Abstract
In this study, the impact of amino acid modifications on the accuracy of the iTRAQ (isobaric tags for relative and absolute quantitation) method was evaluated. MCF-7 breast cancer cells, cultured in the presence of 17 β-estradiol (E2) and tamoxifen (Tam), were used as a model system. The cells were labeled and analyzed by reversed-phase liquid chromatography (RPLC) and pulsed Q dissociation (PQD) ion trap tandem mass spectrometry (MS/MS) detection. Database searching was performed by using various combinations of amino acid modification allowances, i.e, Lys/Tyr/Cys and amino terminal iTRAQ labeling, Lys methylation, acetylation and carbamylation, and Cys/Met oxidation. Other than the intended Lys/amino terminal iTRAQ labeling, such modifications occur as a result of either enzymatic or sample prep related reactions, and are typically ignored in quantitation analysis to minimize the rate of false positive peptide identifications. The study revealed that the modifications with the greatest impact on protein identification and quantitation pertain to Lys and Tyr amino acid residues, that by enabling such modifications the number and type of identified proteins will change (by up to 10 %), and that the rate of false positive protein identifications can be maintained below an upper threshold of 5 % if appropriate data filtering conditions are used. In addition, the interference of possible posttranslational modifications (i.e., phosphorylation) with iTRAQ quantitation was examined.
Introduction
Quantitative profiling of complex samples is a major topic of interest in the field of mass spectrometry-based proteomics. Several quantitation strategies involving covalent attachment of stable isotope tags to specific amino acids in a protein or peptide by metabolic, enzymatic and chemical methods have been developed.1 In addition, label-free quantitation strategies have also evolved. These methods involve an assessment of spectral counts, sequence coverage and normalized ion intensities.2
In recent years, the development of iTRAQ reagents has had a significant impact on label-dependent quantitation.3 This technique consists of chemical labeling of the N-terminus (Nt) and Lys side chains of peptides with unique isobaric tags in up to four or eight different samples (4-plex and 8-plex quantitation, respectively). The tags have three components: a charged reporter group, a balance group and an amine specific peptide reactive group. In the 4-plex iTRAQ kit, such as used in this study, the combined mass of the reporter and the balance groups is 145 Da, however, the mass of each separate group is different for each tag. During MS, tagged identical peptides from different samples have the same mass. After peptide fragmentation, reporter ions at m/z 113, 114, 115 and 116, and peptide fragments with the same mass are generated. Relative quantitation is performed based on reporter ion intensities. Multiplexed quantitation is a major advantage of this approach, as it allows for the simultaneous analysis of samples, and a decrease of total MS analysis times and of experimental/technical variability. Other advantages relate to the comprehensiveness, yet simplicity, of the method.4 Several research groups have explored the potential of iTRAQ for the analysis of a variety of complex samples, in particular of cancer origin,5-10 and have found that the results generated by iTRAQ are complementary to other quantitation methods such as cleavable isotope coded affinity tagging (cICAT) or 2D difference gel electrophoresis.
In a recent study in our lab, we developed an iTRAQ-RPLC-MS/MS strategy using PQD detection on a low-resolution linear ion trap mass spectrometer with the goal of performing differential expression profiling of complex cellular extracts.8 The work evaluated the run-to-run reproducibility of protein identifications and global iTRAQ ratios, as well as the accuracy of the iTRAQ quantitation method when taking into account only peptides labeled on the Lys and N-terminal amino acids. In the present study, we evaluated the impact of some additional amino acid modifications that may interfere and alter the accuracy of protein quantitation with the iTRAQ method. In particular, our study focused on evaluating the impact of Tyr/Cys iTRAQ labeling, Lys carbamylation, Lys methylation, Lys acetylation and Cys/Met oxidation.
Methods
Reagents
MCF-7 breast cancer cells, Eagle's minimum essential medium-EMEM, fetal bovine serum-FBS, Dulbecco's phosphate buffered saline-PBS, and trypsin/EDTA were purchased from ATCC (Manassas, VA). Phenol red-free Dulbecco's modified Eagle's medium-DMEM was obtained from Invitrogen (Carlsbad, CA), charcoal/dextran treated fetal calf serum from Hyclone (Logan, UT), and phenol red free trypsin from SAFC Biosciences (Lenexa, KS). Bovine insulin, E2, Tam, L-glutamine, protease inhibitors, phosphatase inhibitors (NaF, Na3VO4), trifluoroacetic acid, acetic acid, formic acid, TrisHCl, sodium chloride, urea and dithiothreitol-DTT were ordered from Sigma-Aldrich (St. Louis, MO). RIPA lysis buffer was purchased from Upstate (Lake Placid, NY), sequencing-grade modified trypsin from Promega Corporation (Madison, WI), 4-plex iTRAQ reagents from Applied Biosystems (Foster City, CA), HPLC-grade methanol and acetonitrile from Fisher Scientific (Fair Lawn, NJ), and ammonium bicarbonate from Aldrich (Milwaukee, WI). Deionized (DI) water from a MilliQ Ultrapure water system-Millipore (Bedford, MA) was used to prepare all aqueous solutions.
