Abstract
Long-range interactions between transcription regulatory elements play an important role in gene activation, epigenetic silencing, and chromatin organization. Transcriptional activation or repression of developmentally regulated genes is often accomplished through tissue specific chromatin architecture and dynamic localization between active transcription factories and repressive Polycomb bodies. However, the mechanisms underlying the structural organization of chromatin and the coordination of physical interactions are not fully understood. Insulators and Polycomb group proteins form highly conserved multi-protein complexes that mediate functional long-range interactions, and have proposed roles in nuclear organization. In this review, we explore recent findings that have broadened our understanding of the function of these proteins and provide an integrative model for the roles of insulators in nuclear organization.
Keywords: CTCF, Polycomb, TFIIIC, Cohesin, Epigenetics
INTRODUCTION
Eukaryotic genomes are packaged into higher-order chromatin structures and ultimately organized in a manner that functionally relates to gene expression. Understanding the mechanisms and molecular players involved in genome organization is therefore essential to fully comprehend the fundamental relationship between nuclear organization and genome function. Insulators are multi-protein DNA complexes proposed to underlie nuclear architecture based on their ability to facilitate long-range physical interactions, to interact with nuclear substructures, and to cluster into nuclear foci termed insulator bodies. However, spatiotemporal expression and repression of genes pertinent to development and cell-type specification involves the function of additional regulatory elements, such as enhancers and Polycomb response elements (PREs), suggesting additional factors may also play a role in genome organization. It is then possible that the three-dimensional arrangement of chromatin in the nuclear space is in part a consequence of function, i.e. the interaction of regulatory sequences with gene promoters, and in part due to structural elements whose role is to establish interactions between specific sequences to effect particular patterns of gene expression. The recent developments of unbiased, high-throughput methods for mapping protein binding sites and genome-wide interactions have allowed an unprecedented look into the inner-workings of genome biology, and new studies have shed valuable insights into the roles of insulators and chromatin structure in nuclear organization. In this review we highlight the relationship between nuclear organization and genome function and emphasize the dynamic interplay among chromatin insulators, transcription activation and Polycomb (Pc)-mediated repression in creating and/or maintaining a three-dimensional arrangement of the chromatin that is conducive to the establishment of patterns of gene expression required for cell type specification.
NUCLEAR ORGANIZATION AND GENOME FUNCTION
Eukaryotic cells are tasked with packaging the genome several thousand-fold into the confines of the cell nucleus, all while maintaining gene accessibility and chromatin structure that accommodates highly dynamic processes, including gene transcription, replication, and DNA repair. Interphase chromosomes are organized into discrete territories that are distributed non-randomly with respect to the nucleus and with respect to other chromosomes, and whose placement can influence the potential for trans interactions and dictate whether a genomic locus is in an active or repressive nuclear environment (Cremer & Cremer 2010, Fraser & Bickmore 2007). The nucleus also harbors several discrete subnuclear foci, termed nuclear bodies, which are dynamically regulated structures that facilitate greater efficiency of many nuclear processes (Mao et al 2011). For example, active genes can relocate from chromosome territories (Branco & Pombo 2006, Chambeyron & Bickmore 2004), and cluster into subnuclear foci termed transcription factories for gene expression (Chakalova & Fraser 2010). Gene silencing is also accomplished through recruitment to repressive nuclear structures, and most biological processes are similarly compartmentalized into analogous nuclear bodies, indicating an important relationship between nuclear organization and genome function.
DIFFERENTIATION, REPLICATION AND GENOME STABILITY
The nonrandom order and significance of genome organization is perhaps best highlighted by its relationship to cellular differentiation, replication and genome stability. The pathway from pluripotency to differentiated tissues is accompanied by changes in epigenomic landscapes, genome compaction, and some degree of chromosomal reorganization (Ahmed et al 2010, Mikkelsen et al 2007, Vastenhouw et al 2010, Wiblin et al 2005). Developmental genes are differentially targeted to transcriptionally active or transcriptionally repressive loci, and differentiation is associated with restructuring of interactions between chromatin and the nuclear lamina (Peric-Hupkes et al 2010). Increase in genome compaction may accommodate the organization of nuclear foci associated with transcription, DNA repair, replication, and splicing, while restricting the complexity of genome function by concealing irrelevant transcription factor binding sites (Meister et al 2011). Nuclear organization also plays a critical role in organizing DNA replication into discrete subnuclear compartments (Berezney et al 2000), and in maintaining genome integrity. DNA damage gives rise to the accumulation of repair and DNA damage checkpoint proteins concomitant to increased chromatin accessibility (Kruhlak et al 2006, Lisby et al 2003), and studies in both yeast and human cells demonstrate an important relationship between nuclear organization, DNA repeat stability and telomere protection (for reviews see (Mekhail & Moazed 2010, Nagai et al 2011). Characterizing the mechanisms involved in three-dimensional genome organization is therefore essential for understanding the apparent fundamental relationship between nuclear organization and cellular function.
GENOMIC STRATEGIES
Microscopy studies have been invaluable in revealing insights into the distribution and organization of chromosomes in individual cells and their relationship with gene regulation. However, to break down the three dimensional organization of chromosomes and the relationship between nuclear organization and underlying chromatin proteins, new techniques were required that exceeded the resolution and throughput limits imposed by traditional light microscopy. The advent of Chromosome Conformation Capture (3C) described by Dekker et al. (Dekker et al 2002) marked the first approach for effectively mapping physical chromosomal interactions across the genome. While 3C has been useful in identifying locus specific interactions between regulatory elements and target genes (Dekker et al 2002, Tolhuis et al 2002), derivations of the 3C technique have been introduced to extend the approach in an unbiased high-throughput genome-wide fashion. For example, the Hi-C method integrates an extended 3C protocol with massively parallel DNA sequencing, thereby capturing all genome-wide interactions at a resolution limited by the depth of sequencing (Lieberman-Aiden et al 2009). Initial Hi-C analyses in a human lymphoblastoid cell line shed valuable insight into chromatin organization, supporting the fractal globule model in which chromosomes self-organize into a hierarchy of “crumples”, or series of globules governed by topological constraints (Grosberg et al 1988, Lieberman-Aiden et al 2009, Mirny 2011). Subsequent computational modeling supports the existence of chromosome territories and transcriptional foci, and revealed new insights into the relationship between chromatin organization and CTCF, as well as the nuclear lamina (Yaffe & Tanay 2011). However, further conclusions about chromosome topology and nuclear organization of chromatin in human cells will require higher resolution, likely to be obtained in the near future with greater sequencing depth.
MEDIATORS OF NUCLEAR ORGANIZATION
Determining how interphase chromosomes are anchored within the nuclear space, and which proteins mediate structural arrangements conducive to gene regulation and locus plasticity remains a critical hurdle to understanding the mechanisms that regulate genome function. Fortunately, genome-wide mapping of chromatin associated proteins has increased at an extraordinary pace within the past few years thanks to the ENCyclopedia of DNA Elements (ENCODE) Projects (Birney et al 2007, Celniker et al 2009) and the increasingly affordable option of high-throughput sequencing. Analyses combining the three-dimensional organization of interphase chromosomes (Hi-C) with genome-wide binding profiles of chromatin associated factors (ChIP-seq) may ultimately establish which proteins functionally mediate nuclear organization. Information from microscopy and biochemical studies combined with recent genome-wide mapping have implicated multiple factors, including chromatin insulators, Polycomb (Pc) complexes, and non-coding RNAs as having roles in domain formation and chromatin organization that we consider in this review.
