Abstract
Extracellular GRP94 (gp96) can initiate both innate and adaptive immune responses through interactions with antigen presenting cell surface receptors. Following the identification of CD91 as a receptor functioning in the cross-presentation of GRP94-associated peptides, scavenger receptors SR-A and SREC-I were demonstrated to function in GRP94 cell surface binding and endocytosis, lending controversy to the assignment of CD91 as the unique GRP94 endocytic receptor. To assess CD91 function in GRP94 surface binding and endocytosis, these parameters were examined in murine embyronic fibroblast (MEF) cell lines whose expression of CD91 was either reduced via RNAi or eliminated by genetic disruption of the CD91 locus. Reduction or loss of CD91 expression abrogated the binding and uptake of the CD91 ligand receptor-associated protein (RAP); surface binding and uptake of an N-terminal domain of GRP94 (GRP94.NTD) was unaffected. GRP94.NTD surface binding was markedly suppressed following treatment of MEF cell lines with heparin, the sulfation inhibitor sodium chlorate, or heparinase II, demonstrating that heparin sulfate proteoglycans can function in GRP94.NTD surface binding. The role of CD91 in the cross-presentaton of GRP94-associated peptides was examined in the DC2.4 dendritic cell line. In DC2.4 cells, which express CD91, GRP94.NTD-peptide cross-presentation was insensitive to RAP or activated α2-macroglobulin and occurred primarily via a fluid phase uptake pathway. In summary, these data clarify conflicting data on CD91 function in GRP94 surface binding, endocytosis and peptide cross-presentation and identify HSPGs as novel GRP94 cell surface binding sites.
Introduction
GRP94 (gp96, HSP90B1), the endoplasmic reticulum Hsp90 paralog, can initiate innate immune responses, via interacting with Toll-like receptor (TLR) 2 and TLR4, and adaptive immune responses, by directing GRP94-associated peptides into the major histocompatability (MHC) class I cross-presentation pathway of professional antigen presenting cells (pAPCs) (1-11). Although early studies of GRP94-elicited innate immune responses were confounded by concerns regarding the contribution of contaminating bacterial lipopolysaccharide to cell activation, GRP94 has subsequently been demonstrated to function in TLR2 and TLR4 activation (12-16). Additionally, more recent studies have demonstrated that complexes of GRP94 and either Pam3Cys or LPS act synergistically to elicit TLR2 and TLR4 activation, respectively (12). Furthermore, transgenic mice expressing a cell surface form of GRP94 display systemic MyD88-dependent dendritic cell activation, thus establishing a TLR-mediated proinflammatory function for GRP94 (17). With respect to adaptive immune responses, GRP94 can escort antigenic peptides into the MHC class I cross-presentation pathway (1). Thus, GRP94-peptide complexes can be internalized by pAPCs to yield subsequent activation of peptide-specific CD8+ T lymphocytes (18-21). This phenomenon is well established for GRP94-peptide complexes generated in vitro, where complex formation is enabled by co-incubation of GRP94 with synthetic peptides at elevated temperatures (50°C), or for extended time periods at room temperature (20 h at 24°C) (18, 22). It remains to be determined if the GRP94 peptide binding activity observed in vitro reflects an in vivo peptide binding function (23-25).
CD91 (low density lipoprotein receptor-related protein 1, LRP1) was the first endocytic receptor identified to function in the cross-presentation of GRP94-peptide complexes (26). CD91 is a member of the low density lipoprotein (LDL) receptor family and binds numerous structurally distinct and functionally divergent ligands, including apolipoprotein E, PDGF, lactoferrin, tissue factor pathway inhibitor, receptor-associated protein (RAP), and activated α2-macroglobulin (α2M*). Reflecting this diversity of ligands, CD91 displays a wide tissue distribution and participates in a variety of physiological responses, including lipoprotein metabolism, proteinase homeostasis, cell migration, and modulation of the blood-brain barrier (27, 28).
CD91 was first identified as a putative GRP94 receptor by affinity chromatography of cell membrane detergent extracts on a GRP94 affinity matrix (26). These studies, which yielded identification of a 80 kDa fragment of CD91, were further developed in GRP94-peptide cross presentation assays, where it was demonstrated that GRP94-mediated peptide cross-presentation could be inhibited by the CD91 ligands RAP and α2M, as well as a CD91 blocking antibody (26, 29, 30). Subsequent studies extended these findings and identified a broad role for CD91 in the cross-presentation of peptides associated with numerous heat shock proteins (HSPs) and molecular chaperones, including Hsp90, Hsp70, and calreticulin (29).
Given the diversity of cells that express CD91, it is not apparent how CD91-mediated GRP94-peptide uptake would be restricted to pAPCs, as would be expected if CD91 was the primary, immunologically-relevant receptor for Hsps and molecular chaperones. Indeed, following the discovery of CD91 as an HSP/chaperone receptor, a variety of other surface receptors were identified to function in HSP or GRP94 surface recognition and endocytic uptake, including the scavenger receptors SR-A, SREC-I, LOX-1, and FEEL-1 (31-35). In addition to broadening the number of surface receptors that can function in the recognition and endocytic uptake of HSPs, these findings have engendered controversy regarding the identity of CD91 as an endocytic receptor for GRP94 (31, 36, 37).
To directly assess CD91 function in GRP94 surface binding and internalization, these parameters were examined in both CD91 siRNA knock-down and CD91 knock-out fibroblast cell lines. Whereas the reduction or loss of CD91 expression resulted in markedly decreased RAP binding and internalization, surface binding and endocytic uptake of a recombinantly expressed N-terminal domain of GRP94 (GRP94.NTD) was unperturbed. CD91 function in cross-presentation was examined in DC2.4 murine dendritic cells and it was found that molar excess concentrations of RAP or α2M* resulted in either a modest (RAP) or no (α2M*) decrease in the cross-presentation of GRP94.NTD-associated peptides.
Materials and Methods
Cell culture and siRNA transfection
MEF-1 and PEA-13 cells were maintained in DMEM, glucose, L-glutamine, sodium pyruvate (Cellgro), supplemented with 10% fetal bovine serum (FBS; Gibco). DC2.4 cells were maintained in RPMI 1640 with L-glutamine (Cellgro) supplemented with 10% FBS (Gibco) and cultured according to ATCC recommendations. MEF-1 and PEA-13 cells were transfected with 200 nM CD91-directed siRNAs (Qiagen) using Lipofectamine2000 (Invitrogen) and OPTI-MEM serum-free media (Gibco), according to the manufacturer’s protocol. Cells were maintained in DMEM/10% FBS after transfection and examined for CD91 knock-down efficacy at 24, 48, and 72 h post-transfection by RT-PCR. The siRNA sequences used were: LRP1_1 (CAC GTT GGT TAT GCA CAT GAA) and LRP1_2 (CTG CCG GGT GTA CAA ATG TAA), and were directed against murine LRP1 (NM_008512).
Mice
OVA-specific TCR transgenic mice [C57BL/6-Tg(TcraTcrb)1100Mjb/J (OT-1) were purchased from Jackson Laboratories (Bar Harbor, ME, USA). All mice were housed in the Duke University Animal Facility (Durham, NC, USA), an Association for Assessment and Accreditation of Laboratory Animal Care International-approved facility. All experiments were conducted under an Institutional Animal Care and Use Committee-approved protocol.
Protein purification
Recombinant canine GRP94.LREK (residues 69-337) was expressed in E. coli strain BL21 as an N-terminal glutathione S-transferase fusion (pGEX-NB modified pGEX-2T vector), and purified as previously described (38). Following elution from Sepharose 4B glutathione agarose resin (GE Healthcare), the GST-tag was cleaved using thrombin (Haematologic Technologies). Thrombin cleavage was arrested by addition of PMSF and the protein pool was re-chromatographed on a Sepharose 4B glutathione-agarose column. Peak fractions were pooled, loaded onto a HiTrap QHP column (GE Healthcare), and washed with 150 column volumes of cold, sterile depyrogenation buffer (1% Triton X-114, 20 mM Tris, pH 8.0). Detergent was removed by washing the column with sterile 150 mM NaCl, 20 mM HEPES, pH 7.9 until the A230 returned to baseline. Recombinant protein was then eluted with sterile 750 mM NaCl, 20 mM HEPES, pH 8.0. Peak fractions were pooled and concentrated in a YM-10 centrifugal concentrator (Millipore). GRP94.NTD endotoxin content was determined to be ca. 15 EU/mg by the QCL-1000 chromogenic limulus amoebocyte lysate assay (Lonza).
Recombinant canine GRP94.NTD (residues 22-337) was expressed in E. coli strain BL21 as a His-tagged fusion (pET15b vector; Novagen), and purified as described above for RAP. Following depyrogenation, detergent was removed by washing the column with sterile PBS. Recombinant protein was then eluted with sterile 150 mM imidazole in PBS, pH 7.4. Peak fractions were pooled, concentrated in a YM-10 centrifugal concentrator (Millipore) and dialyzed against sterile PBS.
Recombinant rat RAP was expressed in E. coli strain BL21 as a His-tagged fusion (pLE1 modified pET24a vector) (39). Cultures were induced with 0.1mM isopropyl-β-D-galactopyranoside for 3 h and bacteria recovered by centrifugation. Bacterial pellets were resuspended in cold buffer A (50 mM dextrose, 50 mM Tris, pH 8.0, 10 mM imidazole, 300 mM NaCl), and homogenates prepared by French press. Homogenates were incubated with an equal volume of buffer B (10 mM Tris, pH 8.0, 50 mM KCl, 0.5% (v/v) Tween 20, 0.5% (v/v) Triton X-100, 300 mM NaCl, and 10 mM imidazole) for 30 min on ice. Insoluble debris was subsequently removed by centrifugation at 40,000 × g for 30 min at 4°C. The supernatant fraction was applied to nickel-Sepharose resin (GE Healthcare), washed with 30 column volumes of wash buffer (PBS with 0.2% (v/v) Tween 20, 0.2% (v/v) Triton X-100, and 40 mM imidazole, pH 8.0), and washed overnight in sterile depyrogenation buffer (1% Triton X-114 in PBS, pH 7.5). Recombinant protein was eluted using sterile 150 mM imidazole in PBS, pH 7.9, and dialyzed against sterile PBS. To remove excess detergent, protein was incubated with sterilized, depyrogenated Bio-Beads (Bio-Rad) for 1 h at 4°C, and then dialyzed against PBS.
Purification of α2M was performed using endotoxin-free plasma, columns, and buffers and followed a protocol modified from that described previously (40). Mouse plasma was purchased from Harlan. α2M was separated from mouse plasma by consecutive precipitation with 4% and 16% polyethylene glycol (Fluka), followed by DEAE Sephacel fractionation (Sigma-Aldrich) and purified further over a Sephacryl S-300 sizing column (GE Life Sciences). Native α2M was converted to the activated form (α2M*) by incubation in 200 mM ammonium bicarbonate at 37°C overnight. Activated protein was then dialyzed for 4 h into PBS and stored at 4°C. Purified protein contained <10 pg endotoxin/mg protein, as determined by Limulus amoebocyte lysate assay (Cambrex).
Protein labeling
Proteins were conjugated to Alexa Fluor carboxylic acid succinimidyl esters (Molecular Probes) according to the manufacturer’s protocol. Briefly, purified protein samples were diluted with sterile 0.15 M sodium bicarbonate, pH 9.0, and incubated with Alexa Fluor dye (dissolved in sterile DMSO) for 60 min at room temperature in the dark. Free dye was removed using a depyrogenated, PBS-equilibrated Sephadex G-25 column. Proteins were concentrated in a YM-10 or YM-30 centrifugal concentrator (Millipore).