MCF-7 Cell Culture
MCF-7 breast cancer cells were initially cultured in EMEM supplemented with 10 % FBS and 10 μg/mL insulin (i.e., maintenance medium), in a 37 °C, 5 % CO2 incubator, as described in detail elsewhere.8 Experimental media consisted of DMEM supplemented with 10 % charcoal-stripped FBS, 1 μg/mL insulin and 4 mM L-glutamine. For protein differential expression analysis, cells were cultured in maintenance media for approximately 2 weeks, changed to a 3:2 mix of maintenance and experimental media for one day, followed by complete experimental media for 6 days. At ∼35-40 % confluence, cells were divided in two batches and cultured in experimental media supplemented with (A) E2 (1 nM), or (B) E2 (10 pM)/Tam (1 μM) for 3 days. Batch A had a confluence of ∼70-80 %, and batch B had a confluence of ∼45-55 %. For harvesting, the cells were rinsed with PBS (pH 7.4) and incubated in trypsin/EDTA solution (0.25 % trypsin/0.53 mM EDTA) for 5-10 minutes to allow cell detachment. To stop the digestion, maintenance medium was added, the cells were centrifuged, rinsed with PBS, harvested and stored at -80 °C until further analysis.
Cell Lysis and Protein Extract Processing
Cells were lysed in a solution prepared from 1 mL RIPA buffer (500 mM TrisHCl pH 7.4, 1.5 M NaCl, 10 % NP-40, 2.5 % deoxycholic acid, 10 mM EDTA), 100 μL protease inhibitor cocktail (104 mM AEBSF, 0.08 mM aprotinin, 2 mM leupeptin, 4 mM bestatin, 1.5 mM peptatin A, 1.4 mM E-64), 100 μL NaF (∼100 mM) and 50 μL Na3VO4 (∼200 mM) phosphatase inhibitor solutions, and 8.75 mL of ice cold water. Cells were lysed for 2-3 hours while shaking at 4 °C, and centrifuged for ∼15 minutes at 13,000 rpm at 4 °C. The protein content was measured by the Bradford assay using a SmartSpec Plus spectrophotometer (Bio-Rad, Hercules, CA). The protein extract was denatured with 8 M urea and 4.5 mM DTT for 1 hour at 60 °C, diluted 1:10 with 50 mM NH4HCO3, and digested with trypsin (at a ratio of 50:1, substrate:enzyme) for 24 hours at 37 °C. The final concentration in protein extract was 100 μg/mL. The samples were stored in a freezer at -80 °C until further processing.
iTRAQ Labeling
The tryptic digest solutions of the cellular protein extracts were cleaned up from salts and buffer components with SPEC-PTC18 solid-phase extraction pipette tips (Varian, Inc., Lake Forest, CA), concentrated to ∼5-10 μL with an Eppendorf Vacuufuge (Eppendorf AG, Hamburg, Germany), resuspended in 25-30 μL iTRAQ dissolution buffer, and treated with iTRAQ reagent solution for 2 hours at room temperature (4-plex kit). Two experimental replicates of cell condition A [i.e., cells treated with E2 (1 nM)] were labeled with iTRAQ reagents 114 and 115, and two experimental replicates of cell condition B [i.e., cells treated with E2 (10 pM)/Tam (1 μM)] were labeled with iTRAQ reagents 116 and 117. Each replicate contained 100 μg protein digest. The samples were ultimately mixed in a ratio of A:A:B:B of 1:1:1:1 (double 2-plex experiment). Next, the sample mix was cleaned up with SPEC-PTSCX solid-phase extraction pipette tips (Varian, Inc.), dried, and resuspended in LC buffer system A.8
RPLC-ESI-MS/MS
Reversed-phase liquid chromatography tandem mass spectrometry analysis was performed using a micro liquid chromatography system (Agilent Technologies, Palo Alto, CA) and an LTQ ion trap mass spectrometer (Thermo Electron Corporation, San Jose, CA). The LC system and the LTQ were coupled by an on-column/no split injection set up.11 The separation column was a 100 μm i.d. × 12 cm fused silica capillary packed with 5 μm Zorbax SB-C18 particles (Agilent Technologies). A ∼1 cm long capillary (20 μm i.d. × 90 μm o.d.) was inserted into the separation column to generate a nanospray emitter. Mobile phase A was composed of H2O:CH3CN (95:5 vol/vol) and mobile phase B was composed of H2O:CH3CN (20:80 vol/vol), each supplemented with 0.01 % CF3COOH. The volumetric flow rate in the separation column was ∼160-180 nL/min, with a 3-hour long 0 %-100 % separation gradient. MS data were acquired via a data-dependent acquisition method, where each MS event was followed by zoom/MS2 scans on the five most intense peaks. The following parameters were enabled: zoom scan width of ±5 m/z, dynamic exclusion at repeat count of 1, repeat duration of 30 s, exclusion list size of 200, exclusion duration of 60 s, and exclusion mass width of ±1.5 m/z. For PQD detection, an isolation width of 3 m/z, normalized collision energy of 35 %, activation Q of 0.7, activation time of 0.1 ms, and an MS/MS acquisition threshold of 100 counts were used.8
Database Search Parameters
For protein identification, raw data files were searched with the Bioworks 3.3 software (Thermo Electron Corporation, San Jose, CA) using a minimally redundant human protein database (i.e., a database with minimal protein sequence overlaps, yet containing maximally complete sets of proteins) downloaded from the ExPASy/SwissProt website (37,690 entries) that was appended with 10 bovine proteins to facilitate data normalization. Only fully tryptic fragments with up to two missed cleavages were considered in the analysis, and the peptide and fragment ion tolerances were set at 2 u and 1 u, respectively. Five dynamic modifications were allowed for each peptide, and all peptides were assigned to unique protein references. Dynamic (variable) modifications allowed the simultaneous identification of different peptide forms, thus, they were preferred to static modifications. A total of eight conditions were considered with different amino acid modification allowances. In all cases, the iTRAQ related modifications (144.1 Da) at the Nt/Lys residues were allowed. The reference condition included iTRAQ labeling of Nt/Lys residues only. Additional mass shift allowances included: iTRAQ labeling of Tyr and Cys (144 Da); acetylation (42 Da), carbamylation (43 Da) and methylation (14 Da) of Lys; oxidation of Cys to cysteic acid (48 Da); and, oxidation of Met to sulphoxide (16 Da). iTRAQ ratios were extracted by setting the sensitivity threshold to 1 and the mass tolerance to ±0.5. Data were normalized based on a global iTRAQ ratio calculated for each set (i.e., for 116/114 and 117/114). The global iTRAQ ratio was the average of all protein iTRAQ ratios within a given set.8 At the peptide level, mass spectra were filtered with the Xcorr vs. charge state set at a minimum of 1.5, 2.0 and 3.0 for singly, doubly and triply charged peptides, respectively. At the protein level, only proteins with p<0.001 (as calculated by Bioworks) and with three unique peptides per protein were taken into consideration. All peptides that matched a protein with p<0.001 were averaged for the calculation of a protein iTRAQ ratio. When using such data filtering conditions, and considering only iTRAQ modifications on the Lys and N-terminal residues, the rate of false positive protein identifications when searching against the forward/reversed human protein database was zero. The impact of allowing additional amino acid modification on the false positive protein identification rate is described in the results section of the manuscript.