INSULATORS
Chromatin insulators were originally defined as regulatory elements that recruit proteins to establish boundaries between adjacent chromatin domains. Insulators were also shown to block the communication between enhancers and nearby promoters in an orientation-dependent manner, leading to intuitive models proposing that insulators serve to limit the promiscuity of enhancers. However, further characterization of these sequences from yeast to humans have revealed that insulators mediate long-range intra- and inter-chromosomal interactions, co-localize to subnuclear foci called insulator bodies as well as transcription factories, and preferentially cluster in trans as revealed by Hi-C computational modeling in both yeast and humans, together suggesting insulators serve a greater purpose in chromatin organization.
Composition and Evolution
Studies of chromatin insulators in mammals have long been restricted to the highly conserved CCCTC-binding factor (CTCF), which until recently was the only characterized protein capable of insulator activity in humans. However, insulator activity in yeast involves tRNA genes and transcription factor TFIIIC, and this role was recently shown to be conserved in mammals. (Ebersole et al 2011, Raab et al 2011). Concurrent research of insulators in yeast, Drosophila, and mammalian systems therefore suggests that these elements serve an evolutionarily conserved role in gene regulation and nuclear organization.
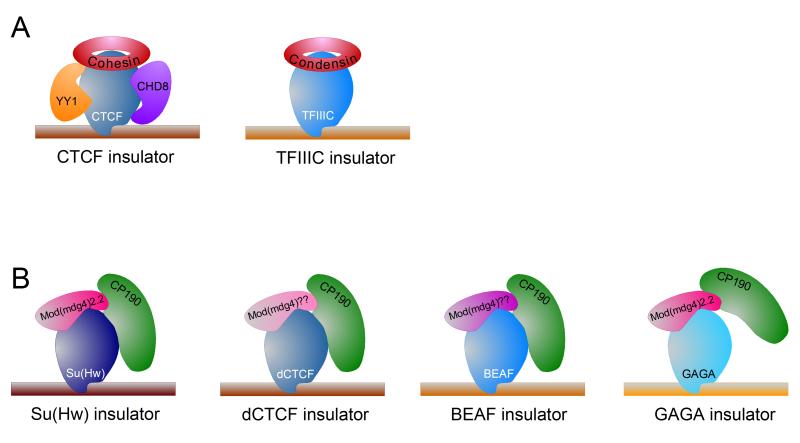
CTCF contains a central domain composed of 11 zinc fingers, and is ubiquitously expressed (Klenova et al 1993). Early biochemical studies demonstrated >93% amino acid identity between human and chicken CTCF proteins (Filippova et al 1996), and studies in Drosophila later identified an orthologous CTCF factor with similar domain structure and conserved insulator function (Moon et al 2005), suggesting this zinc-finger protein plays a vital, highly conserved role in nuclear biology. Remarkably, CTCF primarily targets a highly similar core consensus sequence from Drosophila to humans, despite its ability to bind a variety of DNA sequences (Holohan et al 2007). The proteins with which CTCF associates and the variant sequences it is able to bind have been suggested to underlie the numerous roles in which CTCF has been implicated (Weth & Renkawitz 2011, Zlatanova & Caiafa 2009), such as X-chromosome inactivation, V(D)J rearrangement, and chromatin insulation. Many CTCF binding sites recruit the cohesin complex (Figure 1), which is required for functional CTCF insulator activity (Nativio et al 2009, Wendt et al 2008). The cohesin complex forms a ring-shaped structure and mediates cohesion between sister chromatids from S-phase until mitosis, suggesting that cohesin may specifically stabilize chromatin loops arranged by CTCF through a similar mechanism. CTCF also interacts with Yin and yang 1 (Yy1), a transcription factor involved in X-chromosome inactivation (Donohoe et al 2007) capable of recruiting Pc complexes (Wilkinson et al 2006), and CTCF-mediated insulator activity at the H19/Igf2 has been shown to require the SNF2-like chromodomain helicase protein CHD8 (Figure 1) (Ishihara et al 2006), the DEAD-box RNA helicase p68 and its associated RNA (SRA) (Yao et al 2010).
Figure 1.
Diagram showing the structure of different vertebrate and Drosophila insulators. A. Structure of the vertebrate CTCF and TFIIIC insulators. Indicated are factors associated with CTCF such as cohesin, CHD8 and YY1, and with TFIIIC. B. Each Drosophila insulator subclass contains a different binding protein that may define the specific function of the corresponding subclass. All insulators share the common protein CP190, although the role of this protein in the function of the GAGA insulator has not been demonstrated experimentally. In addition, all subclasses may also have one Mod(mdg4) isoform. The gypsy/Su(Hw) insulator contains Mod(mdg4)2.2. The dCTCF and BEAF insulators lack this isoform but contain a different variant of Mod(mdg4). GAGA has been shown to interact with Mod(mdg4)2.2.
DROSOPHILA INSULATORS have been particularly well characterized thanks to in vivo insulator activity reporter assays made easy by the genetic manipulations available in the fly model system. In addition to the Drosophila homolog of CTCF (dCTCF), several insulator proteins have been identified, including Suppressor of Hairy Wing (Su(Hw)), Boundary Element Associated Factor (BEAF-32), and GAGA factor (GAF) (Figure 1) (Gurudatta & Corces 2009). Contrary to what happens in vertebrates, the Drosophila cohesin complex localizes independently of dCTCF to transcriptionally active genes (Misulovin et al 2008). Instead of cohesin, Drosophila insulator activity relies on fly-specific insulator proteins Centrosomal Protein 190 kDa (CP190) and Modifier of Modg4 (Mod(mdg4), both of which contain BTB/POZ domains and are capable of forming stable multimers in vitro (Bonchuk et al 2011, Gerasimova et al 2007, Ghosh et al 2001). Genome-wide localization studies suggest that dCTCF synergizes with Su(Hw), BEAF-32, and CP190 at many sites throughout the genome, perhaps representing a unifying role in facilitating chromosomal interactions and genome organization (Negre et al 2010, Van Bortle et al 2012). Interestingly, insulator activity in Drosophila also relies on components of the RNAi machinery, (Lei & Corces 2006, Moshkovich et al 2011), though the mechanistic relationship remains poorly understood.
tRNA GENES were first demonstrated to function as boundary elements flanking the repressed HMR locus in S. cerevisiae, and subsequently at the pericentromeric regions of S. pombe (Donze et al 1999, Scott et al 2006). The conservation of tDNA as insulator elements has been further extended by the demonstration that tRNA genes function as barrier and enhancer-blocking insulators in humans (Raab et al 2011). Analysis of the mat locus in S. pombe revealed that a repeat of B-box elements, which are highly conserved intragenic promoter elements in tRNA genes that recruit RNA polymerase III (RNAPIII) transcription initiation factor TFIIIC, were responsible for barrier activity (Figure 1) (Noma et al 2006). Mutations in TFIIIC and TFIIIB, but not RNAPIII, affected insulator activity in S. cerevisiae, suggesting insulator activity occurs independent of RNAPIII transcription (Donze & Kamakaka 2001). Though TFIIIC is sufficient for gene insulation alone, insulator activity is strengthened by TFIIIB and utilizes chromatin remodelers, possibly to evict histones at tDNA insulators (Valenzuela et al 2009). Interestingly, in yeast and humans, TFIIIC binds many regions devoid of RNAPIII, called Extra TFIIIC (ETC) loci (Moqtaderi & Struhl 2004), and these sites are associated with the cohesin complex and localize near CTCF-binding sites in mouse embryonic stem cells and humans (Carriere et al 2012, Moqtaderi et al 2010). TFIIIC sites and tRNA genes also function as loading-sites for the highly conserved condensin complex in yeast, suggesting insulators serve an important role in chromatin architecture during both interphase and mitosis (D’Ambrosio et al 2008).