Analytical ultracentrifugation
The molecular weight/aggregation state of GRP94.NTD preparations were determined by sedimentation equilibrium ultracentrifugation in a Beckman XL-A analytical ultracentrifuge. GRP94.NTD, at concentrations of 0.9, 1.7, and 2.3 mg/mL in 50 mM Tris, pH 8.0, 100 mM NaCl, 1 mM DTT was analyzed at 10,000 rpm. The partial specific volumes (0.7278) and solvent densities (1.015) were calculated from the amino acid and solvent compositions using SEDENTERP.
Reverse-transcriptase PCR
Total RNA was extracted from tissue culture cells using TRIzol Reagent (Invitrogen) according to manufacturer’s specifications and dissolved in nuclease-free water (Ambion). RNA concentrations were determined spectrophotometrically and RNA integrity was assessed using a 1% formaldehyde gel. Samples were treated with RNase-free DNase I (Ambion) at 37°C for 30 min prior to heat-inactivation. cDNA was synthesized from 7.2 μg DNase-treated RNA using Superscript (Invitrogen), according to the manufacturer’s protocol. cDNA was recovered by phenol/chloroform/isoamyl alcohol (25:24:1) extraction and sodium acetate/ethanol precipitation, and resuspended in 10 mM Tris-Cl, pH 8.5.
PCR was performed using Taq polymerase (Qiagen). The following primer sequences were used: CD91 sense: 5′-ATC ACC CTT CCC GGC AGC CCA-3′; CD91 antisense: 5′-ACC CAG AGC CAT CGG CTT TGT-3′; 18S sense: 5′-TCA AGA ACG AAA GTC GGA GG-3′; 18S antisense: 5′-GGA CAT CTA AGG GCA TCA CA-3′ (IDT). Thermocycling was conducted in a PTC-200 Thermal Cycler (MJ Research) using the following parameters: denaturation at 94°C for 3 min; 33 cycles of 94°C for 30 seconds, 62°C for 1 min, 72°C for 1 min; final extension at 72°C for 10 min. PCR products were separated on a 1% agarose gel and visualized using ethidium bromide. Quantification of band intensities was measured using Image J version 1.38x (NIH).
Surface binding and internalization assays
MEF-1 and PEA-13 cultures were rinsed and incubated with pre-warmed PBS++ (PBS with 1% BSA (Gibco), supplemented with 0.22 μm filter-sterilized 0.9 mM CaCl2 and 0.5 mM MgCl2) for 30 min at 37°C. Cells were then rinsed with room temperature PBS without calcium or magnesium (Gibco), lifted with Versene (Gibco), and diluted with an equal volume of cold PBS++. Surface binding and uptake experiments were conducted as previously described (4). Where indicated, cells were incubated with heparin (Sigma) or RAP for 30 min on ice, washed, and then incubated with the specified ligand for an additional 30 min.
Internalization experiments were conducted as previously described, with minor modifications (4). Briefly, cell cultures were rinsed, lifted, incubated with RAP or GRP94.NTD, and washed as described above. Cell suspensions were supplemented with ligands and incubated at 37°C with 5% CO2. Where indicated, cells were pre-incubated with 10 μg/mL unlabeled RAP, and 1 mg/mL unlabeled RAP was included in the internalization buffer (pre-conditioned media). Endocytosis was arrested by the addition of ice cold PBS++ at the specified time points. Cells were then washed with additional cold buffer. To remove residual surface-bound ligand, cells were treated with 0.25% trypsin-EDTA (Gibco) for 15 min on ice and proteolysis was arrested with 50 μg/mL soybean trypsin inhibitor. Cells were washed and resuspended in cold PBS++.
To distinguish between fluid-phase and receptor-mediated endocytosis, cells were incubated at 37°C in the presence of labeled RAP or labeled GRP94.NTD. Where indicated, excess unlabeled RAP or unlabeled GRP94.NTD were included in the internalization buffer. Because GRP94.NTD forms higher-order oligomers following heat-shock, the GRP94.NTD used in these experiments was prepared in the absence of ova20 peptide, as previous described (18). Endocytosis was arrested by the addition of ice cold PBS++ at the specified time points.
For protease inhibition studies, cells were incubated with pre-warmed PBS++ containing 100 μg/mL leupeptin (Sigma) for 30 min. 100 μg/mL leupeptin was also included during surface binding and internalization steps.
All samples were stained with propidium iodide (PI) for 30 min on ice and immediately analyzed using an LSRII flow cytometer (BD). PI-positive cells were excluded from final analyses, and cell autofluorescence was subtracted from the total fluorescence at each data point. All flow cytometry data was analyzed using FlowJo version 8.6.1. All data represents mean fluorescence intensity, and are depicted using biexponential scaling.
All samples analyzed by confocal analysis were fixed with 4% paraformaldehyde, mounted using FluorSave Reagent (Calbiochem), and imaged on a Zeiss LSM510 laser scanning confocal microscope. Labeled GRP94.NTD (22-337) was utilized used for these GRP94.NTD surface binding studies.
Sodium chlorate and heparinase II treatments
MEF-1 and PEA-13 cells were exchanged into media containing 20 mM sodium chlorate (cell culture tested; Sigma), and cells were grown for an additional 24 h. Cells were rinsed and incubated with pre-warmed PBS++ supplemented with 20 mM sodium chlorate for 30 min at 37°C. Cells were then rinsed, lifted, and stained as described above. To examine the effects of heparinase II treatment of cells on GRP94 surface binding, MEF-1 and PEA-13 cells were rinsed with PBS++ and incubated with 0.01 IU/mL heparinase II (Sigma) in PBS++ for 90 min at 37°C. Cells were then rinsed, lifted, and stained as described above. Heparinase II enzymatic activity was assayed at the onset of each experiment. Where indicated, de-sulfation of cell surface HSPGs was confirmed using the monoclonal anti-heparan sulfate antibody 10E4 (Seikagaku) and a goat anti-mouse IgG conjugated to Alexa Fluor 647 (Invitrogen). All experiments included a purified mouse IgM isotype control (Invitrogen).
Trypsin-mediated proteolysis of GRP94
Labeled GRP94.NTD was incubated with trypsin at a 34:1 (w/w) ratio in cytosol/MES buffer (100 mM KCl, 20 mM NaCl, 2 mM Mg(OAc)2, 25 mM MES, pH 6.0). Proteolysis reactions were quenched at the indicated time points by addition of 20% trichloroacetic acid. The precipitated proteins were analyzed by 12.5% SDS-PAGE and visualized with Coomassie Brilliant Blue-G250 staining (BioRad). The relative amounts of intact GRP94 were normalized to the total amount of undigested GRP94 using ImageJ (NIH). To examine the effects of proteolytic cleavage on fluorescence intensity, fluor-conjugated GRP94 was incubated in cytosol buffer until a stable baseline was acquired. Trypsin was then added to the solution and the fluorescence determined at 30 second intervals. Experiments were conducted on a Shimadzu RF-5301 PC spectrofluorophotometer using an excitation wavelength of 647 nm and an emission wavelength of 665 nm. Relative GRP94 fluorescence was normalized to the maximum signal acquired during each analysis.
Cross-presentation of GRP94/ova20 complexes
GRP94.NTD/peptide complexes were prepared using ova20 peptide (SGLEQLESIINFEKLTEWTS) (NeoBioScience) as previously described (18). Free peptide was removed by dialysis against sterile PBS at 4°C. To assay cross-presentation, DC2.4 cells (2.5 × 104 cells/well) were co-incubated with 10 μg/mL GRP94.NTD/ov20 complex in the presence or absence of 300 μg/mL α2M, 300 μg/mL α2M*, 500 μg/mL RAP, or 500 μg/mL GRP94.NTD in pre-conditioned media. These concentrations represent an approximately 15-fold (α2M, α2M*) or 50-fold (RAP, GRP94) molar excess of competitor. GRP94.NTD used in competition experiments was prepared in the absence of ova20 peptide, as previous described (18). Cells were incubated for 4 h under standard conditions, rinsed with PBS containing calcium and magnesium to remove unbound protein, and replenished with 100 μL complete media. OT-1 splenocytes were harvested from MHC class I-restricted OVA-transgenic mice, and single cell suspensions were prepared by repeated subcapsular injection of complete culture media (high glucose phenol red-free DMEM supplemented with 10% FBS, 1% penicillin-streptomycin, 1 mM sodium pyruvate, 2 mM L-glutamine, 10 mM HEPES, 0.1 mM nonessential amino acids, and 50 μM βME). The DC2.4 cells were then co-cultured with 2.5 × 105 OT-1 splenocytes. Cell supernatants were harvested after 24 h and stored at −20°C. IFN-γ levels were quantified using DUO-ELISA kits (R&D Systems).
Statistics
Values reported in all analyses are expressed as the mean ± SD. Differences between groups were analyzed using a paired Student t test. Statistical significance was accepted at p < 0.01.
Results
Expression and biophysical characterization of GRP94.NTD
We previously demonstrated that in vivo expression of a secretable form of the GRP94 N-terminal domain (GRP94.NTD) (residues 22-337) elicited anti-tumor immunity and that highly purified, low endotoxin recombinant GRP94.NTD activated pAPCs (12, 41, 42). Additionally, Biswas et al. demonstrated that GRP94.NTD-synthetic peptide complexes were competent for cross-presentation in vitro and could elicit peptide-specific cytotoxic T cell responses in vivo (43). On the basis of these studies, GRP94.NTD was used to investigate CD91 function in GRP94 surface binding, internalization, and peptide cross-presentation.
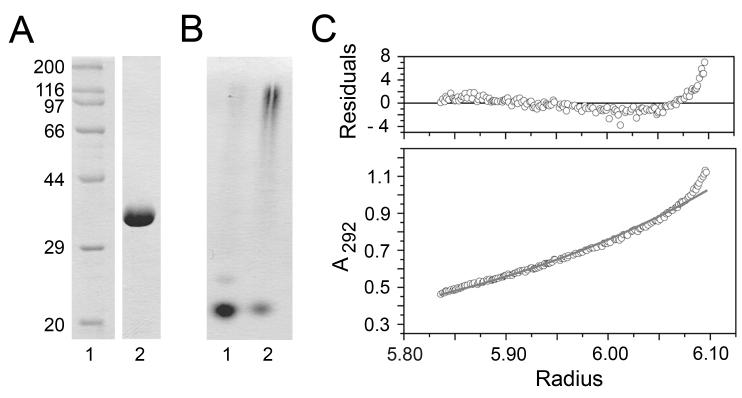
GRP94.NTD (residues 22-337) and GRP94.LREK (residues 69-337) were produced by recombinant expression, purified, and depyrogenated. Both forms were purified to near homogeneity and migrated predominantly as monomers in native PAGE (Figs. 1A and B). Following incubation at 42°C for 30 min, both GRP94.NTD and GRP94.LREK formed higher-order oligomers that displayed reduced mobility in native PAGE, as described in previous publications (44). Analytical ultracentrifugation analyses confirmed that GRP94.LREK behaved as a primarily homogeneous species of 41 kDa at 4°C (Fig. 1C). Because GRP94.NTD and GRP94.LREK behaved comparably in all assays, the term GRP94.NTD will be used interchangeably to describe both constructs.
Figure 1. Expression and biophysical characterization of GRP94.NTD.
Recombinant GRP94.LREK was expressed, purified, and depyrogenated. A. 5 μg GRP94.LREK (69-337) (lane 2) was resolved by 12.5% SDS-PAGE gel and detected by Coomassie Blue staining. B. To examine the oligomeric state of purified GRP94.LREK, 7.5 μg GRP94.LREK was resolved by 6% native PAGE and detected by Coomassie Blue staining (lane 1). Incubation at 42°C for 30 min promotes the formation of higher-order GRP94.LREK oligomers (lane 2). C. Analytical ultracentrifugation was performed on 2.3 mg/mL GRP94.LREK at 4°C. The protein displayed a high degree of homogeneity, and mass average calculations yielded a molecular mass estimation of 41 kDa.
