4, Ccr1 and Il13 (Figure 7B, right side). Interestingly, T-bet bound to all these Th1 cell-specific genes but only one of the negatively regulated Th2 cell-specific genes. Indeed, we have reported that all these Th2 cell-specific genes as well as some of these Th1 cell-specific genes are bound by GATA3 in Th2 cells (Wei et al., 2011). At the genome level, T-bet bound to and positively regulated ~60% of the 44 Th1 cell-specific genes. While T-bet bound to and negatively regulated only ~20% of the 26 Th2 cell-specific genes, GATA3 bound to ~70% of these genes. Thus, it is likely that the up-regulation of these Th2 cell-specific genes in the Tbx21−/− “Th1” cells is due to the enhanced GATA3 function as well as expression that occurs in the absence of T-bet. Indeed, T-bet strongly bound to at least two sites around the Gata3 locus (Figure 7D, upper panel). In keeping with a modest up-regulation of GATA3 expression in T-bet-deficient Th1 cells (~1.7 fold increase), histone H3 tri-methylation at position K27 (H3K27me3), a suppressive histone mark, found in the immediate vicinity of these two T-bet binding sites at the Gata3 locus, was substantially reduced in T-bet-deficient Th1 cells when compared to that of WT Th1 cells (Figure 7D, lower panel). Therefore, T-bet may directly inhibit GATA3 expression in Th1 cells possibly by promoting H3K27me3 modification at the Gata3 locus in addition to the ability of T-bet in inhibiting GATA3 function through protein-protein interaction as previously reported (Hwang et al., 2005).
Figure 7. T-bet directly induces Th1-related genes but indirectly suppresses Th2-related genes through acting on GATA3.
(A) The Venn diagram shows overlap of genes that are Th1 or Th2 cell-specific, positively or negatively regulated by T-bet in Th1 cells.
(B) FPKM values were extracted from RNAseq data in duplicates. Selected Th1- and Th2 cell-specific genes were shown. “*” and “^” mark the genes that are bound by T-bet in Th1 cells and by GATA3 in Th2 cells, respectively.
(C) The plots show the percentages of the genes that are bound by T-bet and/or GATA3 in each category.
(D) UCSC genome browser view of T-bet binding and H3K27me3 modification at the Gata3 locus is shown. Numbers indicate the sums of RPBM (Reads Per Base Per Million reads in library) values in the box. Arrow indicates the direction of the Gata3 gene.
Thus, the balance between T-bet and GATA3 during T cell differentiation plays a critical role in determining the expression of Th1 and Th2 lineage-specific genes and without T-bet induction, GATA3 drives Th2 cell differentiation in the absence of IL-4.
DISCUSSION
We prepared the TBGR indicator mouse strain allowing us to study Th1 cell differentiation both in vitro and in vivo. We showed that ZsG expression faithfully reflects endogenous T-bet expression. Using this T-bet reporter system, we could study the regulation of T-bet at a single cell level with a greater sensitivity both in vitro and in vivo than was previously possible. Either IL-12 or IFN-γ alone was sufficient to induce T-bet expression in cells responding to TCR and CD28 signals. Cells from Ifngr1−/−Stat4−/− TBGR mice cultured under Th1 cell-polarizing conditions showed no induction of ZsG indicating that, under these in vitro conditions, T-bet induction requires at least IL-12 or IFN-γ. Interestingly, the majority of the Ifngr1−/−Stat4−/− TBGR cells were still able to up-regulate ZsG expression in response to T. gondii infection indicating that stimuli other than IFN-γ and IL-12 are capable of inducing T-bet in vivo in activated cells and such stimuli are not present during in vitro culture. We have observed high dose antigen stimulation can induce T-bet in vitro; however, such T-bet induction is largely dependent on endogenous IFN-γ. Thus, it is unlikely that the in vivo T-bet expression by Ifngr1−/− Stat4−/− TBGR cells is due to a strong TCR stimulation. Type I IFNs or IL-27, when exogenously added, was able to induce ZsG in Ifngr1−/−Stat4−/− TBGR cells in vitro; thus, it is possible that these cytokines are responsible for IL-12- and IFN-γ-independent induction of T-bet in vivo. Further investigation is required to confirm such regulation.