Results and Discussion
Our previous study on protein differential expression analysis in cancer cell extracts by iTRAQ-PQD-MS/MS detection has revealed that only 80-90 % of the identified proteins generated peptides with measurable iTRAQ ratios for quantitation, that only 50-60 % of the identified proteins were matched by at least two unique peptides and enabled protein quantitation by multiple iTRAQ measurements, and that ∼80 % of all quantified proteins could be quantified within a range of true value ±50%.8 Quantitation accuracy (measured vs. true) was evaluated by dividing a complex protein cellular extract in multiple aliquots, labeling with different iTRAQ tags, and analyzing the experimental iTRAQ ratios generated at 1:1 mixing ratios. The relative standard deviation of the global iTRAQ used for data normalization was as low as 4-8 % (see definition of the global iTRAQ in the methods section). These data were the outcome of considering the combined results of three-to-five consecutive LC-MS/MS analyses per sample. The possible contributing factors to the variability of protein iTRAQ ratios was discussed broadly in previous work.8,12 Briefly, some of these factors include less than optimal PQD fragmentation of peptides, contamination of the low 114-117 m/z region with fragments other than the iTRAQ reporter ions, sequence redundancy between peptides that match different proteins, low intensity signal, low number of matching tandem mass spectra per protein, and variable number of iTRAQ tags per peptide.
Impact of amino acid modifications on the number of identified peptides and proteins
To gain a better insight into the experimental factors that interfere in protein differential expression analysis, in the present study, we examined the effect of generally dismissed amino acid modifications that may impact the accuracy of the iTRAQ quantitation method. Detailed results are provided in Appendices 1-3 and summarized in Tables 1-5. Table 1 presents the modifications that we took into account: (a) Tyr/Cys labeling by iTRAQ reagents as a side reaction; (b) Lys modifications that can interfere with the iTRAQ labeling reaction (acetylation, carbamylation and methylation); and (c) Oxidation of Cys and Met to cysteic acid and Met sulphoxide, respectively, reactions that occur readily in the presence of reactive oxygen species. As shown in Table 1, these amino acid residue modifications may occur either as a result of enzymatic or sample prep related reactions. The change in mass (Δm, Da) is also provided.
Table 1.
Relevant amino acid modifications that interfere with iTRAQ quantitation.
| Modification | Amino acid | Δm, Da | Reason | Abbreviation |
|---|---|---|---|---|
| iTRAQ | Lys, N terminal | 144 | iTRAQ labeling (intended) | 144K, 144Nt |
| iTRAQ | Tyr | 144 | iTRAQ labeling (side reaction) | 144Y |
| iTRAQ | Cys | 144 | iTRAQ labeling (side reaction, un-blocked Cys) | 144C |
| Acetylation | Lys | 42 | Enzymatic; Sample prep (CH3COOH) | 42K |
| Carbamylation | Lys, Arg | 43 | Sample prep (urea) | 43K, 43R |
| Methylation | Lys | 14 | Enzymatic (methyl transferases) | 14K |
| Oxidation | Cys to cysteic acid | 48 | Enzymatic; Sample prep (reactive oxygen species) | 48C |
| Oxidation | Met to sulphoxide | 16 | Enzymatic; Sample prep (reactive oxygen species) | 16M |
Table 5.
Example of iTRAQ measurements for proteins that are known to carry possible phosphorylation at the indicated Ser and Tyr sites (note “S” and “Y” in bold).