Cell surface binding of GRP94.NTD is independent of CD91 expression
CD91 was the first receptor proposed to function in GRP94 surface binding and endocytosis (26). The initial experimental evidence supporting a physiological function of CD91 in GRP94 surface binding and uptake was primarily indirect, and derived from the observations that GRP94-mediated peptide cross-presentation was inhibited by CD91 ligands (26). More recently, siRNA-mediated knock-down of CD91 expression was shown to markedly suppress cell surface GRP94 binding and GRP94-mediated peptide cross-presentation by RAW264.7 macrophage cells (30). While these data are consistent with an essential role for CD91 in GRP94 surface recognition and endocytic uptake, the observed inhibition was only seen with a GRP94/ova complex; no inhibition was seen with a GRP94/SIINFEKL complex (26).
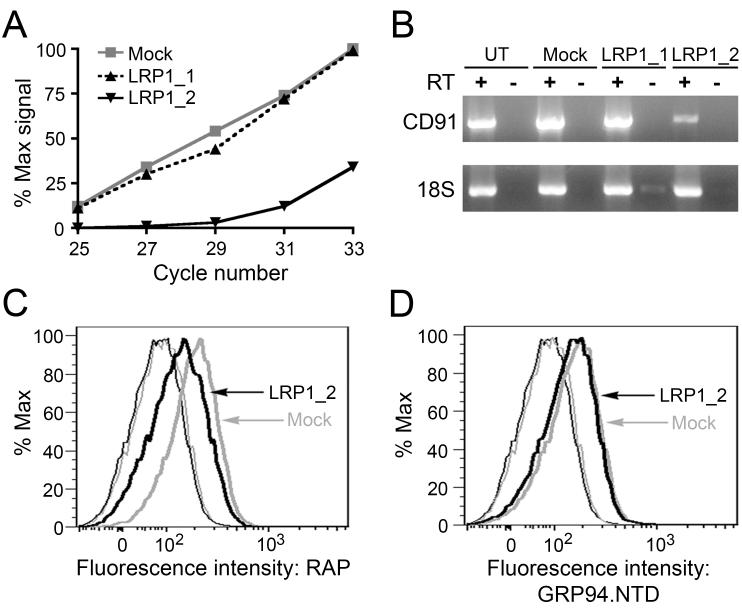
To examine CD91 function in GRP94 surface recognition, binding studies were conducted in CD91-expressing mouse embryonic fibroblast (MEF-1) cells that had been transfected with either of two CD91-directed siRNAs (LRP1_1 or LRP1_2). The effects of LRP1_1 and LRP1_2 on CD91 expression were monitored by RT-PCR. At 24 h post-transfection, cells transfected with LRP1_2 displayed a 75% reduction in CD91 mRNA levels, as compared to untransfected (UT) and mock transfection controls (Figs. 2A and B). Transfection with LRP1_1 had no effect on CD91 mRNA levels, and was used as a control for off-target effects. Plots of relative PCR product intensity versus PCR cycle number indicated that amplification of the CD91 target sequence in the mock and LRP1_1-transfected cells was within the linear range (Fig. 2A). Neither siRNA affected 18S rRNA expression, and genomic DNA contamination was monitored using paired RNA samples in which reverse transcriptase was omitted.
Figure 2. Suppression of CD91 expression does not reduce GRP94.NTD surface binding to MEF-1 cells.
MEF-1 cells were transiently transfected with either of two CD91-targeting siRNAs (LRP1_1 and LRP1_2) or a vector only (mock) control, and examined for changes in CD91 expression 24 h post-transfection. A. Total RNA samples were isolated and CD91 mRNA knockdown efficiency was determined by RT-PCR. CD91 cDNA from mock-transfected cells displayed linear amplification between PCR cycles 25 and 33 (R2 = 0.9974). B. Untransfected (UT), mock-transfected, and siRNA-transfected cells were examined for changes in CD91 mRNA levels using RT-PCR. All samples were tested for DNA contamination using a paired transcriptase-deficient (RT-) reaction, and were examined for off-target effects to 18S rRNA. C & D. MEF-1 cells were transfected with LRP1_2 siRNA or a mock control and analyzed by flow cytometry. CD91 surface expression was determined using 10 μg/mL RAP (C), and GRP94.NTD surface binding was conducted with 25 μg/mL GRP94.NTD (D): unstained cells (thin lines), stained cells (bold lines), mock-transfected cells (grey lines), and LRP1_2 siRNA transfected cells (black lines). Mock-transfected and untransfected cells displayed identical RAP and GRP94.NTD binding (data not shown). The data presented are representative of three independent replicates.
CD91 surface expression was assayed by monitoring the surface binding of fluorescently-labeled receptor-associated protein (RAP) using flow cytometry. RAP, a 39 kDa endoplasmic reticulum resident chaperone, binds multiple sites on the ectodomain of CD91 and is known to inhibit binding of all known CD91 ligands (45). Compared to mock-transfected cells, RAP binding to LRP1_2-transfected cells was reduced by 45%, a finding consistent with prior results (Fig. 2C) (26). These data demonstrate that CD91 expression was significantly reduced following transfection with LRP1_2, yet mock- and LRP1_2-transfected cells bound equivalent levels of fluorescently-labeled GRP94.NTD (Fig. 2D). Transfection with LRP1_1 did not affect RAP or GRP94.NTD binding and served as a negative control (data not shown). These data suggest that GRP94.NTD binds to receptor sites other than CD91.
Although RAP is an antagonist of all established CD91 ligands, it is also known to bind to other LDL family receptors and thus RAP competition studies are not uniquely diagnostic of CD91 function (45). Therefore, additional experimental approaches were utilized to examine CD91-dependent versus CD91-independent GRP94.NTD surface interactions. To this end, RAP and GRP94.NTD surface binding were examined in the CD91 homozygous knockout cell line PEA-13. PEA-13 is a CD91−/− cell line that had been selected from mouse embryonic fibroblasts heterozygous for a disrupted CD91 allele (46). Consistent with this genotype, PEA-13 cells lack CD91 expression at the message level and display very low RAP binding activity (Fig. 3B). Notably, PEA-13 cells displayed higher GRP94.NTD binding activity than MEF-1 cells (Fig. 3C). As an additional means of assessing CD91-GRP94 interactions, RAP surface binding studies were conducted in the presence of excess GRP94.NTD. As can be seen in Fig. 3D, the addition of a 100-fold molar excess GRP94.NTD did not significantly decrease RAP surface binding to MEF-1 cells. The very modest degree of competition seen was also observed in the presence of 100-fold molar excess of bovine serum albumin. In parallel experiments, a 40-fold molar excess unlabeled RAP did not discernibly affect GRP94.NTD surface binding to MEF-1 cells (data not shown). Combined, these data demonstrate that GRP94.NTD binding to MEF and PEA-13 cells is independent of CD91 expression.
Figure 3. GRP94.NTD binds MEF-1 and PEA-13 cells independently of CD91 expression.
A. MEF-1 and PEA-13 cells were analyzed for CD91 expression using RT-PCR. All samples were tested for DNA contamination using a paired transcriptase-deficient (RT-) reaction, and were examined for off-target effects to 18S rRNA. B & C. MEF-1 and PEA-13 cells were incubated with 10 μg/mL RAP (B) or 25 μg/mL GRP94.NTD (C), washed, and analyzed by flow cytometry: unstained cells (thin lines), stained cells (bold lines), MEF-1 cells (black lines), and PEA-13 cells (grey lines). D. MEF-1 cells were incubated with 10μg/mL RAP in the absence or presence of 100-fold molar excess unlabeled RAP, GRP94.NTD, or BSA. Cells were then washed and analyzed by flow cytometry: unstained cells (thin grey line), RAP (bold black line), unlabeled RAP (thin black line), unlabeled GRP94.NTD (bold grey line), and unlabeled BSA (shaded grey line). Data presented are representative of three independent replicates.
Identification of low-affinity/high-capacity RAP and GRP94.NTD binding activities
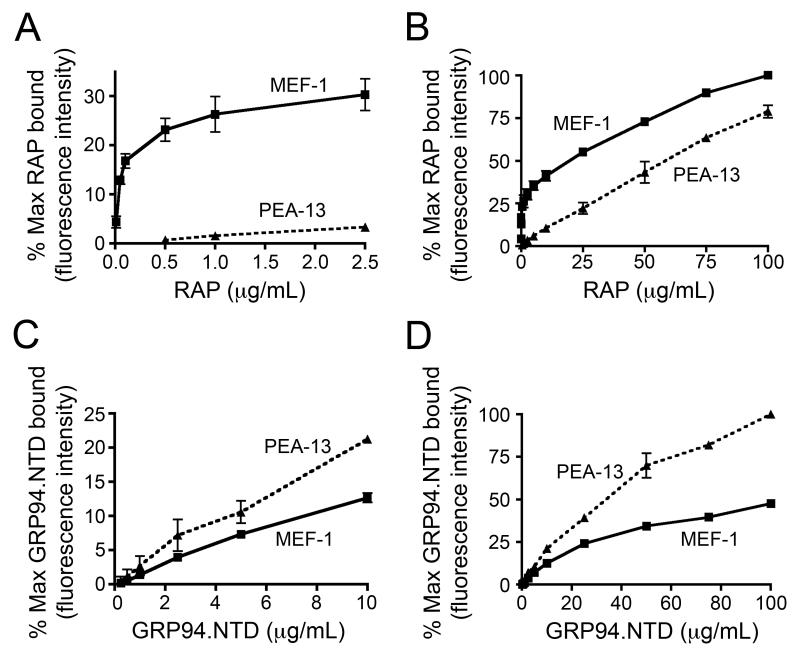
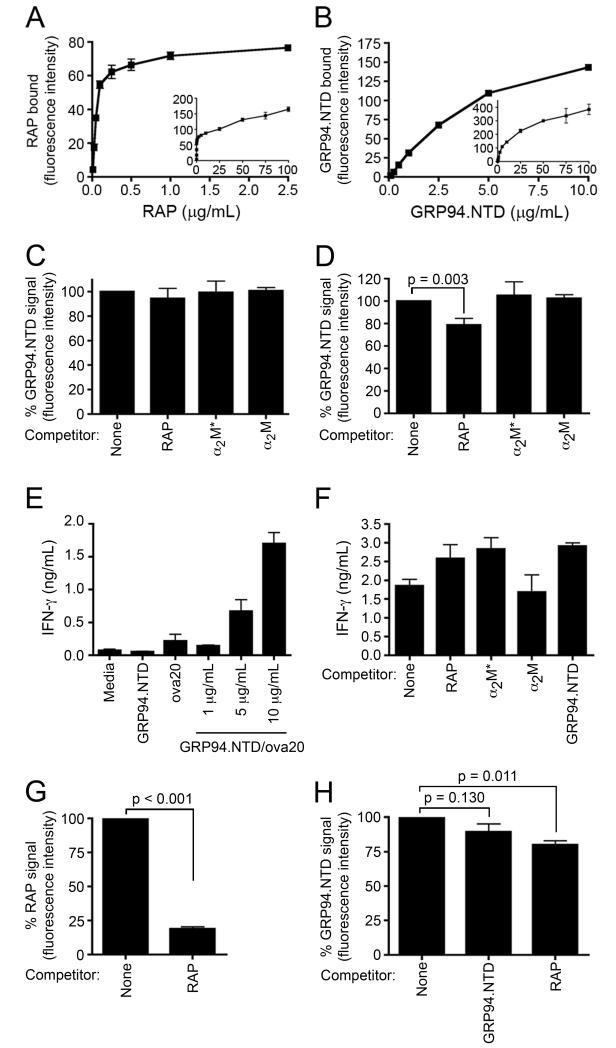
The interaction of RAP and GRP94.NTD with cell surface binding sites expressed by MEF-1 and PEA-13 cells was further characterized in ligand titration studies. In these experiments, RAP displayed high-affinity, saturable binding to MEF-1 cells, with half-maximal and maximal binding observed at ~0.2 and 1.0 μg/ml, respectively (Fig. 4A). Conversely, PEA-13 cells displayed very low RAP binding activity over the same concentration range. Discernible RAP binding to PEA-13 cells was only observed at concentrations that markedly exceeding those necessary to saturate RAP binding to MEF-1 cells. To examine this low-affinity binding phenomenon in greater detail, RAP binding titrations were performed with both cell lines (Fig. 4B). Both MEF-1 and PEA-13 cells displayed linear, low-affinity/high-capacity RAP binding, with no saturation evident at RAP concentrations up to 100 μg/ml. In contrast, GRP94.NTD binding to MEF-1 and PEA-13 cells was linear at concentrations up to 25 μg/ml (Fig. 4C and D), and essentially linear up to 100 μg/ml. As previously observed, PEA-13 cells bound more GRP94.NTD than MEF-1 cells at all concentrations examined. These low-affinity/high-capacity binding interactions indicate that GRP94.NTD interacts with surface sites that are highly expressed by both MEF-1 and PEA-13 cells.