Distribution and Chromatin Structure
Genome-wide localization studies by ChIP-chip and ChIP-seq revealed that insulators are dispersed throughout eukaryotic genomes. CTCF is bound to thousands of independent sites in Drosophila (Bushey et al 2009, Holohan et al 2007, Negre et al 2010) and tens of thousands of sites in human cells lines (Barski et al 2007, Cuddapah et al 2009, Kim et al 2007), consistent with a global role in genome function. ChIP-chip analysis further showed that CTCF co-localizes with cohesin at more than half of its sites (Parelho et al 2008, Rubio et al 2008, Wendt et al 2008), suggesting many CTCF sites are likely capable of functional insulator activity. Meanwhile, recent genome-wide localization of TFIIIC in mouse embryonic stem cells revealed that while < 300 tRNA genes were occupied by all three TFIIIC subunits, as many as 2,200 independent TFIIIC insulators (ETC loci) are dispersed throughout the mouse genome (Carriere et al 2012). Remarkably, as many as 85% of ETC loci lie within 20 kb of CTCF-binding sites, and cohesin subunits Smc1A and Smc3 were enriched at the ETC loci specifically, suggesting CTCF and TFIIIC distribution and insulator activity may be intimately associated.
Though insulators are broadly distributed, studies in yeast, Drosophila, and mammals have demonstrated that a subset of CTCF and tDNAs border repressed chromatin domains, and function as chromatin boundaries to prevent the spread of heterochromatin into flanking chromatin domains. In D. melanogaster, dCTCF and CP190 co-localize at the borders of many H3K27me3 domains, and knockdown of either dCTCF or CP190 results in spreading of repressive chromatin, suggesting chromatin structure at these domain borders relies on insulator activity (Bartkuhn et al 2009). Comprehensive genome-wide mapping of all Drosophila insulators revealed that Su(Hw) and BEAF-32 also localize to repressive domain borders (Negre et al 2010), and recent ChIP-seq results provided sufficient resolution to demonstrate alignment of dCTCF with Su(Hw) and/or BEAF-32 at these boundaries (Van Bortle et al 2012). Knockdown of each insulator component results in upregulation of repressive domain-containing developmental genes, suggesting dCTCF synergizes with Drosophila specific insulator proteins to establish appropriate chromatin domain organization. CTCF and cohesin also form functional barriers at the Wnt4 locus in mammals (Essafi et al 2011), and genome-wide mapping of CTCF by ChIP-seq in human cell lines identified analogous binding at the boundaries of many repressive chromatin domains (Cuddapah et al 2009). CTCF domain border associations in humans are largely tissue specific, whereas DNase-seq and FAIRE-seq mapping across seven human cell lines revealed that CTCF binding sites at or near transcription start sites are common across cell-type (Song et al 2011). In yeast, tDNA insulator activity was first characterized based on the ability of a tRNAThr gene to block the spread of silencing from the HMR locus in S. cerevisiae (Donze et al 1999), and subsequent studies demonstrated analogous barrier activity of a tRNAAla gene at the pericentromeric regions of S. pombe (Haldar & Kamakaka 2006, Scott et al 2006).
Barrier activity appears to be a feature of insulators, but whether these sequences evolved simply to guard active genomic loci from the silencing effects of nearby repressive chromatin domains is questionable. Cavalli and colleagues recently utilized an independent, high-throughput 3C derivative (3C-seq) to explore the three-dimensional folding and functional organization principles of the Drosophila genome (Sexton et al 2012). Their data suggest eukaryotic genomes are partitioned into physical domains that can be clustered based on strong statistical association with linear epigenomic profiles. Physical domains were categorized as active, which correlate with active histone marks, null, which comprise large transcriptionally repressive regions lacking silent chromatin marks, Polycomb (Pc) domains associated with H3K27me3 repression, and HP1/centromeric domains associated with classical heterochromatin. Physical domains are sharply demarcated, and contacts within domains abruptly decay at positions corresponding to physical domain edges. Remarkably, insulators are highly enriched at domain borders, and hierarchical clustering revealed recurrent combinations of insulators and active histone marks that are present at all combinations of physical boundaries (e.g. even between two similarly annotated physical domains, such as Null – Null). The distribution of insulators across the eukaryotic genome may therefore serve to delimit not just repressive chromatin domains, but physical domains that only become distinguishable upon knowledge of chromosomal partitioning. Together, these data suggest a more paramount role for insulators in chromatin organization than classical definitions of enhancer-blocking and barrier activities for which insulators are typically described.
Long-Range Interactions
In addition to forming domain boundaries, insulators have been historically defined by their ability to impede the interaction between promoters and distal enhancers in a direction-dependent manner. The enhancer-blocking capacity of insulators has been experimentally studied using transgene constructs, and observations at genomic loci across species suggest insulators establish chromatin loops through physical interactions. Concurrent models proposed that insulators evolved to ensure the fidelity of enhancers and their target promoters in vivo by establishing chromatin loops, and thereby dictating the potential for enhancer-promoter interactions. However, accumulating data suggest that insulators facilitate long-range inter- and intra-chromosomal interactions across the genome, including directing distant enhancers to the appropriate promoters, suggesting that local determination of enhancer-promoter interaction represents only part of a more significant role in chromosome organization.
The enhancer-blocking activities of CTCF have been best characterized at the chicken β-globin locus and the murine H19/Igf2 imprinted locus, both of which have been extensively reviewed (Phillips & Corces 2009). These loci provide ideal scenarios for studying the role of insulators in allele-specific and developmental cell-type specific gene regulation and chromatin architecture, and 3C experiments suggest CTCF underlies chromatin contacts at both genomic loci (Kurukuti et al 2006, Splinter et al 2006). Recent application of 3C has revealed that CTCF also underlies developmental higher-order architecture at the conserved homeobox gene A (HOXA) locus in mouse and humans (Kim et al 2011). Specifically, CTCF and cohesin were shown to contribute to reorganization and selective gene activation at HOXA by partitioning silenced genes through chromatin loop formation upon differentiation. Furthermore, pluripotency factor OCT4 was shown to antagonize cohesin loading at the CTCF binding site, thereby demonstrating developmental regulation of insulator activity and gene expression by inhibiting chromosome loop formation. Studies in D. melanogaster suggest insulators serve a conserved role in developmental coordination of gene expression and chromatin structure. For example, genome-wide mapping revealed dCTCF and Drosophila specific insulator proteins are regulated through DNA-binding and recruitment of CP190 during the ecdysone hormone-response in Kc cells (Wood et al 2011). 3C analysis at the ecdysone induced Eip75B gene further revealed a developmentally regulated dCTCF site, wherein recruitment of CP190 and enhanced chromatin looping upon ecdysone stimulation suggested alterations in Eip57B locus chromatin organization.