| Protein/peptide | MH+ | ΔM | z | -10lg(p) | Xcorr | ΔCn | RSp | Normalized iTRAQ | |
|---|---|---|---|---|---|---|---|---|---|
| 116/114 | 117/114 | ||||||||
| Q15365|PCBP1_HUMAN Poly(rC)-binding protein 1 (Alpha-CP1) (hnRNP-E1) | |||||||||
| R.L]LMHGK*EVGSIIGK*.K | 1914.2 | 1.9 | 3 | 70 | 4.52 | 0.35 | 1 | ||
| R.L]LMHGK*EVGSIIGK*.K | 1914.2 | 1.1 | 3 | 71 | 4.53 | 0.38 | 1 | 0.87 | 0.73 |
| R.L]LMHGK*EVGSIIGK*.K | 1914.2 | 0.3 | 3 | 71 | 4.01 | 0.30 | 1 | ||
| R.L]LMHGK*EVGSIIGK*.K | 1914.2 | 1.2 | 3 | 66 | 4.14 | 0.38 | 1 | ||
| R.Q]QSHFAMMHGGTGFAGIDSSSPEVK*.G | 2894.4 | 1.1 | 3 | 115 | 5.90 | 0.41 | 1 | 1.33 | 1.96 |
| R.Q]QSHFAMMHGGTGFAGIDSSSPEVK*.G | 2894.4 | 1.1 | 3 | 87 | 4.85 | 0.43 | 1 | 3.66 | 2.87 |
| R.Q]QSHFAMMHGGTGFAGIDSSSPEVK*.G | 2894.4 | 1.1 | 3 | 109 | 5.26 | 0.45 | 1 | 3.44 | 3.81 |
| R.Q]QSHFAMMHGGTGFAGIDSSSPEVK*.G | 2894.4 | 1.1 | 3 | 57 | 4.32 | 0.43 | 1 | 1.77 | 2.03 |
| O95817|BAG3_HUMAN BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene-3) | |||||||||
| R.S]SLGSHQLPR.G | 1225.7 | 0.1 | 2 | 21 | 2.62 | 0.22 | 43 | ||
| K.T]HYPAQQGEYQTHQPVYHK*.I | 2600.3 | 1.0 | 3 | 67 | 3.69 | 0.29 | 1 | 4.21 | 2.06 |
| K.T]HYPAQQGEYQTHQPVYHK*.I | 2600.3 | 0.1 | 3 | 32 | 4.08 | 0.33 | 1 | ||
| K.T]HYPAQQGEYQTHQPVYHK*.I | 2600.3 | 1.0 | 3 | 36 | 3.97 | 0.20 | 1 | 3.39 | 2.76 |
| K.T]HYPAQQGEYQTHQPVYHK*.I | 2600.3 | 1.1 | 3 | 29 | 3.74 | 0.39 | 1 | 1.73 | 2.04 |
| K.T]HYPAQQGEYQTHQPVYHK*.I | 2600.3 | 1.0 | 3 | 47 | 3.88 | 0.36 | 1 | ||
| R.EGHPVYPQLRPGYIPIPVLHEGAENR.Q | 3082.6 | 1.4 | 3 | 51 | 3.90 | 0.17 | 4 | 1.97 | 2.72 |
| R.GYISIPVIHEQNVTRPAAQPSFHQAQK.T | 3304.8 | 1.3 | 3 | 57 | 3.03 | 0.20 | 3 | ||
Note 1: Marks “]” and “*” indicate the iTRAQ tag at the Nt and Lys residues, respectively. S/Y in bold indicate known p-sites.
Note 2: MH+ - molecular weight of protonated molecular ion; ΔM - difference between the theoretical and experimental mass of the protonated molecular ion; p-value - probability of a random match; Xcorr - cross-correlation score between virtual and experimental spectrum; ΔCn - degree by which the lower ranked peptide scores differ from the correlation score of the best match; RSp - rank of preliminary score.
Note 3: Blank iTRAQ ratio cells represent peptides for which iTRAQ ratios were not generated by the LTQ.
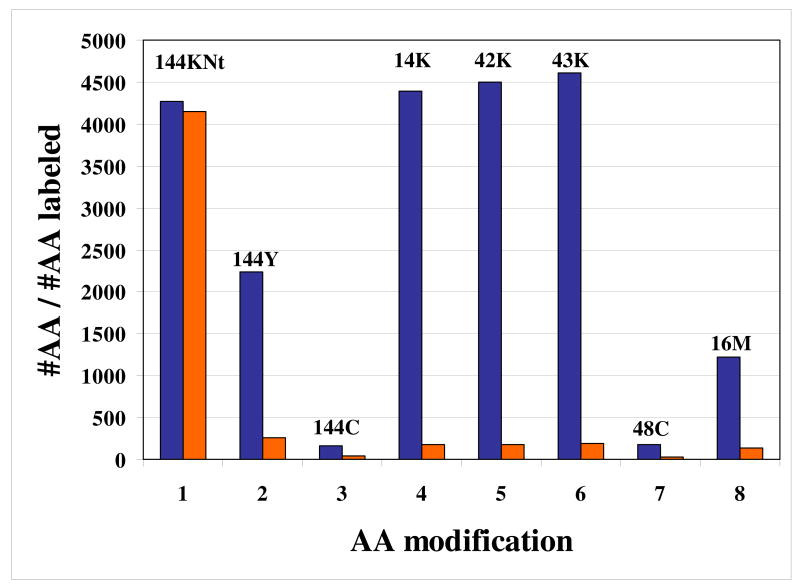
To assess the impact of these modifications on the iTRAQ quantitation accuracy, five replicate LC-PQD-MS/MS analyses (i.e., technical replicates) were conducted on the iTRAQ labeled MCF-7 extracts, and the combined results were evaluated by enabling various amino acid modifications for the database searching process (see experimental section). Two cell states were compared. MCF-7 cells were cultured in the presence of E2, a hormone that is stimulating the proliferation of estrogen receptor positive cancer cells, and Tam, a nonsteroidal drug, commonly prescribed in breast cancer therapy. To increase the confidence in protein identifications and to minimize the rate of false positive matches, only proteins with p<0.001 and matched by at least three peptides were taken into consideration. A threshold of three peptides per protein was chosen as a result of our preliminary findings that demonstrated that after manually eliminating proteins with unreliable iTRAQ ratios (i.e., proteins matched by peptides that did not generate complete sets of iTRAQ reporter ions, proteins matched by peptides that generated contradictory iTRAQ ratios, proteins matched by peptides that generated a broad range of iTRAQ ratios, etc.), most proteins (>93 %) ended up being quantified by ≥3 sets of iTRAQ measurements, and that ∼75 % of these proteins could be quantified within a range of true value ±30%.8 By running a double-duplex experiment, two independent iTRAQ ratios were calculated for each protein (116/114 and 117/115), and the average of the two ratios was used to generate the data in Tables 2-4. In Table 2, we compiled information about the number of identified proteins and peptides, as well as about the number of labeled/non-labeled