Figure 4. Binding characteristics of RAP and GRP94.NTD to MEF-1 and PEA-13 cells.
MEF-1 and PEA-13 cells were incubated with increasing concentrations of RAP (A and B) or GRP94.NTD (C and D). Cells were subsequently washed and analyzed by flow cytometry. Results are expressed as the mean of three independent experiments ± SD.
Cell surface heparan sulfate proteoglycans (HSPGs) are binding sites for GRP94.NTD
The GRP94.NTD binding characteristics reported above are similar to the low-affinity/high-capacity binding interactions observed for numerous cell surface HSPG ligands. HSPGs are also known to bind a diverse array of protein ligands and serve as co-receptors for numerous surface receptors, including CD91 (47-55). Additionally, GRP94 is known to bind heparin, which has a structure similar to HSPG polysaccharide side chains (56-58).
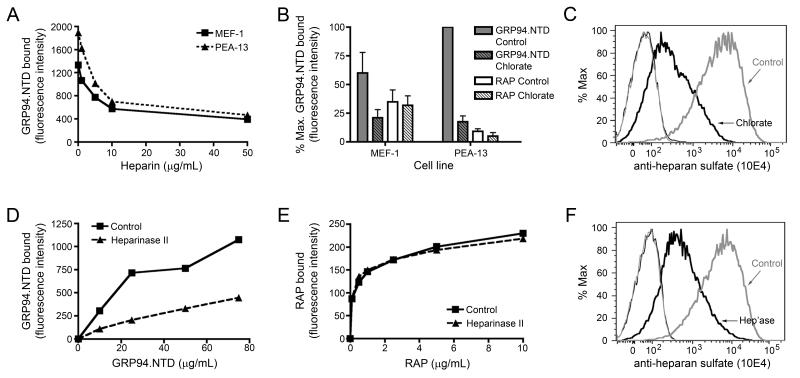
To determine if cell surface HSPGs contribute to GRP94.NTD surface binding to MEF-1 and PEA-13 fibroblasts, three experimental approaches were used. First, MEF-1 and PEA-13 cells were incubated with increasing concentrations of heparin, washed to remove unbound heparin, and examined for GRP94.NTD and RAP binding activity. As shown in Fig. 5A, heparin treatment effectively reduced GRP94.NTD surface binding to MEF-1 and PEA-13 cells. The heparin-dependent inhibition of GRP94.NTD binding was saturable, with near-maximal inhibition occurring at 10 μg/ml heparin. In contrast, RAP binding was insensitive to heparin treatment, even at heparin concentrations two orders of magnitude higher than those needed to inhibit GRP94.NTD binding (data not shown). These data further distinguish the surface binding interactions of RAP and GRP94.NTD.
Figure 5. Modulation of cell surface HSPG structures decreases GRP94.NTD surface binding.
A. MEF-1 and PEA-13 cells were incubated with increasing concentrations of heparin, washed, incubated with 25 μg/mL GRP94.NTD, and analyzed by flow cytometry. B. MEF-1 and PEA-13 cells were grown in the absence (solid) or presence (striped) of 20 mM sodium chlorate. Cells were incubated with 25 μg/mL GRP94.NTD (grey) or 10 μg/mL RAP (white), washed, and analyzed by flow cytometry. Results are expressed as the mean of three independent experiments ± SD. C. To confirm that sodium chlorate treatment reduced HSPG sulfation, MEF-1 cells were stained with the anti-heparan sulfate antibody 10E4, washed, and analyzed by flow cytometry: unstained cells (thin lines), stained cells (bold lines), control cells (grey lines), and chlorate-treated cells (black lines). D & E. MEF-1 cells were incubated with or without 0.01 IU/mL heparinase II (Hep’ase) for 90 min at 37°C. Cells were then incubated with increasing concentrations of GRP94.NTD (D) or RAP (E), and analyzed by flow cytometry. F. To confirm efficient de-sulfation by heparinase II, MEF-1 cells were stained with the anti-heparan sulfate antibody 10E4, washed, and analyzed by flow cytometry: unstained cells (thin lines), stained cells (bold lines), control cells (grey lines), and chlorate-treated cells (black lines). All data presented are representative of at least three independent replicates.
In an alternative experimental approach, MEF-1 and PEA-13 cells were treated with sodium chlorate. Sodium chlorate competitively inhibits the formation of the cellular sulfate donor 3′-phosphoadenosine 5′-phosphosulfate (PAPS) and thereby reduces the sulfation of HSPG glycan chains during their biosynthesis (59-64). After culturing cells in sodium chlorate-supplemented media for 24 h, GRP94.NTD and RAP binding was examined by flow cytometry. This treatment resulted in a nearly complete loss of GRP94.NTD binding activity to both MEF-1 and PEA-13 cells, but had no significant effect on RAP binding (Fig. 5B). Following sodium chlorate treatment, a substantial (>90%) reduction in HSPG sulfation was confirmed using the monoclonal anti-heparan sulfate antibody 10E4 (Fig. 5C).
Lastly, MEF-1 and PEA-13 cells were treated with heparinase II, which cleaves sulfated glycan chains, and then assayed for GRP94.NTD and RAP surface binding. Digestion with heparinase II resulted in a substantial reduction in GRP94.NTD binding at all concentrations examined (Fig. 5D). It did not, however, alter RAP binding (Fig. 5E). Digestion of PEA-13 cells with heparinase II also resulted in a significant reduction in GRP94.NTD surface binding (data not shown). A substantial (>90%) decrease in cell surface HSPG sulfation was confirmed using the anti-heparan sulfate antibody 10E4 (Fig. 5F). These data demonstrate that disruption of HSPG surface structures significantly reduces GRP94.NTD but not RAP binding to MEF-1 and PEA-13 cells, and identify a role for HSPGs in GRP94.NTD cell surface binding. At present, we do not know the identity(s) of the HSPGs involved in GRP94.NTD surface binding.
RAP and GRP94.NTD are internalized via pathways that are kinetically and spatially distinct
It is well established that cell surface HSPGs can facilitate ligand binding to CD91 (52, 54). This is believed to occur through either the formation of ligand-HSPG complexes that are recognized by CD91, or through the “transfer” of HSPG-bound ligands to CD91 for subsequent internalization and/or processing (52, 54). These findings are of particular relevance to the conflicting data regarding the role of CD91 in GRP94 uptake, as they suggest that CD91 could function in GRP94 endocytosis without directly contributing to GRP94 recognition at the cell surface.
To determine if CD91 supports GRP94.NTD internalization, the kinetics of both RAP and GRP94.NTD internalization were examined. MEF-1 cells were incubated with either RAP or GRP94.NTD on ice, washed, and then warmed to 37°C to allow internalization of surface-bound proteins. At the indicated intervals, cells were rapidly cooled and subjected to surface proteolysis to remove residual surface-bound ligands. As would be predicted of a typical CD91 ligand, RAP was rapidly endocytosed by MEF-1 cells, with 80-90% of the total surface-bound RAP becoming resistant to external proteases within 5 min (Fig. 6A). The kinetics of GRP94.NTD internalization were considerably slower than those determined for RAP, with little uptake being observed before 20 min. Because CD91 internalization rates are known to be unusually high, the relatively sluggish rate of GRP94.NTD internalization argues against a significant role for CD91 in GRP94.NTD endocytosis (28, 65). In further support of CD91-independent modes of internalization for GRP94.NTD, MEF-1 and PEA-13 cells internalized GRP94.NTD at nearly identical rates (data not shown).
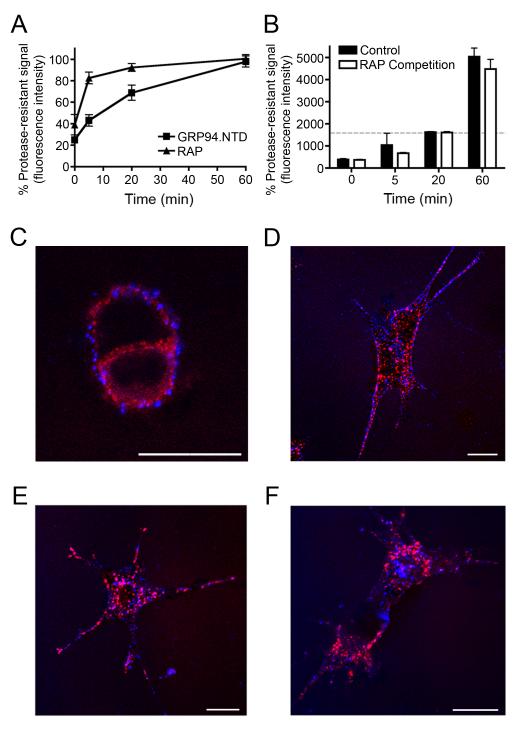
Figure 6. GRP94.NTD and RAP are internalized via spatially and kinetically distinct pathways.
A & B. MEF-1 cells were incubated with either 10 μg/mL RAP or 25 μg/mL GRP94.NTD, washed, warmed to 37°C for 0, 5, 20, or 60 min, and placed on ice to arrest endocytosis. Residual surface-bound ligands were removed by proteolysis on ice. The percent protease-resistant signal was calculated by normalizing the protease resistant signal to the total signal at each time point. Results are expressed as the mean of three independent experiments ± SD B. Comparison of MEF-1 cells treated either as in A (control) or treated with 10 μg/mL RAP prior to GRP94.NTD incubation and excess RAP during internalization (RAP Competition). Residual surface-bound ligands were removed by proteolysis on ice. The dashed gray bar indicates total GRP94.NTD surface binding prior to treatment with extracellular protease. C-F. MEF-1 cells were incubated with 5 μg/mL RAP (red) and 40 μg/mL GRP94.NTD (blue), washed, and warmed to 37°C for 0 (C), 5 (D), 15 (E), or 60 (F) min to allow internalization. Cells were then fixed and processed for confocal microscopy. Scale bars: 20 μm. All data presented are representative of three independent replicates.
To determine if CD91 directly functions in GRP94.NTD internalization, GRP94.NTD uptake experiments were conducted in the presence of excess RAP. In these experiments, cells were pre-incubated with saturating levels of RAP on ice prior to GRP94.NTD surface binding. Because CD91 is rapidly internalized and recycled back to the cell surface, a very large molar excess of unlabeled RAP was included in the internalization buffer to ensure that recycling CD91 molecules would be complexed by extracellular RAP. As expected, these conditions significantly inhibited both RAP surface binding and internalization (data not shown). However, these conditions had no effect on either the quantity or the rate of GRP94.NTD endocytosis (Fig. 6B).
