Abstract
Herein, we demonstrate for the first time the use of hydrogel-in-liposome nanoparticles (lipogels) as a promising drug delivery vehicle for the active encapsulation of the anticancer drug 17-DMAPG, a geldanamycin (GA) derivative. This model drug was chosen due to its improved aqueous solubility (4.6 mg/ml) compared to the parent GA (<0.01 mg/ml), and presence of a tertiary amine which readily protonates at low pH. For the design of lipogels, a PAA hydrogel core was formed inside liposomes through UV-initiated DEAP activation and polymerization of AA and BA. We have demonstrated here that electrostatic interactions between drug and gel are critical for active encapsulation and sustained release of 17-DMAPG. We found that optimal loading conditions could be obtained (88% loading efficiency) through control of pH, temperature and incubation time. Dramatic sustained drug release from lipogels was achieved independent of the external solution pH (ca. 54 h to 50% drug release) and confirmed that the lipid bilayer was intact in the presence of the gel core. In vitro cell culture studies revealed that at the highest concentration tested, which corresponded to approximately 0.4 mg/ml of material, lipogels did not exert cytotoxicity to cells.
Keywords: Liposome, hydrogel, nanogel, geldanamycin, drug delivery, passive, active, drug loading, release kinetics, cytotoxicity
1. Introduction
Liposomes as a template for the preparation of size-controlled hydrogel nanoparticles have recently been reported in the literature [1-14], and show great potential for further development in various biomedical fields [9]. In the few works where such hydrogel nanoparticles have been investigated for drug delivery applications, proteins such as bovine serum albumin (BSA) and lysozyme [13], hemoglobin [6, 7], IL-2 [14], or the small molecule TGF- inhibitor drug [14] relied on the passive encapsulation method for loading into the nanogel systems. Herein, we demonstrate for the first time the use of hydrogel-in-liposome nanoparticles as a promising drug delivery vehicle for the active encapsulation of the anticancer drug 17-DMAPG, a geldanamycin (GA) derivative with enhanced aqueous solubility [15, 16].
Liposomes for drug delivery and their preparation techniques are well known in the literature; briefly, liposomes are nanoparticles with an aqueous core surrounded by one or more outer shell(s) consisting of lipids arranged in a bilayer configuration [17]. Liposomes can be prepared by a variety of methods and are classified into various categories based on size and lamellarity: multilamellar vesicles (MLVs, >500 nm), large unilamellar vesicles (LUVs, 100-500 nm), and small unilamellar vesicles (SUVs, <100 nm) [18]. Hydrophilic small molecule drugs are typically passively loaded into liposomes, meaning that drug encapsulation occurs during liposome formation [19]. On the other hand, amphiphilic drugs can be actively encapsulated into liposomes, meaning after liposome formation. Passive encapsulation relies on the ability of liposomes to entrap aqueous buffer containing the water soluble molecule during vesicle formation. Such hydrophilic molecules typically cannot easily cross the lipid bilayer, and this allows for some control over the release kinetics. A major disadvantage of passive encapsulation is that drug loading is typically limited by the captured volume (20-30% maximum) of liposomes and solubility of the drug of interest [18, 20]. Conversely, active encapsulation relies on the molecule crossing through the lipid bilayer into the aqueous core, where it then becomes entrapped and cannot diffuse back out. For example, some drugs, lipids, and peptides can be actively entrapped into liposomes through the use of pH gradients with very high loading efficiencies [21, 22].
Relatively little is known about the properties of active drug loading in to liposomes containing a hydrogel core. Hydrogels, in general, are cross-linked networks of hydrophilic polymers that can adsorb large amounts of water. The cross-linking density of these polymers creates pores and typically allows for loading and controlled diffusion of water soluble drugs [23]. There have been a variety of hydrogel-based delivery systems reported over the years for loading and release of small molecule and macromolecular drugs in vitro [24-30]. However, hydrogel nanoparticles with unsuitable mechanical properties or poorly-defined dimensions [31] severely limit their usage in vivo due to the requirement for sustained release of drugs and a long circulation time in the body; most hydrogel-based drug delivery systems work better for topical applications [32, 33]. One advantage of hydrogels for controlled drug release includes their responsiveness to external stimuli. For example, pH-sensitive hydrogels are typically characterized by a variable swelling ratio (weight of adsorbed water to the weight of the sample at a dry state) dependent on pH [34]. This on-off swelling state results from electrostatic repulsions between polymer chains in the hydrogel network [35]. For example, poly(acrylic acid) (PAA) was characterized by a larger swelling ratio at higher pH due to the de-protonation of carboxylic groups, which led to electrostatic repulsions between polymer chains and caused the gel to become more porous to water molecules [36]. When BSA was loaded into PAA hydrogel nanoparticles, it was characterized by a pH-dependent release profile [37]. Similarly, when the cationic anticancer drug doxorubicin (DOX) was loaded into hydrogel microspheres composed of anionic poly(methacrylic acid), drug release was triggered by either a pH change or through addition of excess cations to the solution [38]. pH plays a very important role and can therefore favorably influence hydrogel properties, promote drug interactions with the polymer chains, and influence release kinetics.
In this work, PAA hydrogel in liposomes is of particular interest since a pH gradient and electrostatic/hydrophobic interactions between cationic drug and anionic gel in the liposomal core is used for the active encapsulation of 17-DMAPG. As a model drug, 17-DMAPG was chosen due to (1) its improved aqueous solubility (4.6 mg/ml) compared to the parent GA (~0.01 mg/ml) [16], and (2) presence of a tertiary amine which readily protonates at low pH. It was found that under optimized conditions, consistent drug loading and sustained release from these hydrogel-in-liposomes (lipogels) could be achieved. Whereas previous reports removed the lipid bilayer after forming the hydrogel nanoparticles in the liposomes [39, 40], the inclusion of the lipid bilayer here is critical for sustained release and potential surface modifications (i.e. addition of PEG or targeting ligands). This work describes for the first time the preparation of well-defined lipogels for active loading of the model drug 17-DMAPG, its characterizations and release kinetics, and resulting cytotoxicity of the nanoparticles on cancer cells cultured in vitro.
2. Experimental
2.1. Materials
Sodium ascorbate, acrylic acid (AA), N,N’-methylenebis(acrylamide) (BA), 2,2-diethoxyacetophenone (DEAP), 3-(dimethylamino)-1-propylamine and Triton X-100 were purchased from Sigma-Aldrich. Geldanamycin (GA) was obtained from LC Laboratories. The pH-sensitive fluorescent dye Oregon Green 514 was purchased from Life Technologies. 1,2-Dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) and cholesterol were purchased from Avanti Polar Lipids. Thin-layer chromatography was performed using DC-Alufolien Kieselgel 60 F254 plates (EMD Chemicals, Darmstadt, Germany) and visualization was achieved by UV light (254 nm) and a ceric molybdate stain activated by heat. For flash chromatography, silica gel 40-63 μm from EMD Chemicals (Darmstadt, Germany) was used. 1H NMR spectra were acquired on Varian Inova 500 spectrometer. Chemical shifts are reported in parts per million (ppm) relative to tetramethylsilane, and spin multiplicities are given as s (singlet), d (doublet), t (triplet), q (quartet), m (multiplet), or br s (broad singlet). High-resolution mass spectra were obtained on an IonSpec 7 Tesla HiResMALDI FT-Mass Spectrometer at the Analytical Instrumentation Center of University of Wisconsin School of Pharmacy. Dulbecco's Modified Eagle Medium (DMEM), fetal bovine serum (FBA) and 1% penicillin/streptomycin were purchased from Cellgro. The human PC-3 prostate and human MDA-MB-231 cancer cells were obtained from ATCC and cultured according to recommended protocols.