We also found that T-bet was not required for its own induction when IL-12 or IFN-γ are present since ZsG is expressed at the same amounts in both WT and Tbx21−/− TBGR cells in response to T. gondii infection or when cultured in vitro under Th1 cell-polarizing conditions. Nonetheless, T-bet may be involved in directly promoting its own expression when induced by the “IL-12- and IFN-γ-independent” pathway, since the ZsG expression is much lower in Stat4−/−Tbx21−/− TBGR cells, which express no IFNγ, than in Ifngr1−/−Stat4−/− TBGR cells.
Consistent with a redundant role of IFN-γ and IL-12 in inducing T-bet, cells that fail to respond to IFN-γ, specifically Ifngr1−/− cells, were still able to produce IFN-γ at a level comparable to that found in wild type cells, especially in vivo. These results suggest the positive feedback loop mediated by IFN-γ makes only a limited contribution to full Th1 cell differentiation under the circumstance tested although it is possible that such positive feedback will become critical for Th1 responses when limited or no IL-12 is present. On the other hand, although T-bet amounts in Stat4−/− cells were similar to those in Ifngr1−/− cells, Stat4−/− cells expressed much lower IFN-γ than Ifngr1−/− cells, suggesting that Stat4 is critical for collaborating with T-bet in inducing IFN-γ production as previously reported in an in vitro culture system (Thieu et al., 2008). Indeed, Stat4 directly binds to the Ifng locus (Wei et al., 2010). The importance of Stat4 is particularly evident when no exogenous IFN-γ was provided; Stat4−/− cells produced less IFN-γ and therefore, such cells not only had an IL-12 signaling defect but also received less IFN-γ stimulation. Either Stat4 or T-bet was able to induce some IFN-γ production but together they could collaborate to induce maximal IFN-γ production; no IFN-γ production could be detected in Stat4−/−Tbx21−/− cells either in vitro or in vivo. Thus, these two factors both play critical roles in inducing IFN-γ during Th1 responses and Stat4 is responsible for T-bet-independent IFN-γ production.
It is common that during a particular type of Th cell differentiation, the specific program involved also actively inhibits the capability of the differentiating cells to become cells of other Th phenotypes (Zhu and Paul, 2010). Transcription factors induced in each lineage, particularly the critical regulators, are often involved in such cross-regulation. For example, GATA3 not only represses Stat4 expression (Usui et al., 2003), but also suppresses Runx3-mediated IFN-γ induction (Yagi et al., 2010) and modifies the Tbx21 locus during Th2 cell differentiation (Wei et al., 2011). On the other hand, when over-expressed, T-bet suppresses GATA3 expression at the transcriptional level (Usui et al., 2006). T-bet also suppresses GATA3 function through protein-protein interaction (Hwang et al., 2005). T-bet and GATA3 share many target genes when co-expressed (Jenner et al., 2009). Here we have shown that endogenous T-bet is critical for inhibiting GATA3 function during both in vitro and in vivo Th1 cell differentiation, preventing these “Th1-differentiating” cells from activating a “default” Th2 cell differentiation program, a program that they do activate when T-bet is absent.
Although cultured Th2 cells express the highest amounts of GATA3 among Th cells, all CD4+ T cells, including naïve cells, express GATA3 (Wei et al., 2011). The expression of GATA3 is up-regulated during Th2 cell differentiation and down-regulated during Th1 and Th17 cell differentiation in vitro. However, under certain circumstances, Th2 cell differentiation may occur with low amounts of GATA3 expression, although such GATA3 continues to be essential (Zhu et al., 2003). Stat5 activation is a second critical element for IL-4 production and thus Th2 cell differentiation (Cote-Sierra et al., 2004; Zhu et al., 2003). Stat5 is the key signaling molecule for most members of the γc family cytokines, including IL-2 and IL-7 (Rochman et al., 2009).