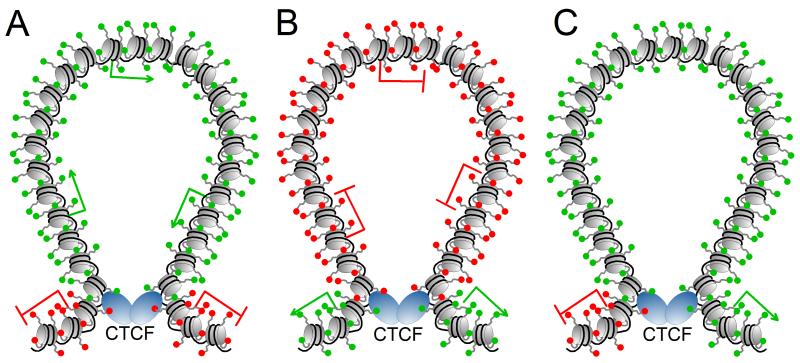
Numerous studies have focused on the role of insulators in locus specific gene regulation and chromatin architecture, yet the emerging picture of insulators in genome-wide nuclear organization requires global analyses of chromatin interactions. The first genome-wide map of CTCF-mediated functional interactions has been obtained by combining chromatin immunoprecipitation with high-throughput sequencing of enriched chromatin interactions (Handoko et al 2011). The authors identified ~1,500 cis- and ~330 trans-interactions facilitated by CTCF in mouse embryonic stem cells, and classified them into five categories based on distinct epigenetic patterns. CTCF interactions were shown to harbor chromatin loops enriched for active or repressive chromatin signatures, suggesting CTCF harnesses clusters of genes with coordinated expression (Figure 2). CTCF interactions were also shown to create chromatin hubs conducive to enhancer and promoter activities, in support of recent evidence suggesting CTCF and cohesin underlie enhancer-promoter interactions at the INFG and MHC-II loci (Hadjur et al 2009, Majumder & Boss 2011). The authors speculate that insulators may instead serve the capacity of “enhancer-bridging”, suggesting CTCF is involved in facilitating cell-type, enhancer-promoter specific interactions. In many cases, CTCF interactions themselves appear to function as chromatin domain barriers, wherein opposing chromatin states flank interacting CTCF loci, though the intervening chromatin loop exhibits no specific epigenetic pattern. Ultimately Handoko et al. provide a genome-wide interrogation of CTCF interactions and reveal several modes through which CTCF functionally organizes the genome.
Figure 2.
Structure of some of the domains created by interactions between CTCF insulators in mouse embryonic stem cells. Actively transcribed genes are represented by a green arrow and silenced genes by a red line; nucleosomes and the histone tails are represented in grey, with active histone modifications indicated as green spheres and repressive modifications as red spheres. DNA is represented in black and CTCF as blue ovals. A. CTCF forms a loop to separate a domain containing active histone modifications and transcribed genes from repressive marks and silenced genes. B. CTCF forms a loop to separate a domain containing repressive histone modifications and silenced genes from active marks and transcribed genes. C. CTCF forms a loop containing nucleosomes enriched in mono and dimethylated H3K4, and trimethylated H3K4 at the boundaries of the loops, whereas the active transcription modification H3K36me3 and repressive H3K27me3 mark are observed outside the loops on opposite sides.
Genome-wide interrogation of intra- and inter-chromosomal interactions in yeast and humans independently demonstrate that tDNA insulators also underlie long-range genomic interactions. Noble and colleagues used a high-throughput 3C-based technique for querying the three-dimensional organization of the S. cerevisiae genome (Duan et al 2010). tRNA genes were significantly enriched for interactions with other tRNA genes, suggesting insulator-to-insulator interactions are a conserved feature of eukaryotic genome organization. Hierarchical clustering revealed two clusters of co-localizing tRNA genes, one consistent with previously described nucleolar localization (Thompson et al 2003), and another with centromeres. Similar mapping of interactions at a tDNA insulator in humans revealed analogous long-range interactions between tDNAs as well as ETC loci, suggesting tRNA genes and TFIIIC play a conserved role in genome organization. Recent findings suggest TFIIIC binding sites facilitate condensin-binding in S. cerevisiae and S. pombe (D’Ambrosio et al 2008) and co-localize with cohesin in mammals (Carriere et al 2012), suggesting TFIIIC recruitment of condensin and cohesin complexes may underlie chromosomal interactions in yeast and humans analogous to CTCF.
Role in Nuclear Organization
Microscopy-based analyses of the physical and functional organization of eukaryotic nuclei have led to the identification of several discrete subnuclear organelles called nuclear bodies, which play host to a variety of nuclear processes, including transcription, splicing, processing, and epigenetic regulation (Mao et al 2011). Nuclear staining of insulator proteins has clearly shown a propensity for insulators to concentrate into distinct nuclear foci, a feature that is conserved in yeast (Noma et al 2006) Drosophila (Gerasimova et al 2000), and mammals (MacPherson et al 2009). Insulators have also been shown to interact with and localize to nuclear substructures, including the nuclear and nucleolar peripheries, suggesting insulators tether associated chromatin to defined nuclear compartments (Gerasimova et al 2000, Yusufzai et al 2004). These findings combined with insulator-mediated functional interactions have led to models proposing that insulators ultimately interact to partition chromatin into structural and functional domains that are physically organized through ‘insulator bodies’. Though tantalizing, the functional importance and molecular underpinnings of insulator bodies remains poorly characterized.
Nuclear organization clearly involves spatial arrangement of chromosomes whose position with respect to the nuclear periphery correlates with chromatin structure and gene expression. Chromatin interactions at the nuclear periphery have recently been mapped in both Drosophila (Pickersgill et al 2006) and humans (Guelen et al 2008), revealing large, sharply-defined lamina-associated domains (LADs) that correlate with low gene density and transcriptional repression. The borders of LADs in humans are enriched for CTCF (Guelen et al 2008), suggesting this protein may serve to separate chromatin environments at the nuclear periphery. Mapping of insulators in D. melanogaster with respect to the nuclear lamina has also revealed a significant enrichment for the Drosophila-specific Su(Hw) insulator at the borders of LADs (van Bemmel et al 2010). Handoko et al independently identified the apparent relationship between lamina and the CTCF interaction network in mouse embryonic stem cells (Handoko et al 2011). Specifically, CTCF loops were depleted within LADs but enriched at LAD borders, supporting a model in which CTCF orchestrates genome organization with respect to the nuclear lamina. An analogous role for TFIIIC in nuclear organization in yeast has been proposed based on perinuclear staining of insulator bodies in S. pombe (Noma et al 2006). In support of this model, perinuclear localization and silencing of the HMR locus in S. cerevisiae was recently shown to rely on nuclear pore proteins that localize to a tDNA barrier insulator (Ruben et al 2011).