2.2. Synthesis of 17-[3’-(dimethylamino)-1’-propyl]-amino-17-demethoxygeldanamycin (17-DMAPG)
The water soluble 17-DMAPG was synthesized from GA with 95% yield [15]. Briefly, GA (56 mg, 0.1 mmol) was dissolved in dichloromethane (4 ml) and 3-(dimethylamino)-1-propylamine (25 μl, 0.2 mmol) was added. The reaction mixture was stirred at room temperature for 24 h and TLC analysis revealed complete conversion of the starting material to product. The reaction mixture was loaded onto a silica gel column and eluted with dichloromethane–methanol (stepwise gradient from 95:5 to 90:10). Fractions containing 17-DMAPG were concentrated, and the resulting purple solid was precipitated with hexane (60 mg, 95%). 1H NMR: 9.21 (s, 1H), 7.77 (s, 1H), 7.24 (s, 1H), 6.96 (d, J = 11.4 Hz, 1H), 6.59 (d, J = 11.4 Hz, 1H), 5.92 (d, J = 9.3, 1H), 5.86 (t, J = 10.8, 1H), 5.19 (s, 1H), 4.86 (br s, 2H), 4.51 (br s, 1H), 4.31 (d, J = 10.1 Hz, 1H), 3.77-3.70 (m, 1H), 3.64-3.56 (m, 2H), 3.48-3.44 (m, 1H), 3.37 (s, 3H), 3.27 (s, 3H), 2.78-2.70 (m, 1H), 2.66 (d, J = 13.8 Hz, 1H), 2.45 (t, J = 6.1 Hz, 2H), 2.43-2.36 (m, 1H), 2.27 (s, 6H), 2.03 (s, 3H), 1.80 (d, J = 1.3 Hz, 3H), 1.84-1.68 (m, 5H), 1.00 (d, J = 6.9 Hz, 3H), 0.97 (d, J = 6.4 Hz, 3H). HRMS (ESI): calcd for C33H51 N4O8[M+H]+ 631.37014, found 631.37199; calcd for C33H50N4O8Na [M+Na]+ 653.35209, found 653.35312.
2.3. Preparation of lipogels and nanogels
To prepare lipogels, 80 mol DPPC and 50 mol cholesterol were dissolved in chloroform. The solvent was evaporated on a rotary evaporator under vacuum. The resulting dried lipid film was rehydrated with 1 ml of hydrogel precursor solution (100 μl AA, 10 mg BA and 1 μl DEAP dissolved with 900 μl ddH2O) in a 55 °C water bath with vigorous shaking for 30 min. The resulting multilamellar vesicles were extruded 11 times through a 100 nm polycarbonate filter via a mini-extruder (Avanti Polar Lipids) to generate LUVs. Sodium ascorbate was added to the solution at a molar ratio of 200:1 relative to DEAP. After 15 min of UV irradiation (Black Ray UV lamp, 365 nm, 100 Watt), polymerization of monomers in liposomes was nearly complete and lipogels of ca. 100-120 nm diameter were obtained. Polymerization outside liposomes was inhibited by the presence of ascorbic acid, a scavenger of free radicals [41]. Next, the lipogel suspension was dialyzed against deionized water to remove un-encapsulated hydrogel precursors and ascorbic acid. Lipogels were concentrated via ultracentrifugation and re-suspended in desired buffer.
To prepare nanogels without the liposomal bilayer, the surfactant Triton X-100 was added to lipogels at a 50:1 surfactant/lipid molar ratio. The solution was heated to 90°C until it became cloudy (indication that the solution reached the cloud point of the surfactant), then allowed to cool to room temperature. DPPC and cholesterol preferentially form micelles with Triton X-100 at this concentration [4, 8], leading to removal of the lipid bilayer from the nanogel core. This mixture was then eluted through a PD-10 column (used as a size exclusion chromatography column) to separate micelles from the resulting nanogels. Fractions were collected for analysis.
2.4. Characterization of lipogels and nanogels
Both Dynamic Light Scattering (DLS) and -potential measurements were conducted with a Zetasizer Nano ZS (Malvern Instruments). DLS was used to determine the size distribution of nanoparticles. The sample sizes were reported as Z-average diameters. For -potential measurements, samples (800 l) in 10 mM Tris-HCl buffer (pH 7.0) were added to a capillary cell and the electrophoretic mobility was converted to -potential by the Henry equation. For scanning electron microscopy (SEM) of liposome and lipogels, nanoparticle samples were dispersed in deionized water, drop-casted onto Silicon substrates, and briefly air dried. The substrates were then mounted onto metallic pucks using double-sided carbon tape and imaged with a LEO Supra 55 VP field emission SEM. For SEM images of nanogels, the whole sample surface was sprayed with a layer of gold before SEM observation. To investigate lipogel and nanogel size as a function of pH, we equilibrated lipogel or nanogel particles in different pH buffered solutions and measured their size distributions with DLS. Different pH solutions (pH 2-10) were prepared from 0.1 M pH 6.0 acetate buffers via addition of 0.5 M HCl for lower pH or 0.5 M NaOH for higher pH. To investigate lipogel and nanogel size in the presence of varying salt concentrations, we equilibrated lipogel or nanogel particles in deionized water containing 0-300 mM NaCl and measured their size distributions with DLS. The pH inside lipogel and liposome cores was measured with a fluorescent probe [42], by passive encapsulation of the pH-sensitive fluorescent dye Oregon Green 514.
2.5. Passive and active encapsulation of 17-DMAPG into liposomes and lipogels
Liposomes with similar lipid compositions to lipogels were prepared as a control to investigate passive encapsulation of 17-DMAPG. Lipids were dissolved in chloroform as before, and the resulting lipid film was rehydrated with 1 ml of 17-DMAPG dissolved in ddH2O (1 mg/ml) and incubated in a 55 °C water bath with vigorous shaking for 30 min. The resulting MLVs were then extruded to generate drug-loaded LUVs. Passive loading of 17-DMAPG into lipogels could not be achieved due to the instability of the drug upon UV irradiation of hydrogel precursors in liposomes (we observed 17-DMAPG degradation products via HPLC).
Active loading of 17-DMAPG into liposomes and lipogels was achieved in a similar manner via the use of a pH-gradient [21, 22]. Control liposomes were prepared by rehydrating the dried lipid film with 10% citric acid solution to maintain an inner acidic pH; citric acid is structurally similar to AA and was utilized because AA released too rapidly out of liposomes in the absence of polymerization. After extrusion to LUVs, the resulting liposomes were dialyzed to exchange the external citric acid solution to pH 7.4 PBS buffer. Next, active loading of 17-DMAPG into liposomes and lipogels was accomplished by incubation in this buffer (e.g. pH 7.4 PBS) at 55 °C for 30 minutes, followed by elution through a PD-10 column to remove un-encapsulated drug molecules. The optimal conditions for active drug loading into lipogels were further investigated below.