Unlike GATA3, the other two key transcription factors, T-bet and RORγt, are not expressed by naïve CD4+ T cells. Similarly, Stat4 and Stat3 activation are usually triggered by cytokines, such as IL-12 and IL-6, produced by antigen presenting cells, whereas Stat5 can be activated by IL-2 that is produced by CD4+ T cells upon activation. Therefore, basal expression of GATA3 as well as continuous autocrine activation of Stat5 in naïve and freshly activated CD4+ T cells provide an internal Th2 cell differentiating setting and thus lead to Th2 cell differentiation without an exogenous source of cytokines. Thus, it could be argued that unless specifically prevented, Th2 cell differentiation is likely to occur without a requirement for specific inducing cytokines produced by antigen presenting cells or other accessory cells.
Over the years, immunologists have struggled to determine how Th2 cell-associated responses are initiated physiologically (Paul and Zhu, 2010). Antigen presenting cells can produce IL-12 that drives Th1 cell differentiation or TGFβ or IL-6 that induce Th17 cell differentiation. However, IL-4 is not produced by conventional antigen presenting cells. Recent reports have raised the possibility that basophils may serve as antigen presenting cells and coupled with their capacity to produce IL-4 and possibly TSLP may act as determinants of Th2 cell differentiation (Perrigoue et al., 2009; Sokol et al., 2008; Sokol et al., 2009; Yoshimoto et al., 2009). However, IL-4 is not always essential for Th2 cell-associated responses in vivo so that these capabilities of basophils, even if they are confirmed as potent APC, may not prove to be essential for the Th2 cell bias they have been reported to exert. Indeed, basophils are not required for Th2 cell differentiation in response to Nippostrongylus brasiliensis infection (Kim et al., 2010). Low dose antigen stimulation or weak antigen presentation have been proposed as another model of Th2 cell differentiation (Steinfelder et al., 2009; Yamane et al., 2005); the Schistosoma mansoni egg product omega-1 has been reported to diminish dendritic cell function, presumably creating a low TCR signal strength environment. Nonetheless, there is as yet no convincing data showing that either mechanism is essential for Th2 cell induction. Our report here clearly shows that even under a strong Th1 cell-biasing stimulation either in vitro or in vivo, evident by the induction of T-bet reporter, in the absence of T-bet, such cells become Th2-like cells. The failure of CD4+ T cells in IL-12-deficient mice to default to a Th2 cell program in a Th1 cell-inducing environment may be explained by induction of T-bet induction by signals other than IL-12.
In conclusion, by using a T-bet reporter mouse strain cross-bred to various gene deficient mouse strains, we have carefully dissected the network of multiple cytokine pathways elicited by IL-12 and IFN-γ, and the role of Stat4 and T-bet in the induction of Th1 and Th2 cell signature molecules during in vivo Th1 responses. Our results also strongly indicate that Th2 cell differentiation may occur without CD4+ T cells receiving differentiation-biasing signals from antigen presenting cells, consistent with this being a “default” pathway.