POLYCOMB
Genome plasticity and selective expression are essential features of multicellular development, and several early regulatory factors involved in body patterning and segmentation need to be strictly regulated to facilitate appropriate developmental decisions. Polycomb group (PcG) proteins are evolutionarily conserved epigenetic transcriptional repressors that play an important role in establishing and maintaining cell fate by influencing the expression status of pertinent genes. PcG proteins specifically mediate the repression of numerous developmental genes through posttranslational modification of histone proteins, and recent studies demonstrate Polycomb (Pc) activity is involved in a broad scope of cellular processes, including differentiation, cell cycle regulation, X-inactivation, and cell signaling (Sawarkar & Paro 2010). Microscopy studies have demonstrated that PcG proteins concentrate into nuclear foci, called Pc bodies, suggesting PcG proteins may also mediate the nuclear organization of their target genes. Here we review recent progress in determining the relationship between Pc and nuclear organization.
Composition and Evolution
The PcG genes were first discovered as chromatin repressors that maintain silencing of the homeotic regulatory genes throughout Drosophila development (Paro 1990). Further studies in the fruit fly demonstrated that PcG proteins are recruited through cis-regulatory elements called Polycomb Response Elements (PREs) (Simon et al 1993), and a recently identified element at the HoxD locus in humans facilitating Pc-dependent transcriptional repression throughout cell differentiation suggests the mechanism of Pc recruitment may be conserved (Woo et al 2010). However PREs are not easily broken into obvious DNA consensus sequences as described for insulator proteins, and the functional mechanism of Pc targeting remains unclear, though it appears to involve numerous players including DNA-binding proteins, histone posttranslational modification binding proteins, RNAi machinery proteins, and non-coding RNAs (Beisel & Paro 2011).
PcG proteins are present in two major complexes, Polycomb repressive complex 1 (PRC1) and 2 (PRC2), whose core components are largely conserved from flies to humans. The PcG family also includes several additional proteins that allow for the formation of diverse Pc chromatin-binding complexes with variable enzymatic activities (Simon & Kingston 2009). PRC2 catalyzes H3K27 di- and tri-methylation (H3K27me3 – associated with transcriptional repression) through SET-domain containing subunit EZH1, as well as EZH2 depending on cellular context (Beck et al 2010, Margueron & Reinberg 2011). PRC2 recruitment and activity is regulated by core components and ancillary subunits, such as PHF1, JARID2, and AEBP2, which stimulate PRC2 enzymatic activity (Beck et al 2010, Kim et al 2009, Li et al 2010, Sarma et al 2008). PRC1 subunits RING1B and BMI1 form a stable heterodimer capable of catalyzing H2AK119 ubiquitylation (H2AK199Ub1) (Cao et al 2005), which likely underlies PRC1 mediated Pc silencing (Wang et al 2004). There is also recent evidence that histone modification-binding proteins containing malignant brain tumor (MBT) modules contribute to Pc function. For example, L3MBTL2 interacts with and is required for Pc-mediated repression by a PRC1-like complex in human cells (Trojer et al 2011). Interestingly, the Drosophila MBT protein L(3)mbt was recently shown to localize specifically to chromatin insulators (Richter et al 2011), and as we discuss below, recent evidence suggests insulator activity may play an important role in Pc repression.
Distribution and Chromatin Structure
The genome-wide localization of PcG proteins has been studied in several independent ChIP-chip and ChIP-seq experiments in both Drosophila and mammals. Pc complexes localize to putative PREs in Drosophila, and correlate with broad repressive H3K27me3 domains that encompass genes involved in major developmental pathways (Schwartz et al 2006, Tolhuis et al 2006). Most PREs are co-occupied by the major Pc complexes PRC1 and PRC2, and the occupancy landscape of PcG proteins changes during development, consistent with its role in mediating cell fate restriction by differential gene silencing (Negre et al 2006, Oktaba et al 2008). Mapping of PRC1 and PRC2 complexes in mammals show similar co-occupation of developmental pathway genes and displacement of PcG proteins during gene activation (Boyer et al 2006, Bracken et al 2006, Lee et al 2006), suggesting Pc repression is a highly conserved feature of multicellular development. However, PcG proteins also localize to bivalent domains characterized by overlapping H3K27me3 and H3K4me3 encompassing genes poised for activation or repression upon cellular differentiation in mammals, a feature that is largely absent in fly embryos (Schuettengruber et al 2009).
Genome-wide mapping of trithorax-group (trxG) proteins, which catalyze H3K4 methylation and antagonize Pc repression through transcriptional activation, suggest a dynamic interplay between Pc repression and Trx activation dependent on overlapping recruitment proteins and the relative levels of Pc- and Trx-associated factors (Kwong et al 2008, Schuettengruber et al 2009, Schwartz et al 2010). Interestingly, PRC2 also functionally associates with numerous ncRNAs (Zhao et al 2010). For example, the 2.2 kb long non-coding RNA (lncRNA) HOTAIR serves as a scaffold for both PRC2 and H3K4 demethylase LSD1 complexes, and thereby coordinates targeting of Pc to chromatin while coupling H3K27 trimethylation and H3K4 demethylation activities for epigenetic repression (Tsai et al 2010). Mapping of the genome-wide occupancy of HOTAIR revealed >800 focal, transcription factor-like binding sites that co-occupied the genomic binding profiles for PRC2 subunits EZH2, SUZ12, and H3K27me3. HOTAIR occupancy was maintained upon EZH2 depletion, suggesting HOTAIR actively binds chromatin and may underlie the nucleation of PRC2 domains (Chu et al 2011).
Long-range Interactions
PcG proteins have long been shown to concentrate into nuclear foci called Pc bodies, whose number and size change upon cellular differentiation (Ficz et al 2005, Grimaud et al 2006), suggesting Pc facilitates genome-wide interactions that are further compartmentalized within the nuclear space. Accumulating data suggest that PcG proteins are indeed involved in long-range interactions essential for Pc-repression. Combinatorial use of 3C and fluorescence in situ hybridization (FISH) showed that PRE-PRE interaction occurs between the bxd and Fab-7 elements separated by ~130 kb in the Drosophila bithorax complex (BX-C), and that co-localization occurs specifically in embryonic tissues and cell lines where both the AbdA and Ubx genes are co-repressed (Lanzuolo et al 2007). Long-range associations dependent on PRC2 core component EZH2 at the mammalian GATA-4 locus, which is silenced in undifferentiated human TERA-2 cells, have also been observed (Tiwari et al 2008b). Tiwari et al subsequently devised a 3C-based approach analogous to ChIA-PET, and demonstrated a limited number of intra- and inter-chromosomal interactions in human TERA-2 cells dependent on EZH2, suggesting PRC2 is involved in mediating long-range interactions in mammals (Tiwari et al 2008a).