Nt, Lys, and other specific amino acid residues. The condition of iTRAQ labeling at only the Nt and Lys residues was used as a reference for all future comparisons (i.e., the 144KNt condition). By allowing additional modifications, the total number of identified proteins and peptides changed by up to 10 %, the average being 188 (relative standard deviation RSD=4.2 %) and 3586 (RSD=2.4%), respectively (Tables 2-3). The efficiency of iTRAQ labeling was evaluated by counting the total number of labeled vs. available Lys, and the total number of labeled Nt vs. the total number of available peptides. Each specific amino acid residue modification was counted separately and referenced to the total number of available residues of interest. By evaluating the data in Table 2, we conclude that: (a) by allowing additional amino acid modifications, the total number of proteins and peptides increased for certain conditions (up to 10 % at the protein level, and up to 7 % at the peptide level), the major contributors to such an increase being Tyr labeling by iTRAQ and Lys acetylation and carbamylation [rows 1-2]; (b) the percentage of labeled Nt amino acid residues was, essentially, not affected (93-95 %) by allowing additional amino acid modifications [rows 3-4]; (c) the percentage of iTRAQ labeled Lys residues was high (92-97 %) [rows 5-7]; Lys methylation, acetylation and carbamylation contributed only 4 % to the pool of labeled Lys residues [rows 8-10]; (d) iTRAQ labeling of internal Tyr/Cys, as well as Cys/Met oxidation, were rather large contributors to the pool of labeled amino acid residues (11-28 %) [rows 8-10]; (e) the number/percentage of peptides carrying additional labeled amino acid residues relative to the total number of peptides was relatively small (0.6-6.5 %) [rows 11-12], the number of high quality peptides with p<0.001 being even smaller except for the case of iTRAQ labeling of Tyr residues [row 13]; and, (f) across all modifications, the RSD of total peptides/proteins identified, of total Lys residues, and of labeled Lys and Nt residues was rather small (2.4-4.2 %), indicating that at the global level such modifications do not have a major impact on iTRAQ quantitation (Table 3). For a better visualization of these effects, Figure 1 displays the number of labeled amino acid residues (light color) relative to the total number of specific residues available for modification (dark color). iTRAQ labeling of N-terminus and Lys residues are clearly the predominant amino acid modifications.
Table 2.
Impact of considering amino acid modifications on the total number of identified proteins, peptides, and iTRAQ labeling.
| 144KNt | 144KNt_144Y | 144KNt_144C | 144KNt_14K | 144KNt_42K | 144KNt_43K | 144KNt_48C | 144KNt_16M | |
|---|---|---|---|---|---|---|---|---|
| 1. Total proteins (p<0.001) # | 183 | 199 | 179 | 182 | 198 | 193 | 183 | 184 |
| 2. Total peptides # | 3526 | 3760 | 3512 | 3498 | 3609 | 3632 | 3536 | 3616 |
| 3. Total Nt labeled # | 3333 | 3529 | 3338 | 3253 | 3372 | 3407 | 3350 | 3417 |
| 4. Nt labeled % | 95 | 94 | 95 | 93 | 93 | 94 | 95 | 94 |
| 5. Total K residues # | 4268 | 4607 | 4252 | 4385 | 4497 | 4602 | 4292 | 4413 |
| 6. K iTRAQ labeled # | 4145 | 4431 | 4127 | 4081 | 4183 | 4250 | 4172 | 4272 |
| 7. K iTRAQ labeled % | 97 | 96 | 97 | 93 | 93 | 92 | 97 | 97 |
| 8. Additional AA residues # | 2230 | 166 | 4385 | 4497 | 4602 | 171 | 1213 | |
| 9. Additional AA labeled # | 254 | 46 | 181 | 171 | 192 | 28 | 131 | |
| 10. Additional AA labeled % | 11 | 28 | 4 | 4 | 4 | 16 | 11 | |
| 11. Peptides w. labeled AA # | 245 | 45 | 139 | 134 | 165 | 21 | 124 | |
| 12. Peptides w. labeled AA % | 6.5 | 1.3 | 3.9 | 3.7 | 4.5 | 0.6 | 3.4 | |
| 13. Peptides w. labeled AA (p<0.001) | 118 | 10 | 2 | 15 | 36 | 0 | 38 |
Note 1: AA stands for amino acid.
Note 2: The 144Y and 144C conditions involve additional labeling of only internal Y and C (N-terminals are assumed to be labeled).
Table 4.
Impact of considering additional amino acid modifications on global and individual protein iTRAQ ratios.