2.6. Optimizing active drug loading conditions and in vitro release studies
To investigate optimal drug loading conditions, lipogels were incubated with 17-DMAPG at various drug/lipogel ratios, pH (4, 5.5 or 7.4), temperature (35-65°C), and drug incubation time (10-60 min). After letting the samples cool down to room temperature, un-encapsulated drug was removed as before by eluting the suspension through a PD-10 column. Then, the concentration of encapsulated 17-DMAPG was determined by adding isopropyl alcohol (IPA) to the suspension to rupture the lipid bilayer and release 17-DMAPG from the hydrogel core. As usual, 17-DMAPG was quantified by reverse phase HPLC (with a C-18 column) and drug loading efficiency was calculated. As comparison, the optimized conditions determined for lipogels were also used for active loading of 17-DMAPG into liposomes containing citric acid.
For in vitro release studies, 3 ml of the drug loaded lipogel (active-loaded) or liposome suspensions (passive and active-loaded) were injected into a Slide-A-Lyzer dialysis cassette (10,000 MWCO, Thermo Scientific) and dialyzed against 1-L pH 7.4 PBS or 1-L pH 5 acetate buffers at 37°C for up to 72 hours. At indicated time points, 50 μl aliquots were withdrawn from the cassettes for HPLC analysis. The release experiments were run in triplicates.
2.7. In vitro cytotoxicity studies
Cytotoxicity of 17-DMAPG in lipogels was assessed in human prostate PC-3 and human breast MDA-MB-231 cancer cells by resazurin-based metabolic assay [43]. Cells were cultured in DMEM medium supplemented with 10% (v/v) heat inactivated FBS, 2 mM L-glutamine, 100 unit/ml penicillin and 1000 g/ml streptomycin, and were maintained at 37°C, 5% CO2 atmosphere for the duration of experiment. For cytotoxicity assay, cells were seeded in 96-well plates at 5,000 cells/well and allowed to adhere for 24 h prior to the assay. The next day, cells were exposed to varying concentrations of free 17-DMAPG (0-35 M), 17-DMAPG in lipogels (at equivalent 17-DMAPG concentrations), or empty lipogels (at equivalent concentrations to that added for 17-DMAPG in lipogels) for 24 h. Cells were subsequently washed with PBS and maintained in fresh DMEM with 10% FBS for an additional 24 h (48 h total). Afterwards, 10 l of resazurin PBS solution (440 M) was added to each well for 2 h and the fluorescence emitted, which corresponds directly to cell viability, was measured with a microplate reader (SpectraMax Gemini XS, Molecular Devices) by exciting at a wavelength of 560 nm and emission of 590 nm. The control reading taken from wells with untreated cells was taken as 100% cell viability (Econtrol), and sample viability was calculated as Esample/Econtrol×100%. The IC50 values were obtained through curve fitting via GraphPad Prism 5 Software (San Diego, California, USA).
3. Results and discussion
As previously reported, 17-DMAPG was synthesized with a yield of about 95% (Fig. 1). The reaction could also be visually confirmed since the original yellow-orange color of GA turned purple upon derivatization to the C-17 position. This particular anticancer drug was chosen due to its favorable aqueous solubility (4.6 mg/ml) and amphiphilic properties.
Fig. 1.
The geldanamycin (GA) derivative 17-DMAPG was synthesized with a yield of about 95%. The original yellow-orange color of GA turned purple upon derivatization to the C-17 position. This particular drug was chosen due to its favorable aqueous solubility (4.6 mg/ml) and amphiphilic properties. At physiologic pH, the tertiary amino group is un-protonated and 17-DMAPG easily crosses the lipid bilayer into the acidic compartment of liposomes and lipogels.
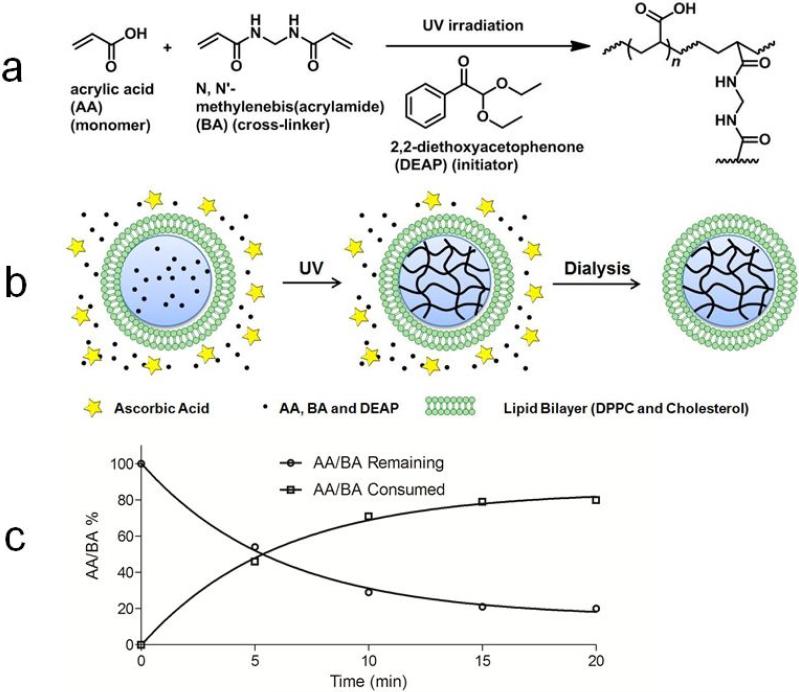
Lipogels with a PAA gel core were formed by selective cross-linking of hydrogel-forming materials inside liposomes upon UV irradiation of DEAP (Fig. 2a) [41]. From analysis with reverse phase HPLC, we estimated that the encapsulation efficiency of precursors AA/BA/DEAP inside liposomes was ca. 15%, with 85% remaining outside of liposomes in solution. To inhibit polymerization of un-encapsulated components outside liposomes, ascorbic acid at a molar ratio of 200:1 relative to DEAP was added to the suspension because it is a free radical scavenger that is impermeable through the lipid bilayer [44] (Fig. 2b). After UV irradiation, excess ascorbic acid and un-polymerized precursors were then removed from the resulting lipogel suspension via dialysis. To investigate optimal UV irradiation times, a thin slab of bulk gel was prepared with scaled-up concentration of precursors, irradiated with UV up to 20 min, and the rate of the reaction was monitored by HPLC. The reaction in the slab proceeded fast, with ca. 80% monomers and cross-linkers used up within 15 min (Fig. 2c). Due to the nanometer size of lipogels, we concluded that 15 min was sufficient time for polymerization inside liposomes to have completed.
Fig. 2.
(a) UV irradiation activates DEAP, generating free radicals which initiate polymerization of AA monomers and BA cross-linkers (b) Addition of ascorbic acid (star) inhibited polymerization of un-encapsulated components (85%) remaining outside liposomes (dots) when added at a molar ratio of 200:1 relative to DEAP; ascorbic acid is a free radical scavenger that is impermeable through the lipid bilayer (c) Upon UV irradiation, 80% of AA monomers and BA cross-linkers were consumed for polymerization within ca.15 min.