The formation of Pc bodies and identification of long-range PRE interactions supports a global role in genome function, yet understanding the role of Pc complexes in multi-gene regulation and nuclear organization requires genome-wide exploration of genomic interactions. To this end, van Steensel and colleagues recently adapted Chromosome Conformation Capture on ChIP (4C), a 3C derivative that determines genome-wide interactions of a given locus (Simonis et al 2006), to explore where Pc domains are able to interact within the Drosophila genome (Tolhuis et al 2011). The authors demonstrate long-range interactions between Pc target genes and independent Pc and/or H3K27me3 domains in larval brain tissue, and further show that Pc interactions are topologically constrained to a single chromosome arm. Accordingly, chromosome inversion dramatically altered the interaction profiles revealed by 4C, yet global gene expression patterns were relatively unchanged. Though specific interaction partners were altered, Pc target genes nevertheless continue to interact with independent Pc domains, suggesting that Pc interactions are flexibly amenable to new partners for gene repression. Genome-wide mapping of chromosomal interactions by Sexton et al independently identified 30 significant pairs of long-range interactions between Pc domains, supporting the role of PcG proteins in mediating specific long-range associations (Sexton et al 2012). However, the mechanisms by which these interactions are established and maintained and whether PcG proteins are directly responsible for mediating physical interactions are not clear.
Role in Nuclear Organization
The correlation of long-range chromatin interactions with Pc domains provides compelling evidence in support of the possibility that Pc complexes orchestrate the nuclear organization of repressed genes, but whether PcG proteins are required for long-range interactions has been recently called into question. Co-staining for insulator protein CTCF and PcG protein Polycomb 2 (Pc2) shows clear co-localization of Pc and insulator bodies in HeLa cells (MacPherson et al 2009), and accumulating evidence suggest that insulators underlie the co-localization and nuclear organization of Pc domains (Pirrotta & Li 2011).
The best studied examples of physical interactions involving Pc targets occur between elements within Drosophila Hox gene clusters that have been shown to harbor insulator activity, and genome-wide localization studies have characterized numerous dCTCF insulator sites within the Antennapedia and bithorax complexes (ANT-C and BX-C) (Holohan et al 2007). Only recently have Pirrotta and colleagues shown that interactions between copies of the BX-C Fab-7 or Mcp elements are not dependent on PREs (Li et al 2011). Importantly, both Fab-7 and Mcp elements have been shown to harbor enhancer-blocking insulator activities (Gruzdeva et al 2005, Zhou et al 1996), and Li et al 2011 now demonstrate that interactions depend on insulators flanking the Mcp and Fab-7 PREs. A concurrent study independently revealed that the Su(Hw) insulator is capable of dictating PRE-target interactions through chromatin looping and topological constraint in D. melanogaster (Comet et al 2011), consistent with earlier findings showing that the Su(Hw) insulator facilitates PRE-PRE contacts in trans (Sigrist & Pirrotta 1997). Interestingly, the maintenance of long-range interactions between copies of the Fab-7 element also require components of the RNAi machinery (Grimaud et al 2006), including those required for insulator activity (Lei & Corces 2006), and recent mapping of RNAi component Argonaute2 (AGO2) shows specific co-localization with insulator proteins dCTCF and CP190 throughout the BX-C (Moshkovich et al 2011).
Though insulators likely play an important role in previously described Pc interactions, Li et al speculate that Pc complexes may somehow contribute to the stability of physical interaction (Li et al 2011). In support of this theory, mutations in PcG proteins significantly reduce the level of Antp and Abd-B co-localization at Pc bodies (Bantignies et al 2011). Meanwhile, current models suggest Pc mechanistically facilitates repression through PRC1-mediated chromatin compaction (Grau et al 2011), and mapping of genome accessibility in D. melanogaster reveals that H3K27me3 domains are indeed the most inaccessible (Bell et al 2010). Together, these findings suggest that insulators mediate the interactions between Pc targets, and that Pc repressive complexes likely strengthen the association and repression of Pc domains through histone modifications and chromatin compaction.
The interrogation of physical interactions between distant Hox loci has provided new and important insight into the regulation of long-range interactions during development. In D. melanogaster, the homeotic genes Antp and Abd-B, which are co-repressed and co-localize to Pc bodies in Drosophila embryo heads, are recruited to separate nuclear compartments when one gene becomes activated (Bantignies et al 2011). In mammals, the HoxD cluster forms a single interaction domain in murine embryonic tissues where all genes are inactive, and transitions to a bimodal state in embryonic tissues where HoxD genes are differentially expressed, with active genes segregated into an active domain (Noordermeer et al 2011b). This suggests that developmentally regulated genes are dynamically targeted to specific nuclear subcompartments, such as Pc bodies and transcription factories, for transcriptional repression or activation. The apparent role of insulators in Pc contacts suggests that insulators are likely involved in gene localization to both transcriptionally repressive and transcriptionally permissive environments.
TRANSCRIPTION FACTORIES
The organization of transcription within eukaryotic nuclei is far more complex than traditional textbook models of polymerase recruitment and gene tracking. Instead, transcription is spatially organized into discernable nuclear structures, where multiple RNA polymerases and active genes dynamically localize into nuclear bodies termed “transcription factories”. The formation of transcriptionally active subcompartments presumably allows for more efficient transcription by concentrating the molecular players, reactants, and DNA substrates within a confined nuclear volume. Recent evidence suggests that transcription factories are highly conserved features of nuclear organization, that long-range chromosomal interactions are a hallmark of gene expression, and that insulators likely play an important role at transcription factories.
Composition and Evolution
Transcription is a fundamental cellular process carried out by highly conserved multi-subunit RNA polymerases that share a high degree of homology in bacteria, archaea, and eukaryotes (Werner & Grohmann 2011). RNA polymerase I transcription of ribosomal genes is organized into a strongly conserved and highly organized nuclear substructure called the nucleolus (Thiry & Lafontaine 2005), which represents the classical example of transcriptional clustering into a ‘factory’ structure. Meanwhile, most protein-coding genes are transcribed by RNA polymerase II (RNAPII), and a number of findings now support models proposing RNAPII transcription occurs at analogous factories. The localization of transcription into discrete sites was initially identified by detection of nascent transcripts and by RNAPII staining, which revealed a limited number of foci unable to account for the number of active genes in human nuclei (Iborra et al 1996). Subsequent studies further revealed that transcription factories are large and relatively immobile proteinacious structures whose numbers vary by cell type and nuclear morphology (Chakalova & Fraser 2010), and that active genes dynamically localize to factories for expression in a transcription-dependent manner (Osborne et al 2004). Recent genome-wide interaction assays described in this review also provide supporting evidence for the existence of transcription factories, and further demonstrate that clustering of active genes is a highly conserved phenomenon. Hi-C modeling of chromosomal contacts reveals preferential clustering and interactions among actively transcribed genes and active chromatin domains in S. pombe, Drosophila, and humans (Sexton et al 2012, Tanizawa et al 2010, Yaffe & Tanay 2011).