EXPERIMENTAL PROCEDURES
Mice and cell culture
Tbx21−/− mice (Line 4648) and Ifngr1−/− (Line 3288) were obtained from Jackson laboratory (Szabo et al., 2002). Stat4−/− mice have been previously described (Kaplan et al., 1996) and they have been backcrossed to C57BL/6 background. C57BL/6 Tcra−/− (Line 98) mice were obtained from Taconic. The C57BL/6 T-bet-ZsGreen reporter (TBGR) mouse strain was generated as described in Supplemental Experimental Procedures. Briefly, the coding region of ZsGreen was inserted into the ATG translational starting site of a genomic fragment encoding T-bet in the BAC clone (RP23-237M14) by recombineering technology using galK replacement method so that T-bet expression from the BAC was replaced by GFP expression. The modified BAC clone, after sequence verification of manipulated region, was used to generate transgenic mice by pronuclear microinjection of fertilized C57BL/6 eggs. The offspring of the founder (B6-Tbet-ZsGreen-E3) that carried only one copy of the transgene were selectively maintained and bred to various mouse strains deficient in different genes. The TBGR mice were genotyped using the following primers: ZsGreen (F), 5′-AAG GGC GAC GTG AGC ATG T-3′ and ZsGreen (R), 5′-CAC GGA CTT GGC CTT GTA CAC-3′. All the mice were bred and maintained in the NIAID specific pathogen free animal facility and the experiments were done when mice were at 8 to 16 weeks of age under protocols approved by the NIAID Animal Care and Use Committee.
Cell preparation and activation
Naïve CD62LhiCD44loCD25negZsGnegCD4+ T cells were sorted by FACSAria (BD Biosciences). For adoptive transfer experiments, naïve CD4 T cells were sorted for ZsGnegCD45RbhiCD25neg[CD8/B220/IAb/CD11b/CD11c/CD24/CD16/CD32/NK1.1]neg fraction. T cell-depleted splenocytes were prepared by incubation with anti-Thy1.2 mAb supernatant and rabbit complement (Cedarlane Laboratories Limited) at 37°C for 45 min followed by irradiation at 30 Gy (3,000 rad). Naïve CD4+ T cells were cultured with irradiated T cell-depleted splenocytes at a ratio of 1:5 in the presence of 1 μg/ml of anti-CD3 (145-2C11) and 3 μg/ml of anti-CD28 (37.51) for 3-4 days with various combinations of antibodies and cytokines: For Th1 conditions, 10 IU/ml of hIL-2, 10 ng/ml of IL-12 and 10 μg/ml anti-IL-4 (11B11), some Th1 cultures include 10 ng/ml IFN-γ as specified; for Th2 conditions, 50 IU/ml of hIL-2, 5000 U/ml of IL-4, 10 μg/ml anti-IL-12 (C17.8) and anti-IFN-γ (XMG1.2); for Th17 conditions, 1 ng/ml TGFβ, 10 ng/ml IL-6, 10 ng/ml IL-1β, 10 μg/ml anti-IL-4, 10 μg/ml anti-IL-12 and anti-IFN-γ. The activated cells were then cultured in RPMI-1640 medium with 10% FBS that contains IL-2 (50 U/ml for Th1 and 100 U/ml for Th2) or cytokine-free medium for Th17 cells.
Toxoplasma gondii infection
T. gondii cysts (the avirulent strain ME-49) were prepared from the brains of infected C57BL/6 mice. Mice were inoculated i.p. with an average of 20 cysts per animal and splenocytes were analyzed one week after infection.
Flow cytometric analysis
Cell surface molecules such as CD4, CD8, CD44, CD25, CD62L were stained in PBS with 0.5% BSA. Cytokine intracellular staining was performed as previously described (Zhu et al., 2004). Briefly, the activated cells were re-stimulated with10 ng/ml phorbol 12-myristate 13-acetate (PMA) and 500 nM ionomycin in the presence of 2 mM monensin for 4 hours. Splenocytes harvested from T. gondii infected mice were harvested and stimulated with plate-bound anti-CD3/anti-CD28 (3 ug/ml of each used for coating) for 4 hours in the presence of monension. At the end of stimulation, cells were washed and fixed with 4% paraformaldehyde for 10 min at room temperature and permeabilized in PBS containing 0.5% Triton X-100 and 0.1% BSA. They were then stained for cytokines together with CD4 and CD44. Data were collected with LSR II (BD Biosciences) and results were analyzed using FlowJo software (Tree Star). All the antibodies for staining cell surface markers or cytokines were purchased from either BD Biosciences or eBiosciences. Staining of the transcription factors were carried out with Foxp3 Staining Buffer Set (eBioscience) according to the manufacturer’s instructions. Anti-GATA3 (L50-823), anti-Foxp3 (FJK-16s) and anti-T-bet (4B10) were purchased from BD Biosciences and e-Biosciences, respectively.