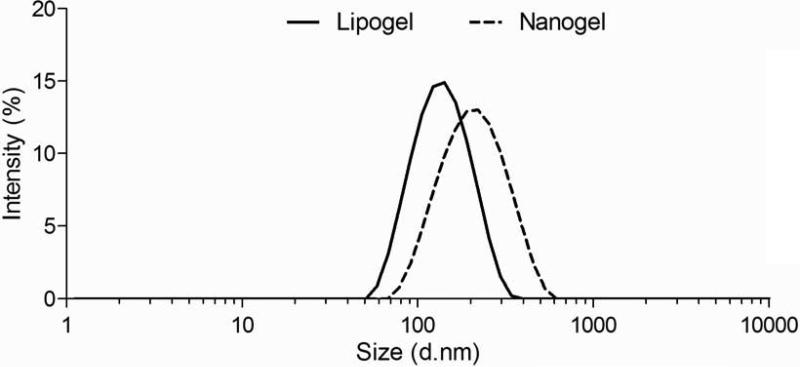
To further confirm a gel formed inside liposomes, we removed the lipid bilayer with the surfactant Triton X-100. In the presence of excess surfactant, DPPC and cholesterol in the lipid bilayer preferentially incorporate into micelles with Triton X-100 [4, 8], and we investigated whether this approach would be sufficient to remove the lipid bilayer from the resulting nanogels. To investigate this, liposomes encapsulating AA/BA/DEAP (but not polymerized by UV irradiation) were mixed with Triton X-100. When the surfactant was added, the ca. 120 nm peak characteristic of liposomes immediately disappeared with the emergence of a ca. 10 nm micelle peak (Supplemental Information, Fig. s1) and validated that this approach would work. Next, a solution of polymerized lipogels was mixed with Triton X-100 and passed through a PD-10 size-exclusion column to separate micelles (ca. 10 nm) from resulting bare nanogels (Supplemental Information, Fig. s2). As seen in Fig. 3, the nanogels collected were larger in size (ca. 320 nm) than lipogels (ca. 120 nm) at pH 7.4. This is because when the lipid bilayer was removed by the surfactant, the higher pH led to de-protonation of carboxylic acid groups in the nanogel, resulting in increased electrostatic repulsions between polymer chains, water adsorption, and a larger overall diameter [37].
Fig. 3.
To obtain pure nanogels, the lipid bilayer on lipogels was removed with the surfactant Triton X-100. The suspension was then passed through a PD-10 size-exclusion column to separate micelles (ca. 10 nm, not shown here) from resulting nanogels. As shown, nanogels average ca. 320 nm compared to lipogels (ca. 120 nm) at pH 7.4.
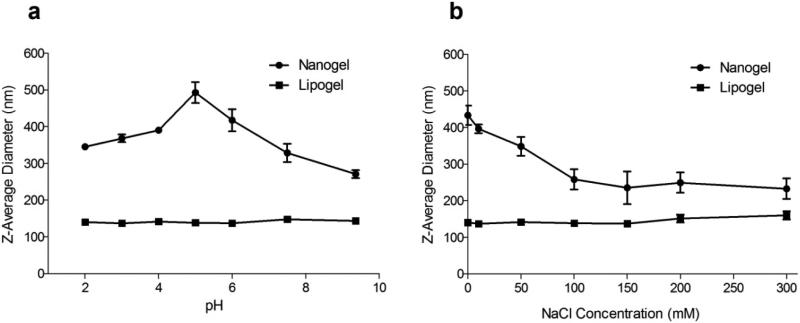
To investigate the effect of pH on particle size, nanogels and lipogels were added to solutions of varying pH. As expected, lipogels incubated in various pH solutions retained an approximate diameter of ca. 120 nm (Fig. 4a). This is expected because protons cannot pass through the lipid bilayer, which shields the gel core from the outside environment. In contrast, nanogel diameter peaked at about 490 nm when incubated in pH 5.0 solution (Fig. 4a). Below pH 5.0, the diameter of nanogels decreased to ca. 345 nm due to increasing protonation of carboxylic groups on PAA. At pH above 5.0, it was expected that the diameter of nanogels would continue to increase and eventually reach a plateau, but contrarily the diameter decreased to ca. 270 nm. This can be explained by the increasing amount of NaOH that was used to titrate the solutions to desired pH, causing excess sodium ions in solution to preferentially form ion pairs with de-protonated carboxylic acid groups and reducing the negative repulsion force. This allowed for closer associations between polymer chains, made nanogels less porous to water molecules (swelling ratio is much lower), and resulted in the smaller overall diameter observed.
Fig. 4.
Nanogels (●) reached a diameter of ca. 490 nm at pH 5.0, ca. 345 nm at pH 2.0, and 270 nm at pH 9.4; in contrast, lipogel (■) diameters did not change significantly, remaining at ca. 120 nm, when the pH was changed (a). When the NaCl concentration was increased from 0-300 mM, nanogel (●) diameters decreased from ca. 430 nm in ddH2O to ca. 230 nm; in contrast, lipogel (■) diameters did not change drastically and increased from ca. 120 nm to ca. 160 nm (b). This is most likely due to some aggregation of lipogels in the presence of increasing salt concentration. The data reported are mean ± SD of triplicates.
To investigate further the effect of NaCl, nanogels and lipogels were incubated in ddH2O containing various concentrations of salt (0-300 mM), and their resulting size was measured via DLS. As expected, nanogels incubated at 0 mM NaCl retained an approximate diameter of ca. 430 nm (Fig. 4b), which agrees with data from Fig. 4a since the pH of ddH2O is ca. 5.5. As the salt concentration was increased to 100 mM, nanogel diameter decreased to ca. 260 nm and then to ca. 230 nm at 300 mM NaCl. This is expected since increasing salt led to increased ion pair formations between Na+ and carboxylic groups, resulting in a smaller overall particle diameter. On the other hand, the diameter of lipogels at 0 mM was ca. 120 nm and did not change drastically, up to ca. 160 nm at 300 mM NaCl. The reason for this slight diameter increase is most likely from aggregation of some lipogels due to their negative surface charge (see below). In agreement, it has previously been reported that hydrogels made from polyelectrolytes such as PAA [45] and polyvinylimidazole [5] can swell and shrink in response to both pH as well as changes in ionic strength.
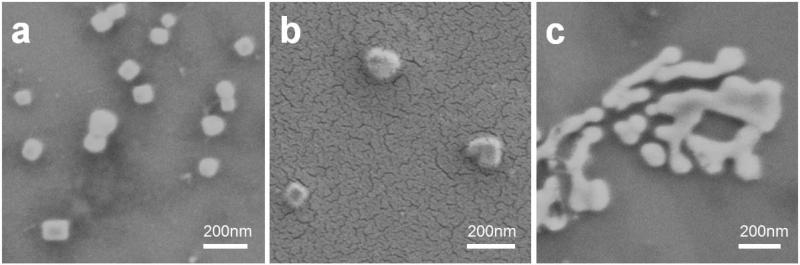
Lipogels, nanogels, and liposomes were dried on silicon substrates and imaged with scanning electron microscopy (SEM). Both lipogels (Fig. 5a) and nanogels (Fig. 5b) were spherical and toroidal shaped as expected, whereas liposomes could not be clearly imaged (Fig. 5c). Lipogels were imaged as-is for SEM. In contrast, the nanogel sample had to be coated with a layer of gold before imaging. Although liposomes prepared by repeated extrusion are usually spherically shaped in aqueous solutions [46], drying of the solution for SEM led to internal collapse of the lipid bilayer and resulted in fuzzy images. The zeta potential of these nanoparticles was measured; as expected, empty liposomes had the most neutral charge at 0.19 mV (the lipid bilayer is composed of DPPC and cholesterol which are neutral lipids) and nanogels had a -58 mV surface charge due to the exposed carboxylic acid groups on the surface of particles. Surprisingly, lipogels were characterized by a −39 mV charge contrary to the expectation that they would possess a neutral charge similar to empty liposomes. It turns out that the negative zeta potential of lipogels is due to short PAA chains that embedded/adhered to the surface of the lipid bilayer (Supplemental Information, Fig. s3). During preparation of lipogels, the presence of ascorbic acid in solution did not completely inhibit polymerization of un-encapsulated AA (i.e. formed short PAA chains that embedded/adhered to the surface of the lipid bilayer), and dialysis could not remove these short PAA chains. Indeed, the ability of PAA to interact with lipid membranes has previously been investigated in the literature.[47] Thus, the - 39 mV charge of lipogels is not due to an incomplete coverage of the gel core, but rather due to the adherence of short negatively-charged PAA chains on the surface of the lipid bilayer.