How transcription factories are physically organized and how genes are dynamically targeted to them remains poorly understood. To this end, Cook and colleagues recently isolated transcription complexes from human nuclei and identified the proteome of RNAPI, II, and III factories by mass spectrometry (Melnik et al 2011). Each complex was shown to harbor a characteristic set of unique proteins, though several proteins involved in DNA or RNA metabolism were shared. Whereas most proteins isolated from RNAPI complexes overlap those characteristic of nucleoli, RNAPII factories contain general transcription factors and CTCF, consistent with the finding that CTCF underlies organization of co-regulated genes in mammals. Interestingly, RNAPII factories also contain repressive histone methyltransferases, including PRC2-core component EZH2, suggesting the transition from epigenetic repression to gene activation may not require the ejection of PcG proteins. In support of this possibility, Polycomb 2 protein (Pc2) was recently shown to relocate from Pc bodies to transcription factories dependent on demethylase KDM4C (Yang et al 2011). Targeting of Pc2 relies on ncRNAs, including TUG1, which interacts with PRC2 and acts as a scaffold at PcG bodies, and NEAT2, which associates with epigenetic regulators involved in gene activation. Taken together, genes are likely directed to repressive Pc bodies or active transcription factories through dynamic interplay of PcG and trxG proteins, whose long-range interactions require the function of ncRNAs and chromatin insulators. This model, recently highlighted by Pirrotta and Li (Pirrotta & Li 2011), is consistent with early findings in D. melanogaster, wherein mutations in both PcG and trxG genes were shown to modulate the activity and nuclear organization mediated by insulators (Gerasimova & Corces 1998).
Enhancer-promoter Interactions
As the name suggests, enhancers are regulatory elements functionally defined by their ability to activate transcription, and do so regardless of their location, distance, or orientation with respect to gene promoters (Banerji et al 1981). Enhancers underlie complex spatiotemporal regulation of tissue-specific gene expression, and have been characterized by chromatin and transcription factor signatures in mammalian cells (Barski et al 2007, Heintzman et al 2007, Wang et al 2008). Enhancers are commonly separated by large genomic distances from their associated promoters, making accurate assignments of enhancer-promoter relationships difficult, and suggesting long-range interactions are a defining feature of gene regulation. Numerous 3C-based interaction studies have supported enhancer looping models wherein enhancers directly contact gene promoters for activation, and recent models suggest chromatin signatures at enhancers may act as epigenetic signals for transcriptional activation (Ong & Corces 2011). Interestingly, a distinct class of enhancer elements are also capable of recruiting RNAPII and are themselves transcribed into enhancer-derived RNAs or ‘eRNAs’ (Kim et al 2010, Wang et al 2011). The transcriptional output of neuronal activity-regulated enhancers positively correlate with the expression levels of associated genes (Kim et al 2010), and alternative models of enhancer function have been proposed (Bulger & Groudine 2011), including ones in which enhancers and associated promoters co-localize by virtue of recruitment to transcription factories.
Enhancer-promoter interactions and transcriptional clustering of active genes consistent with transcription factory models is further supported by ChIA-PET analyses enriching for RNAPII-based chromosomal interactions. (Li et al 2012). As many as 65% of RNAPII binding sites were shown to be involved in a complex network of physical interactions in human cell lines. RNAPII interactions were intergenic (e.g. promoter-promoter), and extragenic (e.g. promoter-enhancer), with most contacts aggregated into ~1,500 interaction complexes. Multigene complexes typically consisted of related and co-regulated genes, suggesting gene families functionally associate for co-transcription, whereas single-gene complexes tended to associate with tissue-specific or developmentally regulated genes and cell-type specific enhancer-promoter interactions. The authors ultimately reveal a complex organization of transcription reflecting the importance of long-range chromatin interactions between co-regulated promoters and between enhancers and promoters, possibly at transcription factories.
Nevertheless, the role of long-range enhancer-promoter interactions in eukaryotic gene activation and how these interactions are organized is not fully understood. Traditional models propose that enhancers underlie recruitment and assembly of the transcription machinery at core promoters (Maston et al 2006). However, in the case of the human FOSL1 gene, histone crosstalk between an enhancer and promoter triggers transcription elongation (Zippo et al 2009), suggesting some enhancers may function by releasing RNAPII from promoter proximal pausing. Meanwhile, recent characterization of chromatin-associated proteins at the human α-globin genes and upstream MCS-R2 enhancer suggest distal enhancers stimulate gene expression by reversing PcG activities (Vernimmen et al 2011). The MCS-R2 enhancer is required for recruitment of H3K27 demethylase JMJD3, though active chromatin marks indicative of Trx activity (H3K4me3) were present in the absence of the enhancer. This supports a model wherein PcG and Trx complexes dynamically associate with target genes, and enhancers promote gene induction by favoring Trx activity. This is consistent with ChIA-PET mapping of RNAPII contacts, which found high enrichment for active chromatin marks H3K4me1 and H3K4me3 coupled with a lack of repressive marks at RNAPII interaction sites (Li et al 2012).
Recent studies have also shed new insight into how enhancers interact with distant target promoters to induce gene transcription, and suggest roles for transcription factors, chromatin insulators, and a unique cohesin complex in enhancer-promoter organization. In the case of the well characterized β-globin locus, whose expression levels are developmentally regulated by a distal upstream locus control region (LCR), long-range enhancer-promoter interactions require transcription factors EKLF and GATA-1 (Drissen et al 2004, Vakoc et al 2005), and genes co-regulated by EKLF preferentially cluster into shared transcription factories (Schoenfelder et al 2010). Ectopically integrated human LCR on a discrete chromosome in transgenic mice preferentially interacts in trans-with EKLF and GATA-1 regulated genes (Noordermeer et al 2011a), suggesting transcription factors coordinate the specificity and organization of enhancer-promoter interactions. Through the powerful combination of 4C and FISH, Noordemeer et al 2011a also demonstrate that inter-chromosomal interactions between the LCR and β-globin genes are limited to specific “jackpot” cells actively transcribing β-globin genes, suggesting interactions are cell-specific and reflect genome conformations that are conducive to enhancer-promoter association. In other words, enhancers preferentially interact with genes through shared transcription factors, but do so stochastically in a restricted nuclear space that varies cell-to-cell.
Insulators also play an important role in facilitating cell-type specific chromatin organization conducive to enhancer-promoter interactions, and recent mapping of CTCF interactions in pluripotent cells supports this possibility (Handoko et al 2011). CTCF-mediated tissue specific chromatin architecture has been characterized at the apolipoprotein gene cluster, the imprinted IGF2-H19 locus, and the developmentally regulated IFNG, β-globin, MHC-II, and CFTR loci (Gillen & Harris 2011, Ong & Corces 2011). However, recent findings suggest that cohesin complexes, which are recruited by CTCF, are also capable of stabilizing enhancer-promoter chromatin looping in the absence of CTCF. For example, cell-type specific enhancer-promoter interactions in murine embryonic stem cells require a unique cohesin complex, including the transcriptional coactivator Mediator and cohesin loading factor nipped B-like protein (NIPBL) (Kagey et al 2010). Cohesin and NIPBL were also recently shown to mediate interactions between the β-globin genes and upstream LCR, and whereas CTCF coordinated organization at the locus does not directly influence gene expression (Splinter et al 2006), the cohesin mediated LCR interaction was shown to regulate globin gene expression in vivo and in vitro (Chien et al 2011).