RNA Purification and Quantitative PCR
Total RNAs were isolated using a combination of TRIzol (Invitrogen) and RNeasy Kit (QIAGEN). cDNAs were prepared using SuperScript III Reverse Transcriptase (Invitrogen). Quantitative PCR was performed on a 7900HT Sequence Detection System (Applied Biosystems) using the following pre-designed primer/probe sets: Il4, Ifng, Tbx21, Cd4 (all purchased from Applied Biosystems) and Gata3 as previously described (Zhu et al., 2004).
ChIP-Seq and RNA-Seq
ChIP-Seq experiments were performed as previously described (Barski et al., 2007). Briefly, chromatin was prepared from the cells cross-linked with 1% formaldehyde for 10min by sonication and immunoprecipitated with the anti-T-bet antibody (sc-21749, Santa Cruz). For histone ChIPseq, cells were treated with MNase to generate approximately 80% mononucleosomes and 20% dinucleosomes. Antibodies against histone H3K4me1 (ab8895, Abcam) and H3K27me3 (07-449, Upstate) were used. The ChIP DNA was blunt-ended, ligated to the Solexa adaptors, amplified and sequenced using an Illumina HiSeq system. Detailed data analyses are described in Supplemental Experimental Procedures.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs. Alan Sher, Ronald Germain, John O’Shea and Pamela Schwartzberg for their critical reading of our manuscript; Julie Edwards for her excellent assistance in cell sorting; Drs. John O’Shea, Yuka Kanno and Mark Kaplan for providing Stat4−/− mice on C57BL/6 background. The work is supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, USA.
Footnotes
ACCESSION NUMBERS All the ChIPseq and RNAseq data are available in the Gene Expression Omnibus (GEO) database under the accession number GSE38808.
SUPPLEMENTAL INFORMATION Supplemental information including three figures, one table, and Supplemental Experimental Procedures can be found with this article online.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no competing financial interests.
REFERENCES
- Afkarian M, Sedy JR, Yang J, Jacobson NG, Cereb N, Yang SY, Murphy TL, Murphy KM. T-bet is a STAT1-induced regulator of IL-12R expression in naive CD4+ T cells. Nat. Immunol. 2002;3:549–557. doi: 10.1038/ni794. [DOI] [PubMed] [Google Scholar]
- Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annu. Rev. Immunol. 2006;24:607–656. doi: 10.1146/annurev.immunol.23.021704.115821. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Cote-Sierra J, Foucras G, Guo L, Chiodetti L, Young HA, Hu-Li J, Zhu J, Paul WE. Interleukin 2 plays a central role in Th2 differentiation. Proc. Natl. Acad. Sci. USA. 2004;101:3880–3885. doi: 10.1073/pnas.0400339101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelman FD, Morris SC, Orekhova T, Mori M, Donaldson D, Reiner SL, Reilly NL, Schopf L, Urban JF., Jr. Stat6 regulation of in vivo IL-4 responses. J. Immunol. 2000;164:2303–2310. doi: 10.4049/jimmunol.164.5.2303. [DOI] [PubMed] [Google Scholar]
- Hatton RD, Harrington LE, Luther RJ, Wakefield T, Janowski KM, Oliver JR, Lallone RL, Murphy KM, Weaver CT. A distal conserved sequence element controls Ifng gene expression by T cells and NK cells. Immunity. 2006;25:717–729. doi: 10.1016/j.immuni.2006.09.007. [DOI] [PubMed] [Google Scholar]