Fig. 5.
Lipogels, nanogels, and liposomes were dried on silicon substrates and imaged with scanning electron microscopy (SEM). Both lipogels (a) and nanogels (b) were spherical and toroidal shaped as expected, whereas liposomes could not be imaged clearly due to collapse of the lipid bilayer upon drying (c). Nanogels were coated with a layer of gold prior to imaging.
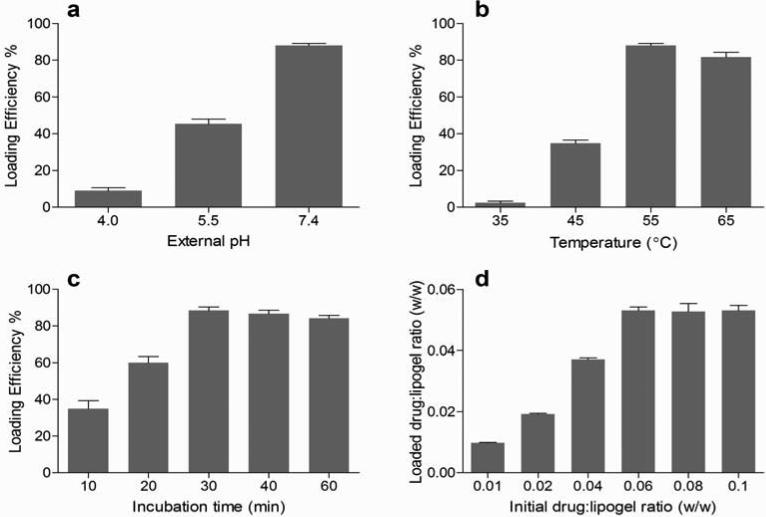
Active loading of 17-DMAPG into lipogels was optimized by varying external pH, temperature, incubation time, and drug to lipogel ratio (w/w). A random set of conditions were initially picked and optimized: external pH 7.4, temperature of 55°C, incubation time of 30 min, and initial drug:lipogel (w/w) ratio of 0.06. Similar to the remote loading of liposomes which commonly utilizes a pH gradient for optimal drug loading [21, 22], it was found that an external pH of 7.4 providing for a larger gradient was essential to effectively load the highest level of 17-DMAPG into lipogels. As shown in Fig. 6a, when the external pH of the solution used to incubate lipogels was increased from 4.0 to 7.4, drug loading drastically increased from 9% to 88%. This is because at low pH of 4 and 5.5, some 17-DMAPG molecules became protonated and could not cross the lipid bilayer; however as the pH was adjusted further to 7.4, a gradient was created whereby neutral 17-DMAPG molecules became more readily available, crossed the lipid bilayer into the acidic environment of lipogels where they picked up a proton, and became entrapped in the gel. At various external pHs and using a calculated pKa of 9.86 for 17-DMAPG (Supplemental Information, Fig. s4), the theoretical encapsulation efficiency (EE%) of drug for liposome and lipogel were calculated using the Henderson-Hasselbalch equation. As expected, the actual EE% were lower than that predicted for liposome and lipogel formulations (Supplemental Information, Table s1). Next, we investigated the effect of temperature on active drug loading into lipogels. As expected, above the phase transition temperature the lipid bilayer became more fluid and permeable to drug loading [48], and so it made sense that as the temperature increased drug loading also increased. As shown in Fig. 6b, at pH 7.4 the drug encapsulation levels were very low when the temperature of the suspension was set to 35oC (<5%), but drug loading drastically increased when the temperature was increased from 45°C to 55°C (88%); this is most likely because the phase transition temperature of the DPPC/cholesterol lipid bilayer was somewhere between 45-55°C. Even though DPPC has a known transition temperature of 41°C, the incorporation of about 38 mol% cholesterol into the lipid bilayer is known to increase this transition temperature. The slight decrease in drug loading observed at 65°C was likely due to degradation of 17-DMAPG. At this temperature we observed presence of a new peak for the drug when the external solution was analyzed by HPLC (data not shown).
Fig. 6.
A random set of condition (pH 7.4, temperature of 55°C, incubation time of 10 min, and drug:lipogel (w/w) ratio of 0.06) was initially chosen and drug encapsulation efficiency into lipogels was optimized by varying (a) pH (b) temperature (c) incubation time and then (d) drug:lipogel ratio (w/w). Results are the average of triplicates, and include standard deviations.
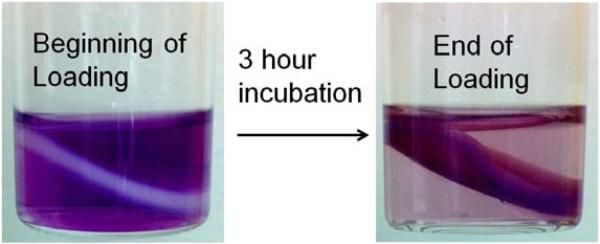
At pH 7.4 and a temperature of 55°C, the optimal incubation time for drug loading was investigated (Fig. 6c). We found that 30 min incubation was enough to reach the maximum loading capacity of lipogels, with minor changes in loading as incubation time was increased. Similarly, a bulk gel representative of the nanogel core was incubated with a scaled up concentration of 17-DMAPG and visually demonstrated the extent of drug loading into the gel slab after 3 h of incubation at room temperature (Fig. 7), with drug molecules moving first from the outer edges of the gel and then into the center. Finally, the optimal drug loading capacity per mg of lipogel was investigated (Fig. 6d). When the drug:lipogel ratio (w/w) was varied from 0.01 to 0.1, the maximal amount of drug loaded (88%) reached a plateau above 0.053; in other words, a maximum 53 g of 17-DMAPG could be successfully loaded into every mg of lipogel. Keeping in mind that the lipid contribution to total weight of lipogel is much greater, with the PAA gel portion only weighing about 16% of the total weight (we lyophilized 5 mg of lipogel to estimate total weight and subtracted from total weight of lyophilized empty liposomes), this means that 53 g of 17-DMAPG was successfully loaded into an equivalent 160 g of PAA gel.
Fig. 7.