| Protein changes relative to the 144KNt condition | 144KNt | 144KNt_144Y | 144KNt_144C | 144KNt_14K | 144KNt_42K | 144KNt_43K | 144KNt_48C | 144KNt_16M |
|---|---|---|---|---|---|---|---|---|
| 1. Protein overlap #(%) | 183(100%) | 174 (95%) | 173 (95%) | 161 (88%) | 163 (89%) | 164 (90%) | 172 (94%) | 173 (95%) |
| 2. Global iTRAQ | 1.6 | 1.5 | 1.6 | 1.6 | 1.6 | 1.5 | 1.6 | 1.5 |
| 3. False positive IDs % | 0 | 1.1 | 1.2 | 3.5 | 1.2 | 3.6 | 3.6 | 2.4 |
| 4. Proteins w. iTRAQ/2 #(%) | 7 (4%) | 8 (5%) | 21 (13%) | 19 (12%) | 17 (10%) | 10 (6%) | 9 (5%) | |
| 5. Protein losses # | 7 | 8 | 20 | 18 | 17 | 9 | 8 | |
| 6. Protein gains #(%) | 23 (13%) | 6 (3%) | 21 (13%) | 34 (21%) | 27 (16%) | 10 (6%) | 10 (6%) | |
| 7. Protein gains w. iTRAQ/2 # | 4 | 1 | 1 | 4 | 6 | 1 | 1 |
Note: The row of “Proteins w. iTRAQ/2” refers to the #(%) of proteins with larger than 2-fold changes in iTRAQ ratios after enabling additional amino acid modifications, as denoted by entries in Appendix 1 with the ratio of iTRAQ ratios≥2 or ≤0.5. The row of “Protein losses #” is denoted by entries in Appendix 1 with the ratio of iTRAQ ratios = 0. The row of “Protein gains #(%)” refers to the #/(%) of proteins that were newly identified after enabling additional amino acid modifications, and are entries with iTRAQ≠0 listed at the bottom of Appendix 1. The row of “Proteins gains w. iTRAQ/2 #“ refers to proteins that were newly identified after enabling additional amino acid modifications, and are entries with iTRAQ≥2 or iTRAQ≤0.5 listed at the bottom of Appendix 1.
Table 3.
iTRAQ statistics derived from enabling additional amino acid modifications. The Mean, SDs and RSDs were calculated by taking into account all 8 conditions considered in this study.
| Mean | SD | RSD, % | |
|---|---|---|---|
| Total proteins (p<0.001) # | 188 | 7.9 | 4.2 |
| Total peptides # | 3586 | 87.1 | 2.4 |
| Total Nt labeled # | 3375 | 80.3 | 2.4 |
| Nt labeled % | 94 | 0.7 | 0.7 |
| Total K residues # | 4415 | 142.8 | 3.2 |
| K iTRAQ labeled # | 4208 | 109.7 | 2.6 |
| K iTRAQ labeled % | 95 | 2.1 | 2.2 |
Note: SD- standard deviation, RSD-relative standard deviation.
Figure 1.
Amino acid labeling for various amino acid modifications. Light color bars represent the number of labeled amino acid residues; dark color bars represent the total number of residues available for labeling.
Impact of amino acid modifications on individual protein iTRAQ ratios
The actual changes related to the nature of identified proteins, as well as the impact of allowing additional modifications on individual protein iTRAQ ratios, are highlighted in Table 4. The protein overlaps with the 144KNt condition ranged from 88 to 95 %, the conditions involving proteins with alternative Lys modifications having the lowest overlap with the reference 144KNt condition [row 1]. The change in the global iTRAQ was minimal, indicating that the allowance of such modifications will not affect the global normalization processes [row 2]. The rate of false positive protein identifications for the reference 144KNt condition was 0 %, when calculated by searching the MS2 scans against a forward/reversed database of proteins, and when considering only proteins with p<0.001 and matched by three peptides [row 3]. By enabling additional amino acid modifications, the rate of false positives increased from 0 % to a range of 1.1 %-3.6 %, but stayed below the 5 % threshold that is commonly used for proteomic data filtering. Statistical evaluation of our iTRAQ data has revealed that at least a 2-fold change in protein iTRAQ ratios is required to consider the change to be of biological relevance.8 With the present mass shift allowances, most changes in iTRAQ ratios for any of the data sets were less than 2-fold. Larger than 2-fold changes were rather the result of losing previously identified proteins after enabling the additional amino acid modifications, than a result of an actual change (i.e., 4-13 % of the proteins identified in the 144KNt condition were lost after enabling additional amino acid modifications, see Table 4, rows 4-5). In addition, protein losses where particularly noteworthy for modification states involving the Lys residues (14K, 42K and 43K). Gains in new proteins that were not detected without modification allowances were 3-21 %, of which, iTRAQ on Tyr and acetylation/carbamylation on Lys generated 4-6 new proteins per set with larger than 2-fold changes in iTRAQ ratios [rows 6-7]. Overall, the greatest impact on the data shown in Tables 2 and 4, whether considering changes in the number of identified peptides/proteins, or changes in iTRAQ ratios, was the result of enabling iTRAQ modifications on Tyr, or taking into account additional Lys modifications. We note, however, that Lys acetylation (+42 mass shift) and carbamylation (+43 mass shift) may not be readily distinguishable on low mass accuracy instruments such as an ion trap, as the corresponding doubly or triply charged peptides would be separated by only 0.5 or 0.3 m/z units, respectively.
A list of all identified proteins is provided in Appendix 1. For all proteins that were present in the reference condition of 144KNt, we provide the SwissProt ID, the name, the MW, -10lg(p-value), the amino acid coverage, the total peptide hits, and the iTRAQ ratio of each protein (i.e., the average of 116/114 and 117/114). In additional columns, we provide the iTRAQ ratios for all proteins that were identified when specific amino acid allowances were enabled, as well as the ratio of the new-to-old iTRAQ ratios for each protein. This ratio is zero for the proteins that disappeared from the reference list. New proteins that were identified after enabling the amino acid allowances are listed at the bottom of Appendix 1, with their corresponding iTRAQ value.