Because the uptake assay used in these experiments measures both internalization and intracellular processing, these data could also be interpreted to indicate that RAP and GRP94.NTD differ significantly in their relative sensitivity to post-internalization processing. The observed time-dependent increase in the fluor-conjugated GRP94.NTD fluorescence would then reflect the kinetics of intracellular processing, rather than the kinetics of ligand uptake. To address this alternative interpretation, the kinetics of RAP and GRP94.NTD endocytosis were visualized by confocal microscopy. MEF-1 cells were incubated with RAP and GRP94.NTD on ice, washed, warmed to 37°C to allow internalization, rapidly cooled at the indicated intervals, fixed, and examined by confocal microscopy (Figs. 6C–F). In agreement with the kinetic analyses of RAP and GRP94.NTD uptake measured by flow cytometry, RAP was fully internalized within 15 min (Fig. 6D). Conversely, GRP94.NTD remained at the cell periphery and was internalized via a substantially slower endocytic route. These micrographs also revealed that RAP and GRP94.NTD seldom co-localize either at the cell surface or during endocytosis. Together, these data demonstrate that RAP and GRP94.NTD are internalized via distinct, independent pathways.
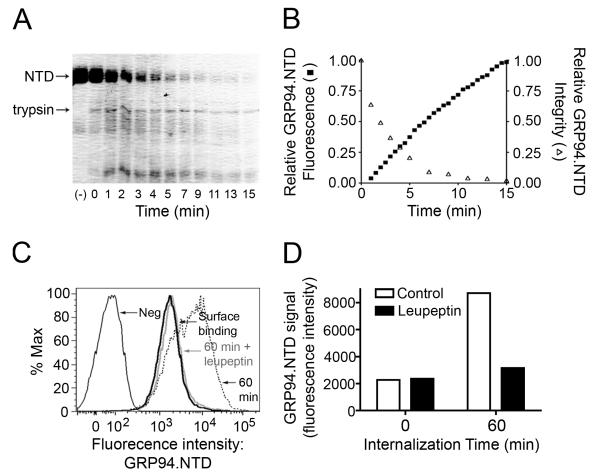
GRP94.NTD that had been internalized by cells for 60 min exhibited a level of fluorescence that exceeded the initial signal from surface-associated protein (Fig. 6B). It has been previously reported that proteolysis of fluor-conjugated proteins can result in the loss of intramolecular quenching and a subsequent increase in fluorescence yield (66, 67). To determine if fluor-conjugated GRP94.NTD behaved in a similar manner, GRP94.NTD integrity and fluorescence were monitored following digestion with tryspin (Figs. 7A and B). Aliquots of the digestion reactions were removed at the indiated time points and GRP94.NTD integrity was visualized by SDS-PAGE. Fractional loss of full length GRP94.NTD was then quantified using ImageJ (Figs. 7A and B). In parallel studies, GRP94.NTD fluorescence intensity was assayed. As depicted in Fig. 7B, the time-dependent proteolytic degradation of fluor-conjugated GRP94.NTD was associated with an increase in total sample fluorescence. This suggests that the time-dependent increase in fluorescence observed following GRP94.NTD endocytic uptake is due to the proteolytic processing of the internalized protein in an intracellular compartment(s). To further test this hypothesis, MEF-1 cell cultures were supplemented with the protease inhibitor leupeptin prior to and during GRP94.NTD uptake (Figs. 7C and D). This treatment effectively suppressed the previously observed increase in fluorescence, reducing the total GRP94.NTD signal to the same level as the initial surface-bound GRP94.NTD signal. Together, these data demonstrate that fluor-conjugated GRP94.NTD undergoes proteolytic processing in endosomal compartments, a finding consistent with previous studies demonstrating the exchange of GRP94-associated peptides onto mature MHC class I molecules in post-ER compartments (36).
Figure 7. Proteolysis enhances the fluorescence yield of fluor-conjugated GRP94.NTD.
A & B. Fluor-conjugated GRP94.NTD was incubated with trypsin at room temperature. At various time points, the reaction was quenched by addition of TCA and precipitated protein was analyzed by SDS-PAGE and Coomassie Blue staining (A). Values were normalized to the untreated control (lane 1). GRP94.NTD fluorescence was determined at thirty second intervals (B). Relative GRP94 fluorescence was normalized to the point of highest fluorescence intensity. C & D. MEF-1 cells were pre-incubated in the absence or presence of 100 μg/mL leupeptin. Cells were then incubated with 25 μg/mL GRP94.NTD on ice, washed, and warmed to 37°C for 0 (bold black line) or 60 min in the absence (dashed line) or presence (bold grey line) of 100 μg/mL leupeptin. D. Data from C represented as a bar graph.
GRP94.NTD processing and GRP94.NTD-mediated peptide cross-presentation by DC2.4 cells is insensitive to exogenous CD91 ligands
The data presented above demonstrate that CD91 does not directly bind GRP94.NTD or contribute significantly to GRP94.NTD internalization by MEF-1 and PEA-13 cells. Although these findings are inconsistent with the observation that CD91 ligands inhibit GRP94-mediated peptide cross-presentation, CD91 could promote GRP94.NTD binding and endocytosis through co-receptor complexes that are not expressed by MEF-1 cells. Alternately, because cross-presentation involves surface binding, internalization, processing and MHC class I-peptide complex assembly, CD91 ligands may indirectly suppress the cross-presentation of GRP94.NTD-peptide complexes, subsequent to their endocytic clearance. To explore these possibilities, GRP4.NTD surface binding, internalization/processing, and GRP94.NTD-mediated peptide cross-presentation were examined using DC2.4 murine dendritic cells.
The interaction of RAP and GRP94.NTD with cell surface binding sites expressed by DC2.4 cells was first characterized using surface binding studies. DC2.4 cells bind RAP with half-maximal and maximal binding observed at ~0.1 and 1.0 μg/ml, respectively (Fig. 8A). In contrast, GRP94.NTD surface binding did not display distinct, high-affinity binding, indicating that GRP94.NTD interacts with low-affinity/high-capacity binding sites on DC2.4 cells (Fig. 8B). Similar to what was observed with the MEF-1 cells, the addition of excess RAP or α2M* did not reduce GRP94.NTD surface binding to DC2.4 cells (Fig. 8C). GRP94.NTD surface binding was also not competed by native α2M, which is not recognized by CD91. CD91 expression by DC2.4 cells was confirmed using RT-PCR (data not shown).
Figure 8. GRP94.NTD internalization and processing efficiencies are independent of exogenous CD91 ligands.
A & B. DC2.4 cells were incubated with increasing concentrations of RAP (A) or GRP94.NTD (B). Cells were subsequently washed and analyzed by flow cytometry. Results are expressed as the mean of two independent experiments ± SD. C. DC2.4 cells were incubated with 10 μg/mL GRP94.NTD in the absence or presence of 500 μg/mL RAP, 300 μg/mL α2M*, or 300 μg/mL native α2M for 30 min on ice. Cells were then washed, and analyzed by flow cytometry. D. DC2.4 cells were prepared as in C, and then incubated in the absence or presence of 500 μg/mL RAP, 300 μg/mL α2M*, or 300 μg/mL native α2M for 60 min at 37°C. E. DC2.4 cells were incubated with media only, 1 μM free ova20 peptide, uncomplexed GRP94.NTD, or increasing concentration of GRP94.NTD/ova20 complex. Cells were then incubated for 4 h at 37°C, washed, and incubated with OT-1 splenocytes for 24 h. IFN-γ secretion was quantified by ELISA. F. DC2.4 cells were incubated with 10 μg/mL GRP94.NTD/ova20 complex in the absence or presence of 500 μg/mL RAP, 300 μg/mL α2M*, 300 μg/mL native α2M, or 500 μg/mL uncompled GRP94.NTD, and treated as in E. G. DC2.4 cells were incubated with 10 μg/mL RAP in the absence or presence of 100 μg/mL unlabeled RAP for 5 min. H. DC2.4 cells were incubated with 10 μg/mL GRP94.NTD in the absence or presence of 250 μg/mL GRP94.NTD or 100 μg/mL RAP for 20 min. Cells were then placed on ice to arrest endocytosis, rinsed with cold buffer, and analyzed by flow cytometry. Samples were normalized to their respective no competitor controls. Results are expressed as the mean of two independent experiments ± SD.
To determine if CD91 facilitates GRP94.NTD internalization/processing by DC2.4 cells, cells were pre-incubated with RAP, α2M*, or native α2M on ice, followed by incubation with labeled GRP94.NTD on ice. The cells were then rinsed and warmed to 37°C in the absence or presence of excess ligand. After 60 min, the cells were chilled to arrest endocytosis and analyzed by flow cytometry. Neither α2M* or native α2M significantly reduced GRP94.NTD internalization/processing (Fig. 8D). RAP modestly (~15%) decreased the overall GRP94.NTD signal.
To determine if CD91 ligands indirectly influence GRP94-mediated peptide cross-presentation, cross-presentation studies were conducted using OT-1 splenocytes. OT-1 mice express a transgenic TCR that recognizes the ovalbumin-based OVA257–264 (SIINFEKL) peptide in the context of an H-2Kb MHC class I receptor. For these studies, GRP94.NTD was complexed to ova20 (SGLEQLESIINFEKLTEWTS) using a previously published protocol (18), and dialyzed to remove uncomplexed ova20 peptide. Because of its length, ova20 must be internalized and processed to yield the SIINFEKL epitope.
DC2.4 cells were pulsed with the GRP94.NTD/ova20 complex in the absence or presence of CD91 ligands for 4 h at 37°C. Cells were then washed, incubated with OT-1 splenocytes, and CTL activation assayed 24 h later by IFN-γ ELISA. GRP94.NTD/ova20-pulsed DC2.4 cells activated OT-1 splenocytes, and DC2.4 cells pulsed with uncomplexed GRP94.NTD were unable to stimulate OT-1 splenocytes (Fig. 8E). In the absence of GRP94.NTD/ova20, DC2.4 cells pulsed with RAP, α2M*, or native α2M did not elicit OT-1 IFN-γ expression (data not shown). In contrast to prior findings, neither RAP, α2M* nor native α2M significantly reduced GRP94-mediated ova20 cross-presentation (Fig. 8F).
As an additional control, excess uncomplexed GRP94.NTD was utilized to compete GRP94.NTD/ova20 cross-presentation (Fig. 8F). Because GRP94-mediated peptide cross-presentation has been reported to occur through receptor-mediated endocytosis, it was expected that GRP94-mediated ova20 cross-presentation would be significantly diminished in the presence of 50-fold molar excess GRP94.NTD. However, GRP94.NTD/ova20 cross-presentation was not significantly reduced under these conditions, suggesting that non-specific fluid phase uptake serves as the primary internalization pathway for GRP94.NTD/ova20 in DC2.4 cells.
To distinguish between the receptor-mediated and fluid-phase components of GRP94.NTD internalization in DC2.4 cells, a modified internalization assay was conducted. In these experiments, fluor-conjugated RAP or GRP94.NTD was added directly to the internalization buffer, and uptake studies were performed at 37°C in the continued absence or presence of excess unlabeled ligand. Endocytosis was then arrested by the addition of ice-cold buffer and the samples analyzed by flow cytometry. As depicted in Fig. 8G, a 10-fold molar excess of unlabeled RAP inhibited labeled RAP internalization/processing by 80% during the first 20 min of internalization. A similar inhibition was observed after 5 min of internalization (data not shown). These data confirm that DC2.4 cells internalize extracellular RAP almost exclusively through receptor-mediated endocytosis. In contrast, a 25-fold molar excess of unlabeled GRP94.NTD suppressed GRP94.NTD internalization/processing by <30% after 20 min of internalization (Fig. 8H). A similar inhibition was observed after 5 min of internalization (data not shown). These data distinguish the mechanisms of GRP94.NTD and RAP internalization by DC2.4 cells and are consistent with the RAP and GRP94.NTD binding characteristics depicted in Figs. 8A and 8B, respectively. The inclusion of ten-fold excess unlabeled RAP inhibited GRP94.NTD internalization by approximately 20% in DC2.4 cells. This parallels the RAP-dependent decrease in GRP94.NTD internalization/processing observed in Fig. 8D.