Loading of 17-DMAPG (purple) into a PAA hydrogel slab after 3 h incubation at room temperature. Drug concentration used was 1 mg/ml in ddH2O (pH of the solution containing bulk gel and drug was ca. 5). Monomer/cross-linker/initiator ratios were scaled up from lipogel ratios. Image reveals that drug molecules in the buffer gradually diffuse into the PAA hydrogel slab, moving from the outer edges of the gel toward the center.
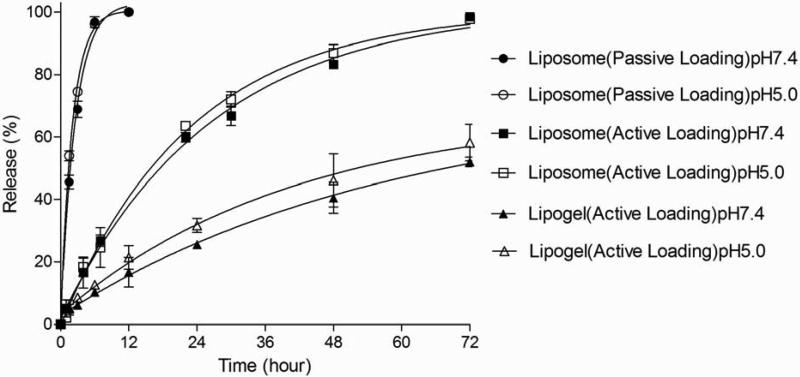
At optimal conditions of pH 7.4, temperature of 55 °C, incubation time of 30 min, and drug:lipogel ratio of 0.053 (w/w), the pH inside lipogels was measured via the use of a fluorescent probe (Oregon green) passively loaded into lipogels and was confirmed to be ca. pH 3. Similarly, active loading of 17-DMAPG into liposomes encapsulating citric buffer resulted in an acidic core with ca. pH 3, as determined by the fluorescent probe, and a max ca. 91% drug encapsulation. In contrast, passive loading of 17-DMAPG into liposomes only yielded ca. 17% drug loading. We were not able to passively load 17-DMAPG into lipogels due to degradation of the drug upon UV irradiation and passive drug loading into lipogels was not investigated further. For release kinetic studies, it is important to note that although active and passive loaded liposomes could load variable amounts of 17-DMAPG (91% vs 17% respectively) compared to lipogels (88%), an equivalent amount of drug for each formulation was used. The release study was done in pH 7.4 and pH 5.0 buffers at 37°C and results are shown in Fig. 8.
Fig. 8.
In vitro release kinetics at 37°C for 17-DMAPG loaded into liposomes (passive/active methods) and lipogels (active method) at pH 7.4 and 5.0. There was no significant release difference at pH 7.4 and 5.0 within the liposome (passive/active-loaded) or lipogel (active-loaded) formulations (p>0.05), confirming that release kinetics were pH-independent. However, there were significant release differences between the formulations at pH 7.4 and 5.0 for all time points >6 h (p<0.01); passive-loaded liposomes exhibited the fastest release (ca. 1.5 h to 50% release), active-loaded liposomes exhibited moderate sustained release (ca. 16 h to 50% release), and lipogels exhibited the slowest sustained release (ca. 54 h to 50% release). The data reported are mean ± SD of triplicates.
Although it isn't clear whether the drug is fully solubilized inside the core of lipogels and liposomes, or whether it remains partially solubilized in the lipid bilayer and partially exposed to the aqueous center, what is evident is that the tertiary amino group becomes protonated under the acidic conditions present in the core and slows down drug release. For example, the slow release of active-loaded 17-DMAPG from liposomes (characterized by an acidic core) compared to passive-loaded 17-DMAPG (characterized by a ddH2O core) confirms that protonation alone significantly slows down release of drug molecules across the lipid bilayer. Un-protonated drug molecules encapsulated by the passive method easily passed through the lipid bilayer and resulted in rapid release of 17-DMAPG from liposomes, with a time to 50% release of ca. 1.5 h for both external pH. In contrast, an acidic liposomal core allowed for protonation of drug molecules and the time to 50% release increased drastically to ca. 16 h. Performing the release study at two different external pH conditions was important to confirm that there was no exchange of protons through the lipid bilayer between the external environment and the inner compartment of liposomes. Although active drug loading encapsulation levels into liposomes and lipogels were similar (91% vs 88%), the drug release kinetics from lipogels were drastically slower, with an average time to 50% release of ca. 54 h. Statistically, the release data for lipogels at external pH 7.4 and 5.0 were not significantly different. This confirms that drug release from liposome or lipogel is not pH-dependent, meaning that there is no interaction between the outer environment and inner lipogel compartment. Therefore, although PAA nanogels can swell or shrink in response to pH and salt (Fig. 4), lipogels do not drastically respond to pH or salt due to an intact lipid bilayer. The interaction between cationic drug and cross-linked anionic polymer chain further slows down drug release, resulting in a drastic 3.4 fold increase in the time to 50% drug release compared to actively loaded liposomes. This study confirms that high drug encapsulation levels observed for liposomes and lipogels was mainly dependent on the ability of the un-protonated drug to pass through the lipid bilayer where it then became entrapped by picking up a proton (via interactions with citric acid or the PAA gel core); similarly, release kinetics depend on this mechanism.
In addition, in contrast to active-loaded liposomes, active-loaded lipogels possess an extra diffusion barrier (i.e. the cross-linked PAA gel core) that further slows down drug release. It is therefore important to note that release kinetics observed is not rate-limited by transport across the dialysis membrane but rather by the physical properties of lipogels; additional experiments supporting this conclusion can be found in Supplemental Information, Fig. s5. Briefly, the release of free 17-DMAPG from the dialysis cassette was very fast, with 100% release in <5 h, whereas drug release kinetics from lipogels upon dilution and at variable drug loading levels were similar. It is also significant to point out that pH gradient systems in liposomes (that entrap either an acid or a base) often suffer from a limited shelf life due to change of the pH-gradient over time. In contrast, the gel portion of lipogels is much more stable and does not change over time through diffusion; short-term investigations into the stability of lipogels upon their storage up to 7 days at 4°C revealed that lipogels exhibit similar drug loading levels and release kinetics to freshly prepared lipogels (Supplemental Information, Fig. s6)
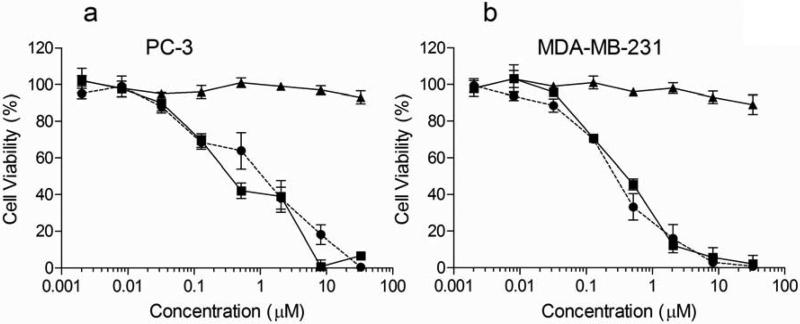
The cytotoxicity of 17-DMAPG and lipogel encapsulated 17-DMAPG was evaluated in two different cancer cell lines, the PC-3 human prostate and the MDA-MB-231 human breast cancer cells (Fig. 9). The IC50 values after 48 h incubation with the formulations were generated by fitting the viability data through GraphPad Prism Software. The IC50 for free drug was calculated to be 807 nM and 152 nM, for PC-3 and MDA-MB-231 cells respectively. The IC50 obtained for 17-DMAPG is similar to what was previously reported in another human breast cancer cell line SK-Br-23 [16]. In comparison, the IC50 for 17-DMAPG in lipogels was found to be 466 nM and 330 nM in PC-3 and MDA-MB-231 cells respectively. Although these IC50's differ numerically, they are not drastically different in terms of magnitude; therefore we conclude that both free drug and lipogel drug formulation exert similar levels of cytotoxicity to cancer cells. Most importantly, we found that empty lipogels incubated with cells at identical concentrations to drug loaded lipogels did not exert cytotoxicity to cells at the highest concentration used for the study (35 M 17-DMAPG in lipogels corresponded to approximately 0.4 mg/ml of lipogels), with 95% and 93% cell viability in PC-3 and MDA-MB-231 cells respectively. At concentrations of empty lipogels above 0.4 mg/ml, namely 1 and 2 mg/ml, proliferation in both cell lines was inhibited up to 30% (data not shown). This toxicity may be due to the non-biodegradable nature of PAA, and lipogels designed with more biocompatible gel cores can help circumvent this issue.