- Ho IC, Tai TS, Pai SY. GATA3 and the T-cell lineage: essential functions before and after T-helper-2-cell differentiation. Nat. Rev. Immunol. 2009;9:125–135. doi: 10.1038/nri2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang ES, Szabo SJ, Schwartzberg PL, Glimcher LH. T helper cell fate specified by kinase-mediated interaction of T-bet with GATA-3. Science. 2005;307:430–433. doi: 10.1126/science.1103336. [DOI] [PubMed] [Google Scholar]
- Jankovic D, Kullberg MC, Feng CG, Goldszmid RS, Collazo CM, Wilson M, Wynn TA, Kamanaka M, Flavell RA, Sher A. Conventional T-bet+Foxp3− Th1 cells are the major source of host-protective regulatory IL-10 during intracellular protozoan infection. J. Exp. Med. 2007;204:273–283. doi: 10.1084/jem.20062175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankovic D, Kullberg MC, Hieny S, Caspar P, Collazo CM, Sher A. In the absence of IL-12, CD4+ T cell responses to intracellular pathogens fail to default to a Th2 pattern and are host protective in an Il10−/− setting. Immunity. 2002;16:429–439. doi: 10.1016/s1074-7613(02)00278-9. [DOI] [PubMed] [Google Scholar]
- Jankovic D, Kullberg MC, Noben-Trauth N, Caspar P, Paul WE, Sher A. Single cell analysis reveals that IL-4 receptor/Stat6 signaling is not required for the in vivo or in vitro development of CD4+ lymphocytes with a Th2 cytokine profile. J. Immunol. 2000;164:3047–3055. doi: 10.4049/jimmunol.164.6.3047. [DOI] [PubMed] [Google Scholar]
- Jenner RG, Townsend MJ, Jackson I, Sun K, Bouwman RD, Young RA, Glimcher LH, Lord GM. The transcription factors T-bet and GATA-3 control alternative pathways of T-cell differentiation through a shared set of target genes. Proc. Natl. Acad. Sci. USA. 2009;106:17876–17881. doi: 10.1073/pnas.0909357106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan MH, Sun YL, Hoey T, Grusby MJ. Impaired IL-12 responses and enhanced development of Th2 cells in Stat4-deficient mice. Nature. 1996;382:174–177. doi: 10.1038/382174a0. [DOI] [PubMed] [Google Scholar]
- Kim S, Prout M, Ramshaw H, Lopez AF, LeGros G, Min B. Cutting edge: basophils are transiently recruited into the draining lymph nodes during helminth infection via IL-3, but infection-induced Th2 immunity can develop without basophil lymph node recruitment or IL-3. J. Immunol. 2010;184:1143–1147. doi: 10.4049/jimmunol.0902447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarevic V, Chen X, Shim JH, Hwang ES, Jang E, Bolm AN, Oukka M, Kuchroo VK, Glimcher LH. T-bet represses TH17 differentiation by preventing Runx1-mediated activation of the gene encoding RORγt. Nat. Immunol. 2011;12:96–104. doi: 10.1038/ni.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman LA, Banica M, Reiner SL, Hunter CA. STAT1 plays a critical role in the regulation of antimicrobial effector mechanisms, but not in the development of Th1-type responses during toxoplasmosis. J. Immunol. 2004;172:457–463. doi: 10.4049/jimmunol.172.1.457. [DOI] [PubMed] [Google Scholar]
- Lighvani AA, Frucht DM, Jankovic D, Yamane H, Aliberti J, Hissong BD, Nguyen BV, Gadina M, Sher A, Paul WE, O’Shea JJ. T-bet is rapidly induced by interferon-gamma in lymphoid and myeloid cells. Proc. Natl. Acad. Sci. USA. 2001;98:15137–15142. doi: 10.1073/pnas.261570598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min B, Prout M, Hu-Li J, Zhu J, Jankovic D, Morgan ES, Urban JF, Jr., Dvorak AM, Finkelman FD, LeGros G, Paul WE. Basophils produce IL-4 and accumulate in tissues after infection with a Th2-inducing parasite. J. Exp. Med. 2004;200:507–517. doi: 10.1084/jem.20040590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen AC, High FA, Hutchins AS, Lee HW, Villarino AV, Livingston DM, Kung AL, Cereb N, Yao TP, Yang SY, Reiner SL. Role of T-bet in commitment of TH1 cells before IL-12-dependent selection. Science. 2001;292:1907–1910. doi: 10.1126/science.1059835. [DOI] [PubMed] [Google Scholar]