Discussion
In this study, we report two primary findings. 1) CD91 is dispensable for GRP94.NTD binding and endocytosis (29). Comparisons of surface binding and uptake functions of CD91+/+ and CD91−/− mouse embryonic fibroblasts demonstrated that CD91 expression was required for the surface binding and endocytic uptake of the CD91 ligand RAP, but not GRP94.NTD. Additionally, when present in molar excess, the CD91 ligands RAP or α2M* displayed little to no competition for either surface binding of GRP94.NTD or cross-presentation of GRP94.NTD/ova20 complexes by DC2.4 cells. 2) HSPGs function as GRP94.NTD cell surface binding sites on mouse embryonic fibroblast cell lines. This latter observation extends the diversity of cellular binding interactions that participate in the surface recognition of Hsps and molecular chaperones, and suggests that HSPG-dependent interactions may contribute to the biology of GRP94-elicited immune responses.
HSPGs are ubiquitously expressed, structurally diverse proteoglycans that contain highly heterogeneous, sulfated polysaccharide side chains. Their cell surface expression patterns vary by cell/tissue type and developmental stage, and can be altered in response to extracellular stimuli such as tissue injury (61). Additionally, the HSPG structure itself can be modified during its biosynthesis and through interactions with extracellular proteases. These characteristics make HSPGs uniquely poised to play a broad variety of physiological roles in ligand binding, processing, and signal transduction (61). The included data demonstrate that cell surface HSPGs can serve as binding sites for extracellular GRP94.NTD. This conclusion was derived from three distinct experimental approaches. First, disrupting HSPG surface structures with heparin resulted in diminished GRP94.NTD binding to both MEF-1 cells and CD91-deficient PEA-13 cells. Second, modulating HSPG sulfation by culturing cells in sodium chlorate-supplemented media resulted in the dramatic loss of GRP94.NTD surface binding activity. Third, altering cell surface HSPG structures by digestion with exogenous heparinase II resulted in decreased GRP94.NTD surface binding by both MEF-1 and PEA-13 cell lines. Thus, by multiple independent methods, HSPGs have been implicated in the direct surface recognition of GRP94.NTD.
These experiments demonstrate that cell surface HSPGs are necessary for a significant portion of GRP94.NTD surface binding to MEF-1 and PEA-13 cells. With such a prominent contribution to GRP94.NTD surface binding, it is curious that this interaction was not previously reported. As depicted in Figs. 4 and 8, HSPG-dependent GRP94.NTD surface binding is a low-affinity/high-capacity binding process. This is in contrast to the high-affinity interactions observed between GRP94 and the scavenger receptors SR-A and SREC-I (31, 32). Thus, if the cells used in previous studies expressed high-affinity GRP94 receptors such as SR-A and SREC-I, the contributions of low-affinity/high-capacity receptors such as cell surface HSPGs could have gone unnoticed. Alternatively, but not exclusively, the immunological cell lines used in previous studies may not have expressed the specific class(es) of HSPGs that contribute to GRP94.NTD surface binding on MEF-1 and PEA-13 cells.
CD91 was the first endocytic receptor proposed to function in GRP94 recognition and uptake (26). In these studies, Binder et al. demonstrated that GRP94-mediated peptide cross-presentation was almost completely suppressed by α2-macroglobulin (α2M), receptor-associated protein (RAP), and an anti-CD91 blocking antibody (26, 29, 30). In a subsequent study, it was shown that siRNA-mediated knock-down of CD91 expression ablated GRP94-mediated peptide cross-presentation by RAW264.7 cells (30). Together, these studies directly implicate CD91 in the cross-presentation of GRP94-associated peptides and suggest that CD91 functions as a GRP94 endocytic receptor. To further examine the role of CD91 as a GRP94 endocytic receptor, our studies focused on the ability of CD91 to mediate three processes that are required for GRP94-mediated peptide cross-presentation: surface binding, endocytosis, and intracellular processing. We first characterized the contribution of CD91 to GRP94.NTD surface binding by utilizing both siRNA-mediated knock-down and CD91 knock-out systems. Reduction or loss of CD91 expression did not diminish GRP94.NTD surface binding and excess GRP94.NTD did not compete for RAP surface binding. These findings are inconsistent with a direct role for CD91 in the surface binding of GRP94.NTD. It is difficult to reconcile these data with prior reports demonstrating that siRNA-mediated loss of CD91 expression blocked GRP94 surface binding and cross-presentation activity (30). However, it is not clear how these investigators obtained a complete silencing of CD91 expression without selecting for the siRNA-transfected RAW264.7 cells; in the absence of selection, siRNA-dependent silencing of CD91-expression would be limited to that fraction of the cell population undergoing productive transfection.
Our findings are in agreement with several past reports concluding that CD91 does not contribute to the surface recognition of GRP94, calreticulin, or Hsp70 (37, 68, 69). In this regard, it should be noted that both Berwin et al. and Binder and Srivastava demonstrated that neither a 100-fold molar excess RAP nor 100-fold molar excess α2M* inhibited GRP94 binding to RAW264.7 mouse macrophage cells (30, 37). However, Binder and Srivastava also demonstrated that a partial (~60%) competition between these ligands could be achieved if cells were chemically fixed prior to ligand binding. A later study demonstrated that a 50-fold molar excess of α2Mreduced GRP94 surface binding by 35% to fixed primary human PBMCs (70). Because chemical fixation was required to observe the binding competition, the physiological relevance of these results remains unclear.
Although CD91 does not directly bind GRP94.NTD at the cell surface, CD91 could influence GRP94.NTD endocytosis through alternative mechanisms. In fact, it is well established that CD91 can facilitate ligand internalization and metabolism without significantly contributing to ligand recognition at the cell surface (50, 52, 54). The ability of CD91 to mediate GRP94 internalization was thus examined by studying the uptake kinetics of RAP and GRP94.NTD by MEF-1 cells. Our results demonstrated that GRP94.NTD and RAP were internalized via kinetically and spatially distinct pathways, indicating that these ligands have divergent mechanisms of uptake and intracellular sorting. These observations are in agreement with past studies by Berwin et al., who demonstrated that GRP94 and the CD91 ligand Pseudomonas exotoxin A did not co-localize following 30 minutes of internalization by RAW264.7 cells (37). Additionally, GRP94.NTD uptake kinetics were nearly identical in MEF-1 and PEA-13 cells (data not shown), and the continuous presence of RAP during internalization had no significant effect on either the quantity or rate of GRP94.NTD endocytosis by MEF-1 cells. Together, these data demonstrate that CD91 does not directly contribute to GRP94.NTD endocytosis by MEF-1 cells. It remains to be determined if CD91 forms co-receptor complexes that are unique to RAW264.7 cells, and in this way contribute to GRP94 surface binding and endocytosis.
Finally, we examined if CD91 ligands indirectly influenced the cellular processing of GRP94.NTD. Our experiments demonstrate that α2M* did not significantly reduce GRP94.NTD surface binding, intracellular processing, or GRP94.NTD-mediated ova20 cross-presentation by DC2.4 cells. While RAP had a modest (~15%) effect on GRP94.NTD processing, it had no effect on GRP94.NTD/ova20 cross-presentation, a finding which contrasts with previous reports demonstrating that a 50-fold molar excess of RAP suppressed GRP94/ova20 cross presentation by ca. 70% (30). Our observations are, however, in agreement with Berwin et al., who demonstrated that α2M* did not inhibit the cross-presentation of GRP94/SIINFEKL complexes by RAW264.7 cells (37). Conversely, our data contradict reports that identify CD91 as having an essential role in GRP94-mediated peptide cross-presentation (26, 30). Interestingly, these past studies utilized a commercial source of native α2M that was reported to contain a mixture of activated and native α2M, though such preparations are primarily comprised of native α2M, which is not a ligand for CD91 (30). Because α2M is a broad-spectrum protease inhibitor, we also determined if native α2M could affect GRP94-mediated peptide cross-presentation independently of CD91, perhaps via suppression of intracellular proteolytic processing events. We conclude that in DC2.4 cells, α2M does not inhibit GRP94.NTD surface binding, internalization/processing, or GRP94.NTD-mediated ova20 cross-presentation.
It is important to note that GRP94.NTD-mediated ova20 cross-presentation was not inhibited in the presence of 50-fold excess GRP94.NTD. Combined with the experiments that measured the continuous internalization of GP94.NTD, these studies demonstrate that DC2.4 cells can internalize and process GRP94.NTD/ova20 through a fluid-phase pathway. Although it is well established that GRP94-mediated peptide cross-presentation can occur through receptor-mediated endocytosis, the contribution of fluid-phase uptake to GRP94-mediated peptide cross-presentation has not been widely studied. Several studies have established that pAPCs can internalize extracellular GRP94 though fluid-phase endocytosis (4, 36). Additionally, it is well established that exogenous antigens can access the cross-presentation pathway via fluid-phase uptake (71-74). In light of these findings, it is unclear how CD91 ligands could efficiently ablate GRP94-mediated peptide cross-presentation in RAW264.7 cells (30). RAW264.7 cells are known to internalize exogenous ligands through fluid-phase uptake and would be expected to be competent for cross-presentation of GRP94/peptide complexes internalized via this pathway (75-77). Indeed, in the assay conditions used by these investigators, cells are cultured in the presence of GRP94 and CD91 ligands for 20 hours and thus uptake via fluid phase pathways would be expected to contribute a significant fraction of the internalized GRP94 (26, 29, 30).
Combined, our data clarify the existing controversies regarding CD91 function in GRP94 surface binding and endocytic uptake. The challenge now confronting the field is to understand the precise molecular mechanism(s) of GRP94-mediated peptide cross-presentation, as well as its physiological implications.
Acknowledgements
We thank Dr. Zachary Hartman, Dr. Qi-Quan Huang, and Dr. Richard Pope for assistance with determination of endotoxin levels, Dr. Harvey Sage for assistance with analytical ultracentrifugation, and Dr. Jason Maynard, Joshua Lacsina, and Sujatha Jagannathan for critical comments. The GRP94.NTD (69-337) plasmid was a kind gift from the laboratory of Dr. Daniel T. Gewirth. Confocal imaging was conducted at the Duke Cell Biology confocal facilities with the aid of Dr. Tim Oliver. Flow cytometry was performed in the Duke Human Vaccine Institute Flow Cytometry Core Facility, which is supported by the National Institutes of Health (AI-51445).
Glossary
3. Abbreviations
- α2M*
activated α2-macroglobulin
- GRP94
glucose regulated protein of 94 kDa
- gp96
glycoprotein of 96 kDa
- HSPG
heparan sulfate proteoglycan
- Hsp
heat shock protein
- LDL
low density lipoprotein
- LRP1
low density lipoprotein-related protein 1
- MEF
mouse embryonic fibroblast
- NTD
amino terminal domain
- PEA-13
Pseudomonas exotoxin A-resistant MEF
- RAP
receptor-associated protein
Footnotes
This work was supported by NIH grants CA-104392 and GM-077382 (CVN) and HL-24066 (SVP).