A close evaluation of the reasons for changes in iTRAQ ratios, as a result of enabling amino acid modifications, revealed that such changes were induced mainly by the disappearance or appearance of new peptides with rather poor Sequest scores that were mistakenly assigned to a protein (however, new peptides did not always carry the enabled modification). Slight changes in the p-value of proteins (with border line p-value) resulted occasionally in these proteins to not pass the data filtering thresholds, and to be eliminated from the list. Occasionally, peptides with good Sequest scores were not observable any longer when the database searching was performed with new search parameters. All new proteins that displayed a larger than 2-fold increase in iTRAQ ratio after enabling new modifications were inspected manually [Table 4, row 7]. These proteins are displayed in Appendix 2, including the corresponding peptide/protein Sequest scores. In addition, Appendix 2 lists all identified peptides with p<0.001 in a data set that carried the newly enabled modification. A set of tandem mass spectra, relevant for each enabled modification (144Y, 144C, 42K, 43K and 16M), is provided in the Supplemental Figure. Such tandem mass spectra were confirmed by the m/z of the parent ion, relevant a, b or y ions indicative of the amino acid sequence, and good Sequest scores (Xcorr, ΔCn, Sp-preliminary score, and RSp-rank of preliminary score). However, the exact modification site could not be always confirmed, as ions indicative of the amino acid sequence at, or in the immediate right/left vicinity of the modification site, were either missing or overlapping with multiply charged b/y ions. The 14K and 48C conditions did not generate reliable tandem mass spectra. Full Sequest reports for each data set are provided in Appendix 3 (only the proteins that matched the data filtering criteria described in the methods section are listed; iTRAQ ratios are raw, non-normalized values).
Up/down regulation or change in posttranslational modifications (PTMs)?
Research studies focused on protein differential expression analysis have often revealed that not all peptides that match a given protein show the same change in expression levels. For example, some peptides may display up-regulation, while others down-regulation, or no-change at all. Such results are confusing, and are often attributed to random errors associated with the quantitation method. However, even if contradictory, such results could also have a very reasonable explanation. Careful evaluation of the peptide-level raw data, in this study, has revealed that some of the peptides that displayed differential expression according to the iTRAQ measurements were also peptides that are known to carry PTMs. We present the case of PCBP1 Poly(rC) (Q15365) and BAG 3 (O95817) proteins identified in the 144KNt condition, for which, the peptides that contributed to the up-regulation status carried several known possible phosphorylation sites (p-sites) at Ser 246, 262, 263 and 264, and at Tyr 240 and 247, respectively [ExPAsy proteomics server, http://www.expasy.ch/]. The Sequest report for these two proteins is provided in Table 5 [note that the Poly(rC) protein was matched by only two unique peptides, thus, it was not included in the 144KNt dataset provided in Appendix 3]. Poly(rC) was matched by 8 spectral counts (corresponding to two unique peptides), of which, only five generated measurable iTRAQ ratios. We attributed the lack of measurable reporter ions for these tandem mass spectra to be the outcome of less than optimal fragmentation of peptides via PQD-MS. Of the two unique peptides, the non-phosphorylated peptide (L]LMHGK*EVGSIIGK*) did not display a significant change in abundance. The peptide that carried four possible Ser p-sites (Q]QS(246)HFAMMHGGTGFAGIDS(262)S(263)S(264)PEVK*) displayed a ∼2-3-fold increase in abundance for four independent tandem MS events and two independent iTRAQ ratio measurements (116/114 and 117/114). The marks “]” and “*” indicate the iTRAQ tag at the Nt or Lys residues, respectively. BAG3 was matched by 8 spectral counts (corresponding to four unique peptides), of which, four generated measurable iTRAQ ratios. Of these, three measurements were on a peptide that carries two known possible p-sites (T]HYPAQQGEY(240)QTHQPVY(247)HK*), and one measurement was on a peptide that carries no known p-site (EGHPVYPQLRPGYIPIPVLHEGAENR). The other two peptides did not generate measurable iTRAQ ratios. In the case of BAG3, all iTRAQ measurements displayed a ∼2-4 fold increase in abundance, including for the peptide that carries no known p-site. However, an RSp value of 4 for this peptide is an indicator that the peptide may not be a match for BAG3.
As no Ser/Thr/Tyr phosphorylations were allowed during database searching of the iTRAQ labeled samples, if in the E2 treated cells these peptides were completely or partially phosphorylated (thus being transparent to the search), and in the Tam treated cells were not, then the iTRAQ ratio measurements will display an up-regulation in the Tam condition as a larger abundance of non-phosphorylated peptides will be detected. We note that phosphorylated peptides could not be observed in this dataset even if the modification was enabled for database searching, as such peptides are hard to detect in the presence of their non-phosphorylated counterparts without proper enrichment techniques. However, as a variety of signaling pathways during cell proliferation and cell cycle arrest are mediated by protein phosphorylation/dephosphorylation events, it is worth pinpointing the biological significance of these proteins and of their phosphorylation status. For example, the Poly(rC) protein is a nucleic acid binding protein that functions as an accelerator of mRNA metabolic processes. The phosphorylation of Poly(rC) results in a marked decrease of its binding activity, and such an alteration of binding properties relates signal transduction pathways to nucleic acid dependent processes (transcription, translation, RNA processing).13,14 Likewise, the BAG family of proteins regulates a variety of cellular functions that include cell survival, cell proliferation, and cell motility. BAG3 was recently found to have anti-apoptotic activity and to sustain cell survival in response to stress inducing factors.15 In response to Tam treatment, which is inducing apoptosis in cells,16 dephosphorylation of the Poly(rC) could activate the protein to accelerate nucleic acid metabolic processes and prevent cell death, and up-regulation of BAG 3 or a change in it's phosphorylated status could inhibit apoptosis. Thus, the ability to place an experimental outcome in the proper biological context can clearly strengthen the validity of proteomic data interpretation.