Fig. 9.
Resulting cell viability for free 17-DMAPG (●), 17-DMAPG in lipogels (■) and empty lipogels (▲) on the PC-3 human prostate (a) and MDA-MB-231 human breast cancer cell lines (b) at 48 h. The IC50 was obtained by fitting the data with GraphPad Prism Software. In PC-3 cells, the IC50 was found to be 466 nM for 17-DMAPG and 330 nM for 17-DMAPG in lipogels. In MDA-MB-231 cells, the IC50 was found to be 807 nM for 17-DMAPG and 152 nM for 17-DMAPG in lipogels. Most importantly, empty lipogels did not exert cytotoxicity to PC-3 cells (95% cell viability) and MDA-MB-231 cells (93% cell viability) at the highest drug concentration tested (35 M 17-DMAPG) which corresponded to approximately 0.4 mg/ml of lipogels.
4. Conclusions
We have successfully prepared hydrogel-in-liposome nanoparticles (lipogels) for active encapsulation of 17-DMAPG, a GA derivative. This drug was chosen due to (1) its improved aqueous solubility (4.6 mg/ml) compared to the parent GA (<0.01 g/ml), and (2) presence of a tertiary amine which readily protonates at low pH. For the design of lipogels in particular, a PAA hydrogel core was formed inside liposomes through UV-initiated DEAP activation and polymerization of AA and BA. We have demonstrated that a pH gradient and electrostatic interactions between drug and gel were critical for the active encapsulation and controlled-release of 17-DMAPG. More specifically, we found that optimal loading conditions could be obtained (88% drug loading efficiency, drug:lipogel (w/w) ratio of 0.053) through control of the external pH, temperature, and incubation time, with optimum conditions being pH 7.4, temperature of 55°C, incubation time of 30 min. Consistent drug loading and dramatic sustained drug release from lipogels could be achieved independent of the external solution pH (ca. 54 h to 50% drug release) due to an intact lipid bilayer. In vitro cell culture studies for 17-DMAPG and 17-DMAPG in lipogels revealed similar levels of cytotoxicity to PC-3 and MDA-MB-231 cells. Most importantly, lipogels alone did not exert cytotoxicity to cells at the highest concentration of drug tested (35 M), corresponding to approximately 0.4 mg/ml of material. This work describes for the first time the preparation of well-defined lipogels for active loading of a model drug 17-DMAPG, characterizations and release kinetics, and finally resulting cytotoxicity of the nanoparticles on cancer cells cultured in vitro.
Supplementary Material
Highlights.
Hydrogel-in-liposomes (lipogels) were prepared by UV irradiation
The model drug used was 17-DMAPG, a geldanamycin derivative
Passive and active drug loading, and release properties were characterized
The in vitro cytotoxicity of the nanoparticles and materials were investigated
Acknowledgments
This research was supported by NIH grant R00 CA136970 and startup funds from the University of Wisconsin-Madison, School of Pharmacy. We would also like to thank Fei Meng (Dept. of Chemistry, University of Wisconsin-Madison) for guidance and assistance with the SEM studies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- 1.Torchilin VP, Klibanov AL, Ivanov NN, Ringsdorf H, Schlarb B. Die Makromolekulare Chemie, Rapid Communications. 1987;8:457. [Google Scholar]
- 2.Monshipouri M, Rudolph AS. J Microencapsul. 1995;12:117. doi: 10.3109/02652049509015282. [DOI] [PubMed] [Google Scholar]
- 3.Jin T, Pennefather P, Lee PI. FEBS Lett. 1996;397:70. doi: 10.1016/s0014-5793(96)01021-6. [DOI] [PubMed] [Google Scholar]
- 4.Kazakov S, Kaholek M, Teraoka I, Levon K. Macromolecules. 2002;35:1911. [Google Scholar]
- 5.Kazakov S, Kaholek M, Kudasheva D, Teraoka I, Cowman MK, Levon K. Langmuir. 2003;19:8086. [Google Scholar]
- 6.Patton JN, Palmer AF. Biomacromolecules. 2005;6:2204. doi: 10.1021/bm050144b. [DOI] [PubMed] [Google Scholar]
- 7.Patton JN, Palmer AF. Biomacromolecules. 2005;6:414. doi: 10.1021/bm049432i. [DOI] [PubMed] [Google Scholar]
- 8.Van Thienen TG, Lucas B, Flesch FM, van Nostrum CF, Demeester J, De Smedt SC. Macromolecules. 2005;38:8503. [Google Scholar]
- 9.Kazakov S, Levon K. Curr Pharm Design. 2006;12:4713. doi: 10.2174/138161206779026281. [DOI] [PubMed] [Google Scholar]
- 10.An SY, Bui MPN, Nam YJ, Han KN, Li CA, Choo J, Lee EK, Katoh S, Kumada Y, Seong GH. J Colloid Interf Sci. 2009;331:98. doi: 10.1016/j.jcis.2008.11.022. [DOI] [PubMed] [Google Scholar]
- 11.Hong JS, Stavis SM, Lacerda SHD, Locascio LE, Raghavan SR, Gaitan M. Langmuir. 2010;26:11581. doi: 10.1021/la100879p. [DOI] [PubMed] [Google Scholar]
- 12.Saleem Q, Liu BX, Gradinaru CC, Macdonald PM. Biomacromolecules. 2011;12:2364. doi: 10.1021/bm200266z. [DOI] [PubMed] [Google Scholar]
- 13.Van Thienen TG, Raemdonck K, Demeester J, De Smedt SC. Langmuir. 2007;23:9794. doi: 10.1021/la700736v. [DOI] [PubMed] [Google Scholar]
- 14.Park J, Wrzesinski SH, Stern E, Look M, Criscione J, Ragheb R, Jay SM, Demento SL, Agawu A, Licona Limon P, Ferrandino AF, Gonzalez D, Habermann A, Flavell RA, Fahmy TM. Nat Mater. 2012;11:895. doi: 10.1038/nmat3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schnur RC, Corman ML, Gallaschun RJ, Cooper BA, Dee MF, Doty JL, Muzzi ML, Moyer JD, DiOrio CI, Barbacci EG, et al. J Med Chem. 1995;38:3806. doi: 10.1021/jm00019a010. [DOI] [PubMed] [Google Scholar]