References
- 1.Srivastava P. Roles of heat-shock proteins in innate and adaptive immunity. Nat Rev Immunol. 2002;2:185–194. doi: 10.1038/nri749. [DOI] [PubMed] [Google Scholar]
- 2.Nicchitta CV. Re-evaluating the role of heat-shock protein-peptide interactions in tumour immunity. Nat Rev Immunol. 2003;3:427–432. doi: 10.1038/nri1089. [DOI] [PubMed] [Google Scholar]
- 3.Binder RJ, Harris ML, Menoret A, Srivastava PK. Saturation, competition, and specificity in interaction of heat shock proteins (hsp) gp96, hsp90, and hsp70 with CD11b+ cells. J Immunol. 2000;165:2582–2587. doi: 10.4049/jimmunol.165.5.2582. [DOI] [PubMed] [Google Scholar]
- 4.Wassenberg JJ, Dezfulian C, Nicchitta CV. Receptor mediated and fluid phase pathways for internalization of the ER Hsp90 chaperone GRP94 in murine macrophages. J Cell Sci. 1999;112(Pt 13):2167–2175. doi: 10.1242/jcs.112.13.2167. [DOI] [PubMed] [Google Scholar]
- 5.Arnold-Schild D, Hanau D, Spehner D, Schmid C, Rammensee HG, de la Salle H, Schild H. Cutting edge: receptor-mediated endocytosis of heat shock proteins by professional antigen-presenting cells. J Immunol. 1999;162:3757–3760. [PubMed] [Google Scholar]
- 6.Singh-Jasuja H, Scherer HU, Hilf N, Arnold-Schild D, Rammensee HG, Toes RE, Schild H. The heat shock protein gp96 induces maturation of dendritic cells and down-regulation of its receptor. Eur J Immunol. 2000;30:2211–2215. doi: 10.1002/1521-4141(2000)30:8<2211::AID-IMMU2211>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 7.Srivastava PK, Udono H, Blachere NE, Li Z. Heat shock proteins transfer peptides during antigen processing and CTL priming. Immunogenetics. 1994;39:93–98. doi: 10.1007/BF00188611. [DOI] [PubMed] [Google Scholar]
- 8.Srivastava PK, Menoret A, Basu S, Binder RJ, McQuade KL. Heat shock proteins come of age: primitive functions acquire new roles in an adaptive world. Immunity. 1998;8:657–665. doi: 10.1016/s1074-7613(00)80570-1. [DOI] [PubMed] [Google Scholar]
- 9.Binder RJ, Anderson KM, Basu S, Srivastava PK. Cutting edge: heat shock protein gp96 induces maturation and migration of CD11c+ cells in vivo. J Immunol. 2000;165:6029–6035. doi: 10.4049/jimmunol.165.11.6029. [DOI] [PubMed] [Google Scholar]
- 10.Zheng H, Dai J, Stoilova D, Li Z. Cell surface targeting of heat shock protein gp96 induces dendritic cell maturation and antitumor immunity. J Immunol. 2001;167:6731–6735. doi: 10.4049/jimmunol.167.12.6731. [DOI] [PubMed] [Google Scholar]
- 11.Singh-Jasuja H, Toes RE, Spee P, Munz C, Hilf N, Schoenberger SP, Ricciardi-Castagnoli P, Neefjes J, Rammensee HG, Arnold-Schild D, Schild H. Cross-presentation of glycoprotein 96-associated antigens on major histocompatibility complex class I molecules requires receptor-mediated endocytosis. J Exp Med. 2000;191:1965–1974. doi: 10.1084/jem.191.11.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Warger T, Hilf N, Rechtsteiner G, Haselmayer P, Carrick DM, Jonuleit H, von Landenberg P, Rammensee HG, Nicchitta CV, Radsak MP, Schild H. Interaction of TLR2 and TLR4 ligands with the N-terminal domain of Gp96 amplifies innate and adaptive immune responses. J Biol Chem. 2006;281:22545–22553. doi: 10.1074/jbc.M502900200. [DOI] [PubMed] [Google Scholar]
- 13.Huang Q, S. R., Jockheck-Clark AR, Shi B, Mandelin AM, II, Tak PP, Haines GK, Nicchitta CV, Pope RM. Heat Shock Protein 96 Is Elevated in Rheumatoid Arthritis and Activates Macrophages Primarily via TLR2 Signaling. J Immunol. 2009:182. doi: 10.4049/jimmunol.0801563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vabulas RM, Braedel S, Hilf N, Singh-Jasuja H, Herter S, Ahmad-Nejad P, Kirschning CJ, Da Costa C, Rammensee HG, Wagner H, Schild H. The endoplasmic reticulum-resident heat shock protein Gp96 activates dendritic cells via the Toll-like receptor 2/4 pathway. J Biol Chem. 2002;277:20847–20853. doi: 10.1074/jbc.M200425200. [DOI] [PubMed] [Google Scholar]
- 15.Reed RC, Berwin B, Baker JP, Nicchitta CV. GRP94/gp96 elicits ERK activation in murine macrophages. A role for endotoxin contamination in NF-kappa B activation and nitric oxide production. J Biol Chem. 2003;278:31853–31860. doi: 10.1074/jbc.M305480200. [DOI] [PubMed] [Google Scholar]
- 16.Tsan MF, Gao B. Cytokine function of heat shock proteins. Am J Physiol Cell Physiol. 2004;286:C739–744. doi: 10.1152/ajpcell.00364.2003. [DOI] [PubMed] [Google Scholar]
- 17.Liu B, Dai J, Zheng H, Stoilova D, Sun S, Li Z. Cell surface expression of an endoplasmic reticulum resident heat shock protein gp96 triggers MyD88-dependent systemic autoimmune diseases. Proc Natl Acad Sci U S A. 2003;100:15824–15829. doi: 10.1073/pnas.2635458100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suto R, Srivastava PK. A mechanism for the specific immunogenicity of heat shock protein-chaperoned peptides. Science. 1995;269:1585–1588. doi: 10.1126/science.7545313. [DOI] [PubMed] [Google Scholar]
- 19.Blachere NE, Li Z, Chandawarkar RY, Suto R, Jaikaria NS, Basu S, Udono H, Srivastava PK. Heat shock protein-peptide complexes, reconstituted in vitro, elicit peptide-specific cytotoxic T lymphocyte response and tumor immunity. J Exp Med. 1997;186:1315–1322. doi: 10.1084/jem.186.8.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lammert E, Arnold D, Nijenhuis M, Momburg F, Hammerling GJ, Brunner J, Stevanovic S, Rammensee HG, Schild H. The endoplasmic reticulum-resident stress protein gp96 binds peptides translocated by TAP. Eur J Immunol. 1997;27:923–927. doi: 10.1002/eji.1830270418. [DOI] [PubMed] [Google Scholar]
- 21.Singh-Jasuja H, Hilf N, Scherer HU, Arnold-Schild D, Rammensee HG, Toes RE, Schild H. The heat shock protein gp96: a receptor-targeted cross-priming carrier and activator of dendritic cells. Cell Stress Chaperones. 2000;5:462–470. doi: 10.1379/1466-1268(2000)005<0462:thspga>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vogen S, Gidalevitz T, Biswas C, Simen BB, Stein E, Gulmen F, Argon Y. Radicicol-sensitive peptide binding to the N-terminal portion of GRP94. J Biol Chem. 2002;277:40742–40750. doi: 10.1074/jbc.M205323200. [DOI] [PubMed] [Google Scholar]
- 23.Demine R, Walden P. Testing the role of gp96 as peptide chaperone in antigen processing. J Biol Chem. 2005;280:17573–17578. doi: 10.1074/jbc.M501233200. [DOI] [PubMed] [Google Scholar]
- 24.Ying M, Flatmark T. Binding of the viral immunogenic octapeptide VSV8 to native glucose-regulated protein Grp94 (gp96) and its inhibition by the physiological ligands ATP and Ca2+ FEBS J. 2006;273:513–522. doi: 10.1111/j.1742-4658.2005.05084.x. [DOI] [PubMed] [Google Scholar]
- 25.Nicchitta CV, Carrick DM, Baker-Lepain JC. The messenger and the message: gp96 (GRP94)-peptide interactions in cellular immunity. Cell Stress Chaperones. 2004;9:325–331. doi: 10.1379/CSC-62.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Binder RJ, Han DK, Srivastava PK. CD91: a receptor for heat shock protein gp96. Nat Immunol. 2000;1:151–155. doi: 10.1038/77835. [DOI] [PubMed] [Google Scholar]
- 27.Lillis AP, Van Duyn LB, Murphy-Ullrich JE, Strickland DK. LDL receptor-related protein 1: unique tissue-specific functions revealed by selective gene knockout studies. Physiol Rev. 2008;88:887–918. doi: 10.1152/physrev.00033.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herz J, Strickland DK. LRP: a multifunctional scavenger and signaling receptor. J Clin Invest. 2001;108:779–784. doi: 10.1172/JCI13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Basu S, Binder RJ, Ramalingam T, Srivastava PK. CD91 is a common receptor for heat shock proteins gp96, hsp90, hsp70, and calreticulin. Immunity. 2001;14:303–313. doi: 10.1016/s1074-7613(01)00111-x. [DOI] [PubMed] [Google Scholar]
- 30.Binder RJ, Srivastava PK. Essential role of CD91 in re-presentation of gp96-chaperoned peptides. Proc Natl Acad Sci U S A. 2004;101:6128–6133. doi: 10.1073/pnas.0308180101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berwin B, Hart JP, Rice S, Gass C, Pizzo SV, Post SR, Nicchitta CV. Scavenger receptor-A mediates gp96/GRP94 and calreticulin internalization by antigen-presenting cells. EMBO J. 2003;22:6127–6136. doi: 10.1093/emboj/cdg572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berwin B, Delneste Y, Lovingood RV, Post SR, Pizzo SV. SREC-I, a type F scavenger receptor, is an endocytic receptor for calreticulin. J Biol Chem. 2004;279:51250–51257. doi: 10.1074/jbc.M406202200. [DOI] [PubMed] [Google Scholar]
- 33.Delneste Y, Magistrelli G, Gauchat J, Haeuw J, Aubry J, Nakamura K, Kawakami-Honda N, Goetsch L, Sawamura T, Bonnefoy J, Jeannin P. Involvement of LOX-1 in dendritic cell-mediated antigen cross-presentation. Immunity. 2002;17:353–362. doi: 10.1016/s1074-7613(02)00388-6. [DOI] [PubMed] [Google Scholar]
- 34.Theriault JR, Adachi H, Calderwood SK. Role of scavenger receptors in the binding and internalization of heat shock protein 70. J Immunol. 2006;177:8604–8611. doi: 10.4049/jimmunol.177.12.8604. [DOI] [PubMed] [Google Scholar]
- 35.Calderwood SK, Mambula SS, Gray PJ., Jr. Extracellular heat shock proteins in cell signaling and immunity. Ann N Y Acad Sci. 2007;1113:28–39. doi: 10.1196/annals.1391.019. [DOI] [PubMed] [Google Scholar]
- 36.Berwin B, Rosser MF, Brinker KG, Nicchitta CV. Transfer of GRP94(Gp96)-associated peptides onto endosomal MHC class I molecules. Traffic. 2002;3:358–366. doi: 10.1034/j.1600-0854.2002.30505.x. [DOI] [PubMed] [Google Scholar]
- 37.Berwin B, Hart JP, Pizzo SV, Nicchitta CV. Cutting edge: CD91-independent cross-presentation of GRP94(gp96)-associated peptides. J Immunol. 2002;168:4282–4286. doi: 10.4049/jimmunol.168.9.4282. [DOI] [PubMed] [Google Scholar]
- 38.Soldano KL, Jivan A, Nicchitta CV, Gewirth DT. Structure of the N-terminal domain of GRP94. Basis for ligand specificity and regulation. J Biol Chem. 2003;278:48330–48338. doi: 10.1074/jbc.M308661200. [DOI] [PubMed] [Google Scholar]