Conclusions
A detailed examination of iTRAQ quantitation results, generated by allowing various combinations of amino acid modifications during database searching, revealed that the allowance of such modifications will impact mainly the number and type of identified peptides and proteins, but not the iTRAQ ratios of identified proteins, per se. Overall, we conclude that: (a) iTRAQ labeling of Tyr residues, Lys acetylation/carbamylation and Met oxidation generated the largest number of new peptides with good Sequest scores and p-values<0.001, of which Tyr and Lys labeling also generated the largest changes in protein losses or new protein identifications (up to 13 % or 21 %, respectively); the protein overlaps with the reference KNt condition were, however, in excess of 88 %; for practical purposes, conducting the database search with Tyr enabled iTRAQ modifications will generate the most reliable results (smallest number of protein losses, largest number of protein gains, and minimal increase in false rate of protein identifications); (b) Larger then 2-fold changes for proteins that were identified in the reference KNt condition were mainly the result of protein losses as a result of new database search parameters, than actual changes in iTRAQ ratios; as for the newly identified proteins with larger than 2-fold changes in iTRAQ ratios, manual examination of the modified peptide tandem mass spectra and their associated scores was necessary to confirm that such peptides were correct assignments and that their contribution to the protein iTRAQ ratios was legitimate; (c) To avoid an increase in the rate of false positive identifications, iTRAQ data generated on low-resolution mass spectrometers should not be searched simultaneously with many additional peptide modifications; in addition, it should be noted that all possible modifications on a peptide cannot be anticipated a priori, and it is unpractical to simultaneously allow for a large number of modifications due to a need for excessive computing power; high resolution/high mass accuracy mass spectrometers that can accurately confirm the modification type and site will be beneficial for generating high-confidence results; (d) The possible interference of changes in protein PTMs with the iTRAQ quantitation should not be ignored, but rather carefully assessed and confirmed by complementary analysis techniques; the larger the number of quantified peptides for a given protein, and the smaller the number of peptides that carry eventual PTMs, the smaller will be the impact of such peptides on the quantitation outcome; if at all possible, the peptides that are used for the quantitation of a protein should be minimally affected by PTMs, regardless of the chosen method for performing the quantitation; overall, changes in protein expression level should be also evaluated in a relevant biological context; (e) The results generated in this study will support the efforts invested in establishing guidelines for improved proteomic quantitation and differential expression analysis.
Supplementary Material
Acknowledgments
This project was partially supported by Award Number R21CA126669-01A1 from the National Cancer Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
References
- 1.Lill J. Mass Spectrom Rev. 2003;22:182–194. doi: 10.1002/mas.10048. [DOI] [PubMed] [Google Scholar]
- 2.Chen EI, Yates JR., 3rd Mol Oncol. 2007;1:144–159. doi: 10.1016/j.molonc.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ. Mol Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 4.Aggarwal K, Choe LH, Lee KH. Brief Funct Genomic Proteomic. 2006;5:112–120. doi: 10.1093/bfgp/ell018. [DOI] [PubMed] [Google Scholar]
- 5.DeSouza L, Diehl G, Rodrigues MJ, Guo J, Romaschin AD, Colgan TJ, Siu KW. J Proteome Res. 2005;4:377–386. doi: 10.1021/pr049821j. [DOI] [PubMed] [Google Scholar]
- 6.Wu WW, Wang G, Baek SJ, Shen RF. J Proteome Res. 2006;5:651–658. doi: 10.1021/pr050405o. [DOI] [PubMed] [Google Scholar]
- 7.Keshamouni VG, Michailidis G, Grasso CS, Anthwal S, Strahler JR, Walker A, Arenberg DA, Reddy RC, Akulapalli S, Thannickal VJ, Standiford TJ, Andrews PC, Omenn GS. J Proteome Res. 2006;5:1143–1154. doi: 10.1021/pr050455t. [DOI] [PubMed] [Google Scholar]
- 8.Armenta JM, Hoeschele I, Lazar IM. J Am Soc Mass Spectrom. 2009;20:1287–1302. doi: 10.1016/j.jasms.2009.02.029. [DOI] [PubMed] [Google Scholar]
- 9.Chaerkady R, Kerr CL, Kandasamy K, Marimuthu A, Gearhart JD, Pandey A. Proteomics. 2010;10:1–15. doi: 10.1002/pmic.200900483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kolla V, Jeno P, Moes S, Tercanli S, Lapaire O, Choolani M, Hahn S. J Biomed Biotechnol. 2010;2010:952047. doi: 10.1155/2010/952047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sarvaiya HA, Yoon JH, Lazar IM. Rapid Commun Mass Spectrom. 2006;20:3039–3055. doi: 10.1002/rcm.2677. [DOI] [PubMed] [Google Scholar]
- 12.Ow SY, Salim M, Noirel J, Evans C, Rehman I, Wright PC. J Proteome Res. 2009;8:5347–55. doi: 10.1021/pr900634c. [DOI] [PubMed] [Google Scholar]
- 13.Makeyev AV, Liebhaber SA. RNA. 2002;8:265–278. doi: 10.1017/s1355838202024627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leffers H, Dejgaard K, Celis JE. Eur J Biochem. 1995;230:447–453. [PubMed] [Google Scholar]
- 15.Rosati A, Ammirante M, Gentilella A, Basile A, Festa M, Pascale M, Marzullo L, Belisario MA, Tosco A, Franceschelli S, Moltedo O, Pagliuca G, Lerose R, Turco MC. Int J Biochem Cell Biol. 2007;39:1337–1342. doi: 10.1016/j.biocel.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 16.Stackievicz R, Drucker L, Radnay J, Beyth Y, Yarkoni S, Cohen I. Clin Cancer Res. 2001;7:415–420. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.