- Murphy KM, Reiner SL. The lineage decisions of helper T cells. Nat. Rev. Immunol. 2002;2:933–944. doi: 10.1038/nri954. [DOI] [PubMed] [Google Scholar]
- Oestreich KJ, Weinmann AS. Transcriptional mechanisms that regulate T helper 1 cell differentiation. Cur. Opin. Immunol. 2012;24:191–195. doi: 10.1016/j.coi.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai SY, Truitt ML, Ho IC. GATA-3 deficiency abrogates the development and maintenance of T helper type 2 cells. Proc. Natl. Acad. Sci. USA. 2004;101:1993–1998. doi: 10.1073/pnas.0308697100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul WE, Zhu J. How are TH2-type immune responses initiated and amplified? Nat. Rev. Immunol. 2010;10:225–235. doi: 10.1038/nri2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrigoue JG, Saenz SA, Siracusa MC, Allenspach EJ, Taylor BC, Giacomin PR, Nair MG, Du Y, Zaph C, van Rooijen N, et al. MHC class II-dependent basophil-CD4+ T cell interactions promote TH2 cytokine-dependent immunity. Nat. Immunol. 2009;10:697–705. doi: 10.1038/ni.1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochman Y, Spolski R, Leonard WJ. New insights into the regulation of T cells by γc family cytokines. Nat. Rev. Immunol. 2009;9:480–490. doi: 10.1038/nri2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol CL, Barton GM, Farr AG, Medzhitov R. A mechanism for the initiation of allergen-induced T helper type 2 responses. Nat. Immunol. 2008;9:310–318. doi: 10.1038/ni1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol CL, Chu NQ, Yu S, Nish SA, Laufer TM, Medzhitov R. Basophils function as antigen-presenting cells for an allergen-induced T helper type 2 response. Nat. Immunol. 2009;10:713–720. doi: 10.1038/ni.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinfelder S, Andersen JF, Cannons JL, Feng CG, Joshi M, Dwyer D, Caspar P, Schwartzberg PL, Sher A, Jankovic D. The major component in schistosome eggs responsible for conditioning dendritic cells for Th2 polarization is a T2 ribonuclease (omega-1) J. Exp. Med. 2009;206:1681–1690. doi: 10.1084/jem.20082462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–669. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- Szabo SJ, Sullivan BM, Stemmann C, Satoskar AR, Sleckman BP, Glimcher LH. Distinct effects of T-bet in TH1 lineage commitment and IFN-γ production in CD4 and CD8 T cells. Science. 2002;295:338–342. doi: 10.1126/science.1065543. [DOI] [PubMed] [Google Scholar]
- Thieu VT, Yu Q, Chang HC, Yeh N, Nguyen ET, Sehra S, Kaplan MH. Signal transducer and activator of transcription 4 is required for the transcription factor T-bet to promote T helper 1 cell-fate determination. Immunity. 2008;29:679–690. doi: 10.1016/j.immuni.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui T, Nishikomori R, Kitani A, Strober W. GATA-3 suppresses Th1 development by downregulation of Stat4 and not through effects on IL-12Rβ2 chain or T-bet. Immunity. 2003;18:415–428. doi: 10.1016/s1074-7613(03)00057-8. [DOI] [PubMed] [Google Scholar]