- 16.Tian ZQ, Liu Y, Zhang D, Wang Z, Dong SD, Carreras CW, Zhou Y, Rastelli G, Santi DV, Myles DC. Bioorg Med Chem. 2004;12:5317. doi: 10.1016/j.bmc.2004.07.053. [DOI] [PubMed] [Google Scholar]
- 17.Torchillin VP, Weissig V. Liposomes: A practival approach. Oxford University Press; Oxford: 2003. [Google Scholar]
- 18.Hope MJ, Bally MB, Mayer LD, Janoff AS, Cullis PR. Chemistry and Physics of Lipids. 1986;40:89. doi: 10.1016/0009-3084(86)90077-0. [DOI] [PubMed] [Google Scholar]
- 19.Kulkarni SB, Betageri GV, Singh M. J Microencapsul. 1995;12:229. doi: 10.3109/02652049509010292. [DOI] [PubMed] [Google Scholar]
- 20.Perkins WR, Minchey SR, Ahl PL, Janoff AS. Chem Phys Lipids. 1993;64:197. doi: 10.1016/0009-3084(93)90066-c. [DOI] [PubMed] [Google Scholar]
- 21.Cullis PR, Hope MJ, Bally MB, Madden TD, Mayer LD, Fenske DB. Biochim Biophys Acta. 1997;1331:187. doi: 10.1016/s0304-4157(97)00006-3. [DOI] [PubMed] [Google Scholar]
- 22.Cullis PR, Bally MB, Madden TD, Mayer LD, Hope MJ. Trends Biotechnol. 1991;9:268. doi: 10.1016/0167-7799(91)90088-y. [DOI] [PubMed] [Google Scholar]
- 23.Kim SW, Bae YH, Okano T. Pharm Res. 1992;9:283. doi: 10.1023/a:1015887213431. [DOI] [PubMed] [Google Scholar]
- 24.Andrade-Vivero P, Fernandez-Gabriel E, Alvarez-Lorenzo C, Concheiro A. J Pharm Sci. 2007;96:802. doi: 10.1002/jps.20761. [DOI] [PubMed] [Google Scholar]
- 25.Bos GW, Jacobs JJ, Koten JW, Van Tomme S, Veldhuis T, van Nostrum CF, Den Otter W, Hennink WE. Eur J Pharm Sci. 2004;21:561. doi: 10.1016/j.ejps.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 26.Bouhadir KH, Kruger GM, Lee KY, Mooney DJ. J Pharm Sci. 2000;89:910. doi: 10.1002/1520-6017(200007)89:7<910::AID-JPS8>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 27.Cai S, Liu Y, Zheng Shu X, Prestwich GD. Biomaterials. 2005;26:6054. doi: 10.1016/j.biomaterials.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 28.Cho KY, Chung TW, Kim BC, Kim MK, Lee JH, Wee WR, Cho CS. Int J Pharm. 2003;260:83. doi: 10.1016/s0378-5173(03)00259-x. [DOI] [PubMed] [Google Scholar]
- 29.Galeska I, Kim TK, Patil SD, Bhardwaj U, Chatttopadhyay D, Papadimitrakopoulos F, Burgess DJ. AAPS J. 2005;7:E231. doi: 10.1208/aapsj070122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J, Lin S, Li L, Liu E. Int J Pharm. 2005;298:117. doi: 10.1016/j.ijpharm.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 31.Liu Y, Ibricevic A, Cohen JA, Cohen JL, Gunsten SP, Frechet JM, Walter MJ, Welch MJ, Brody SL. Mol Pharm. 2009;6:1891. doi: 10.1021/mp900215p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murphy DJ, Sankalia MG, Loughlin RG, Donnelly RF, Jenkins MG, PA MCC. Int J Pharm. 2011 doi: 10.1016/j.ijpharm.2011.11.018. [DOI] [PubMed] [Google Scholar]
- 33.Kim M, Jung B, Park JH. Biomaterials. 2012;33:668. doi: 10.1016/j.biomaterials.2011.09.074. [DOI] [PubMed] [Google Scholar]
- 34.Lele BS, Hoffman AS. J Biomater Sci Polym Ed. 2000;11:1319. doi: 10.1163/156856200744354. [DOI] [PubMed] [Google Scholar]
- 35.Falamarzian M, Varshosaz J. Drug Dev Ind Pharm. 1998;24:667. doi: 10.3109/03639049809082369. [DOI] [PubMed] [Google Scholar]
- 36.Park H, Robinson JR. Pharm Res. 1987;4:457. doi: 10.1023/a:1016467219657. [DOI] [PubMed] [Google Scholar]
- 37.Argentiere S, Blasi L, Ciccarella G, Barbarella G, Cingolani R, Gigli G. Macromol Symp. 2009;281:69. [Google Scholar]
- 38.Kiser PF, Wilson G, Needham D. Nature. 1998;394:459. doi: 10.1038/28822. [DOI] [PubMed] [Google Scholar]
- 39.An SY, Bui MP, Nam YJ, Han KN, Li CA, Choo J, Lee EK, Katoh S, Kumada Y, Seong GH. J Colloid Interface Sci. 2009;331:98. doi: 10.1016/j.jcis.2008.11.022. [DOI] [PubMed] [Google Scholar]
- 40.Kazakov S, Kaholek M, Kudasheva D, Tereoka I, Cowman M, Levon K. Macromolecules. 2003;35:1911. [Google Scholar]
- 41.Schillemans JP, Flesch FM, Hennink WE, van Nostrum CF. Macromolecules. 2006;39:5885. [Google Scholar]
- 42.Chemin C, Pean JM, Bourgaux C, Pabst G, Wuthrich P, Couvreur P, Ollivon M. Biochim Biophys Acta. 2009;1788:926. doi: 10.1016/j.bbamem.2008.11.017. [DOI] [PubMed] [Google Scholar]
- 43.O'Brien J, Wilson I, Orton T, Pognan F. Eur J Biochem. 2000;267:5421. doi: 10.1046/j.1432-1327.2000.01606.x. [DOI] [PubMed] [Google Scholar]
- 44.Schillemans JP, Flesch FM, Hennink WE, Nostrum C.F.v. Macromolecules. 2006;39:5885. [Google Scholar]
- 45.Elliott JE, Macdonald M, Nie J, Bowman CN. Polymer. 2004;45:1503. [Google Scholar]
- 46.Bibi S, Kaur R, Henriksen-Lacey M, McNeil SE, Wilkhu J, Lattmann E, Christensen D, Mohammed AR, Perrie Y. Int J Pharmaceut. 2011;417:138. doi: 10.1016/j.ijpharm.2010.12.021. [DOI] [PubMed] [Google Scholar]
- 47.Fujiwara M, Grubbs RH, Baldeschwieler JD. J Colloid Interface Sci. 1997;185:210. doi: 10.1006/jcis.1996.4608. [DOI] [PubMed] [Google Scholar]
- 48.Chen J, Cheng D, Li J, Wang Y, Guo JX, Chen ZP, Cai BC, Yang T. Drug Dev Ind Pharm. 2013;39:197. doi: 10.3109/03639045.2012.668912. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.