- 39.Laskowitz DT, Thekdi AD, Thekdi SD, Han SK, Myers JK, Pizzo SV, Bennett ER. Downregulation of microglial activation by apolipoprotein E and apoE-mimetic peptides. Exp Neurol. 2001;167:74–85. doi: 10.1006/exnr.2001.7541. [DOI] [PubMed] [Google Scholar]
- 40.Bowers EV, Horvath JJ, Bond JE, Cianciolo GJ, Pizzo SV. Antigen delivery by alpha(2)-macroglobulin enhances the cytotoxic T lymphocyte response. J Leukoc Biol. 2009;86:1259–1268. doi: 10.1189/jlb.1008653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baker-LePain JC, Sarzotti M, Fields TA, Li CY, Nicchitta CV. GRP94 (gp96) and GRP94 N-terminal geldanamycin binding domain elicit tissue nonrestricted tumor suppression. J Exp Med. 2002;196:1447–1459. doi: 10.1084/jem.20020436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang QQ, Sobkoviak R, Jockheck-Clark AR, Shi B, Mandelin AM, 2nd, Tak PP, Haines GK, 3rd, Nicchitta CV, Pope RM. Heat shock protein 96 is elevated in rheumatoid arthritis and activates macrophages primarily via TLR2 signaling. J Immunol. 2009;182:4965–4973. doi: 10.4049/jimmunol.0801563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Biswas C, Sriram U, Ciric B, Ostrovsky O, Gallucci S, Argon Y. The N-terminal fragment of GRP94 is sufficient for peptide presentation via professional antigen-presenting cells. Int Immunol. 2006;18:1147–1157. doi: 10.1093/intimm/dxl049. [DOI] [PubMed] [Google Scholar]
- 44.Rosser MF, Trotta BM, Marshall MR, Berwin B, Nicchitta CV. Adenosine nucleotides and the regulation of GRP94-client protein interactions. Biochemistry. 2004;43:8835–8845. doi: 10.1021/bi049539q. [DOI] [PubMed] [Google Scholar]
- 45.Bu G, Schwartz AL. RAP, a novel type of ER chaperone. Trends Cell Biol. 1998;8:272–276. doi: 10.1016/s0962-8924(98)01283-5. [DOI] [PubMed] [Google Scholar]
- 46.Willnow TE, Herz J. Genetic deficiency in low density lipoprotein receptor-related protein confers cellular resistance to Pseudomonas exotoxin A. Evidence that this protein is required for uptake and degradation of multiple ligands. J Cell Sci. 1994;107(Pt 3):719–726. [PubMed] [Google Scholar]
- 47.Bishop JR, Schuksz M, Esko JD. Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature. 2007;446:1030–1037. doi: 10.1038/nature05817. [DOI] [PubMed] [Google Scholar]
- 48.Spijkers PP, Denis CV, Blom AM, Lenting PJ. Cellular uptake of C4b-binding protein is mediated by heparan sulfate proteoglycans and CD91/LDL receptor-related protein. Eur J Immunol. 2008;38:809–817. doi: 10.1002/eji.200737722. [DOI] [PubMed] [Google Scholar]
- 49.Wilsie LC, Orlando RA. The low density lipoprotein receptor-related protein complexes with cell surface heparan sulfate proteoglycans to regulate proteoglycan-mediated lipoprotein catabolism. J Biol Chem. 2003;278:15758–15764. doi: 10.1074/jbc.M208786200. [DOI] [PubMed] [Google Scholar]
- 50.Sarafanov AG, Ananyeva NM, Shima M, Saenko EL. Cell surface heparan sulfate proteoglycans participate in factor VIII catabolism mediated by low density lipoprotein receptor-related protein. J Biol Chem. 2001;276:11970–11979. doi: 10.1074/jbc.M008046200. [DOI] [PubMed] [Google Scholar]
- 51.Mikhailenko I, Kounnas MZ, Strickland DK. Low density lipoprotein receptor-related protein/alpha 2-macroglobulin receptor mediates the cellular internalization and degradation of thrombospondin. A process facilitated by cell-surface proteoglycans. J Biol Chem. 1995;270:9543–9549. doi: 10.1074/jbc.270.16.9543. [DOI] [PubMed] [Google Scholar]
- 52.Mahley RW, Ji ZS. Remnant lipoprotein metabolism: key pathways involving cell-surface heparan sulfate proteoglycans and apolipoprotein E. J Lipid Res. 1999;40:1–16. [PubMed] [Google Scholar]
- 53.Bernfield M, Gotte M, Park PW, Reizes O, Fitzgerald ML, Lincecum J, Zako M. Functions of cell surface heparan sulfate proteoglycans. Annu Rev Biochem. 1999;68:729–777. doi: 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]
- 54.Nykjaer A, Willnow TE. The low-density lipoprotein receptor gene family: a cellular Swiss army knife? Trends Cell Biol. 2002;12:273–280. doi: 10.1016/s0962-8924(02)02282-1. [DOI] [PubMed] [Google Scholar]
- 55.Wang S, Herndon ME, Ranganathan S, Godyna S, Lawler J, Argraves WS, Liau G. Internalization but not binding of thrombospondin-1 to low density lipoprotein receptor-related protein-1 requires heparan sulfate proteoglycans. J Cell Biochem. 2004;91:766–776. doi: 10.1002/jcb.10781. [DOI] [PubMed] [Google Scholar]
- 56.Reed RC, Zheng T, Nicchitta CV. GRP94-associated enzymatic activities. Resolution by chromatographic fractionation. J Biol Chem. 2002;277:25082–25089. doi: 10.1074/jbc.M203195200. [DOI] [PubMed] [Google Scholar]
- 57.Menoret A, Bell G. Purification of multiple heat shock proteins from a single tumor sample. J Immunol Methods. 2000;237:119–130. doi: 10.1016/s0022-1759(00)00137-x. [DOI] [PubMed] [Google Scholar]
- 58.Riera M, Roher N, Miro F, Gil C, Trujillo R, Aguilera J, Plana M, Itarte E. Association of protein kinase CK2 with eukaryotic translation initiation factor eIF-2 and with grp94/endoplasmin. Mol Cell Biochem. 1999;191:97–104. [PubMed] [Google Scholar]
- 59.Baeuerle PA, Huttner WB. Chlorate--a potent inhibitor of protein sulfation in intact cells. Biochem Biophys Res Commun. 1986;141:870–877. doi: 10.1016/s0006-291x(86)80253-4. [DOI] [PubMed] [Google Scholar]
- 60.Burnell JN, Roy AB. Purification and properties of the ATP sulphurylase of rat liver. Biochim Biophys Acta. 1978;527:239–248. doi: 10.1016/0005-2744(78)90273-5. [DOI] [PubMed] [Google Scholar]
- 61.Conrad HE. Heparin-Binding Proteins. Academic Press; San Diego, CA: 1998. [Google Scholar]
- 62.Farley JR, Nakayama G, Cryns D, Segel IH. Adenosine triphosphate sulfurylase from Penicillium chrysogenum equilibrium binding, substrate hydrolysis, and isotope exchange studies. Arch Biochem Biophys. 1978;185:376–390. doi: 10.1016/0003-9861(78)90180-7. [DOI] [PubMed] [Google Scholar]
- 63.Keller KM, Brauer PR, Keller JM. Modulation of cell surface heparan sulfate structure by growth of cells in the presence of chlorate. Biochemistry. 1989;28:8100–8107. doi: 10.1021/bi00446a021. [DOI] [PubMed] [Google Scholar]
- 64.Safaiyan F, Kolset SO, Prydz K, Gottfridsson E, Lindahl U, Salmivirta M. Selective effects of sodium chlorate treatment on the sulfation of heparan sulfate. J Biol Chem. 1999;274:36267–36273. doi: 10.1074/jbc.274.51.36267. [DOI] [PubMed] [Google Scholar]
- 65.Li Y, Lu W, Marzolo MP, Bu G. Differential functions of members of the low density lipoprotein receptor family suggested by their distinct endocytosis rates. J Biol Chem. 2001;276:18000–18006. doi: 10.1074/jbc.M101589200. [DOI] [PubMed] [Google Scholar]
- 66.Pimpaneau V, Midoux P, Durand G, De Baetselier P, Monsigny M, Roche AC. Endocytosis of alpha 1-acid glycoprotein variants and of neoglycoproteins containing mannose derivatives by a mouse hybridoma cell line (2C11-12). Comparison with mouse peritoneal macrophages. Glycoconj J. 1989;6:561–574. doi: 10.1007/BF01053778. [DOI] [PubMed] [Google Scholar]
- 67.Anderson GP, Nerurkar NL. Improved fluoroimmunoassays using the dye Alexa Fluor 647 with the RAPTOR, a fiber optic biosensor. J Immunol Methods. 2002;271:17–24. doi: 10.1016/s0022-1759(02)00327-7. [DOI] [PubMed] [Google Scholar]
- 68.Walters JJ, Berwin B. Differential CD91 dependence for calreticulin and Pseudomonas exotoxin-A endocytosis. Traffic. 2005;6:1173–1182. doi: 10.1111/j.1600-0854.2005.00351.x. [DOI] [PubMed] [Google Scholar]
- 69.Theriault JR, Mambula SS, Sawamura T, Stevenson MA, Calderwood SK. Extracellular HSP70 binding to surface receptors present on antigen presenting cells and endothelial/epithelial cells. FEBS Lett. 2005;579:1951–1960. doi: 10.1016/j.febslet.2005.02.046. [DOI] [PubMed] [Google Scholar]
- 70.De Filippo A, Binder RJ, Camisaschi C, Beretta V, Arienti F, Villa A, Della Mina P, Parmiani G, Rivoltini L, Castelli C. Human plasmacytoid dendritic cells interact with gp96 via CD91 and regulate inflammatory responses. J Immunol. 2008;181:6525–6535. doi: 10.4049/jimmunol.181.9.6525. [DOI] [PubMed] [Google Scholar]
- 71.Sallusto F, Cella M, Danieli C, Lanzavecchia A. Dendritic cells use macropinocytosis and the mannose receptor to concentrate macromolecules in the major histocompatibility complex class II compartment: downregulation by cytokines and bacterial products. J Exp Med. 1995;182:389–400. doi: 10.1084/jem.182.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brossart P, Bevan MJ. Presentation of exogenous protein antigens on major histocompatibility complex class I molecules by dendritic cells: pathway of presentation and regulation by cytokines. Blood. 1997;90:1594–1599. [PMC free article] [PubMed] [Google Scholar]
- 73.Norbury CC, Hewlett LJ, Prescott AR, Shastri N, Watts C. Class I MHC presentation of exogenous soluble antigen via macropinocytosis in bone marrow macrophages. Immunity. 1995;3:783–791. doi: 10.1016/1074-7613(95)90067-5. [DOI] [PubMed] [Google Scholar]
- 74.Swanson JA, Watts C. Macropinocytosis. Trends Cell Biol. 1995;5:424–428. doi: 10.1016/s0962-8924(00)89101-1. [DOI] [PubMed] [Google Scholar]
- 75.Gerondopoulos A, Jackson T, Monaghan P, Doyle N, Roberts LO. Murine norovirus-1 cell entry is mediated through a non-clathrin-, non-caveolae-, dynamin- and cholesterol-dependent pathway. J Gen Virol. 91:1428–1438. doi: 10.1099/vir.0.016717-0. [DOI] [PubMed] [Google Scholar]
- 76.Peppelenbosch MP, DeSmedt M, Pynaert G, van Deventer SJ, Grooten J. Macrophages present pinocytosed exogenous antigen via MHC class I whereas antigen ingested by receptor-mediated endocytosis is presented via MHC class II. J Immunol. 2000;165:1984–1991. doi: 10.4049/jimmunol.165.4.1984. [DOI] [PubMed] [Google Scholar]
- 77.Yao W, Li K, Liao K. Macropinocytosis contributes to the macrophage foam cell formation in RAW264.7 cells. Acta Biochim Biophys Sin (Shanghai) 2009;41:773–780. doi: 10.1093/abbs/gmp066. [DOI] [PubMed] [Google Scholar]