- Usui T, Preiss JC, Kanno Y, Yao ZJ, Bream JH, O’Shea JJ, Strober W. T-bet regulates Th1 responses through essential effects on GATA-3 function rather than on Ifng gene acetylation and transcription. J. Exp. Med. 2006;203:755–766. doi: 10.1084/jem.20052165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voehringer D, Shinkai K, Locksley RM. Type 2 immunity reflects orchestrated recruitment of cells committed to IL-4 production. Immunity. 2004;20:267–277. doi: 10.1016/s1074-7613(04)00026-3. [DOI] [PubMed] [Google Scholar]
- Wei G, Abraham BJ, Yagi R, Jothi R, Cui K, Sharma S, Narlikar L, Northrup DL, Tang Q, Paul WE, et al. Genome-wide analyses of transcription factor GATA3-mediated gene regulation in distinct T cell types. Immunity. 2011;35:299–311. doi: 10.1016/j.immuni.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Vahedi G, Sun HW, Watford WT, Takatori H, Ramos HL, Takahashi H, Liang J, Gutierrez-Cruz G, Zang C, et al. Discrete roles of STAT4 and STAT6 transcription factors in tuning epigenetic modifications and transcription during T helper cell differentiation. Immunity. 2010;32:840–851. doi: 10.1016/j.immuni.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi R, Junttila IS, Wei G, Urban JF, Jr., Zhao K, Paul WE, Zhu J. The transcription factor GATA3 actively represses RUNX3 protein-regulated production of interferon-gamma. Immunity. 2010;32:507–517. doi: 10.1016/j.immuni.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane H, Zhu J, Paul WE. Independent roles for IL-2 and GATA-3 in stimulating naive CD4+ T cells to generate a Th2-inducing cytokine environment. J. Exp. Med. 2005;202:793–804. doi: 10.1084/jem.20051304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Ochando JC, Bromberg JS, Ding Y. Identification of a distant T-bet enhancer responsive to IL-12/Stat4 and IFN-γ/Stat1 signals. Blood. 2007;110:2494–2500. doi: 10.1182/blood-2006-11-058271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto T, Yasuda K, Tanaka H, Nakahira M, Imai Y, Fujimori Y, Nakanishi K. Basophils contribute to TH2-IgE responses in vivo via IL-4 production and presentation of peptide-MHC class II complexes to CD4+ T cells. Nat. Immunol. 2009;10:706–712. doi: 10.1038/ni.1737. [DOI] [PubMed] [Google Scholar]
- Zhang DH, Cohn L, Ray P, Bottomly K, Ray A. Transcription factor GATA-3 is differentially expressed in murine Th1 and Th2 cells and controls Th2-specific expression of the interleukin-5 gene. J. Biol. Chem. 1997;272:21597–21603. doi: 10.1074/jbc.272.34.21597. [DOI] [PubMed] [Google Scholar]
- Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–596. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- Zhu J, Cote-Sierra J, Guo L, Paul WE. Stat5 activation plays a critical role in Th2 differentiation. Immunity. 2003;19:739–748. doi: 10.1016/s1074-7613(03)00292-9. [DOI] [PubMed] [Google Scholar]
- Zhu J, Min B, Hu-Li J, Watson CJ, Grinberg A, Wang Q, Killeen N, Urban JF, Jr., Guo L, Paul WE. Conditional deletion of Gata3 shows its essential function in TH1-TH2 responses. Nat. Immunol. 2004;5:1157–1165. doi: 10.1038/ni1128. [DOI] [PubMed] [Google Scholar]
- Zhu J, Paul WE. Peripheral CD4+ T-cell differentiation regulated by networks of cytokines and transcription factors. Immunol. Rev. 2010;238:247–262. doi: 10.1111/j.1600-065X.2010.00951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Yamane H, Paul WE. Differentiation of effector CD4+ T cell populations. Annu. Rev. Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.