PERSPECTIVES
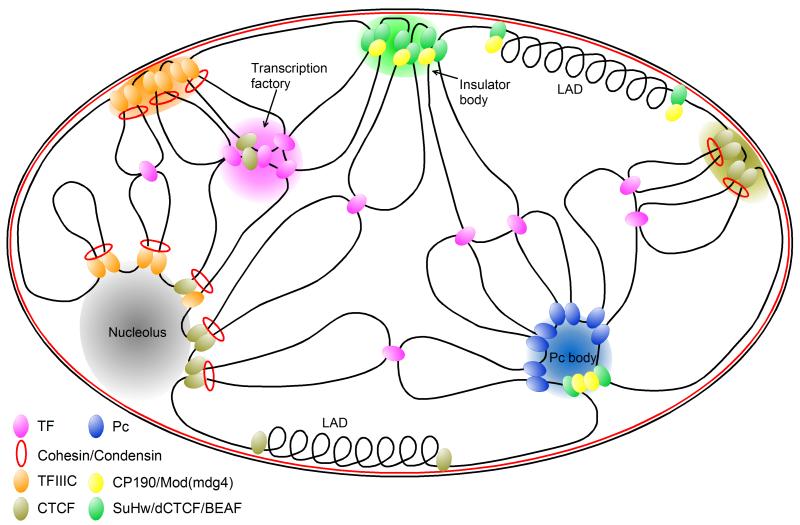
The power of chromatin profiling and 3C-based genomic strategies for exploring genome-wide interactions have led to substantial progress in our understanding of nuclear organization over the past few years. Meanwhile, the recent identification of tDNA insulator activity in humans (Raab et al 2011), mapping of CTCF mediated chromosomal interactions in mammals (Handoko et al 2011), and identification of genome folding principles in human cells (Lieberman-Aiden et al 2009, Yaffe & Tanay 2011) and Drosophila embryos (Sexton et al 2012) have significantly expanded our knowledge of insulator function and their highly conserved role in genome organization. The requirement for insulators in mediating long-range interactions essential for Pc repression (Li et al 2011) and their localization to transcription factories (Melnik et al 2011) suggest insulators underlie the dynamic interplay between epigenetic gene repression and gene induction associated with developmental gene regulation. These observations can be used to derive a comprehensive model for the role of insulators and chromatin structure in nuclear organization (Figure 3), with emphasis on the evolutionary conserved role of insulators in yeast, Drosophila, and mammals, the molecular players involved in each, and their role in gene localization to PcG bodies and transcription factories. Despite substantial progress, several important questions remain, including how chromosomal associations and underlying insulator activities are regulated. The discovery of tDNA insulator activity in mammals raises the question of whether the highly conserved tDNA and CTCF insulators functionally cooperate. Finally, studies have only begun to characterize the new roles for ncRNAs in gene regulation and nuclear organization.
Figure 3.
Comprehensive model for the highly conserved role of insulators in nuclear organization. Insulators in yeast (TFIIIC – orange), Drosophila (dCTCF, Su(Hw), BEAF – green, CP190, Mod(mdg4) – yellow), and mammals (CTCF – olive green, TFIIIC - orange) mediate long-range inter- and intra-chromosomal interactions important for gene regulation, and cluster into subnuclear foci called insulator bodies. Insulators underlie interactions necessary for Pc body repression (blue) and localize with general transcription factors (pink) to transcription factories. Insulators localize to subnuclear structures, including the nuclear lamina (red) where they are enriched at the borders of Lamina-associated domains (LAD), and the nucleolus (grey). CTCF insulator activity in mammals requires cohesin (red), and TFIIIC insulator sites are associated with both cohesin and condensin (red). Insulator activity in Drosophila relies on recruitment of fly-specific proteins CP190 and Mod(mdg4).
Interactions mediated by insulators, PcG proteins and enhancer/promoter-bound factors result in the creation of a three-dimensional arrangement of the DNA that must represent a fingerprint of the functional status of the nucleus. Therefore, a detailed understanding of all inter- and intra-chromosomal interactions in the nucleus together with information on the nature and function of the interacting loci can lead to the establishment of structure-based functional maps of nuclear output that are a representation of cell identity. Some of these interactions may be a consequence of genome function while others may be established during cell differentiation in order to elicit specific patterns of gene expression. As a consequence, the three-dimensional architecture of the genetic material may carry epigenetic information in addition to that written into the 10 nm chromatin fiber. Understanding how this information is maintained during the cell cycle and how the three-dimensional arrangement of chromosomes during interphase relates to their structure during mitosis remains some of the major challenges for the near future.
ACKNOWLEGMENTS
Work in the author’s lab is supported by U.S. Public Health Service Award GM35463 from the National Institutes of Health.
REFERENCES
- Ahmed K, Dehghani H, Rugg-Gunn P, Fussner E, Rossant J, Bazett-Jones DP. Global chromatin architecture reflects pluripotency and lineage commitment in the early mouse embryo. PloS one. 2010;5:e10531. doi: 10.1371/journal.pone.0010531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerji J, Rusconi S, Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27:299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- Bantignies F, Roure V, Comet I, Leblanc B, Schuettengruber B, et al. Polycomb-dependent regulatory contacts between distant Hox loci in Drosophila. Cell. 2011;144:214–26. doi: 10.1016/j.cell.2010.12.026. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bartkuhn M, Straub T, Herold M, Herrmann M, Rathke C, et al. Active promoters and insulators are marked by the centrosomal protein 190. The EMBO journal. 2009;28:877–88. doi: 10.1038/emboj.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck DB, Bonasio R, Kaneko S, Li G, Margueron R, et al. Chromatin in the nuclear landscape. Cold Spring Harbor symposia on quantitative biology. 2010;75:11–22. doi: 10.1101/sqb.2010.75.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisel C, Paro R. Silencing chromatin: comparing modes and mechanisms. Nature reviews. Genetics. 2011;12:123–35. doi: 10.1038/nrg2932. [DOI] [PubMed] [Google Scholar]
- Bell O, Schwaiger M, Oakeley EJ, Lienert F, Beisel C, et al. Accessibility of the Drosophila genome discriminates PcG repression, H4K16 acetylation and replication timing. Nature structural & molecular biology. 2010;17:894–900. doi: 10.1038/nsmb.1825. [DOI] [PubMed] [Google Scholar]
- Berezney R, Dubey DD, Huberman JA. Heterogeneity of eukaryotic replicons, replicon clusters, and replication foci. Chromosoma. 2000;108:471–84. doi: 10.1007/s004120050399. [DOI] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonchuk A, Denisov S, Georgiev P, Maksimenko O. Drosophila BTB/POZ domains of “ttk group” can form multimers and selectively interact with each other. Journal of molecular biology. 2011;412:423–36. doi: 10.1016/j.jmb.2011.07.052. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–53. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes & development. 2006;20:1123–36. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco MR, Pombo A. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS biology. 2006;4:e138. doi: 10.1371/journal.pbio.0040138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulger M, Groudine M. Functional and mechanistic diversity of distal transcription enhancers. Cell. 2011;144:327–39. doi: 10.1016/j.cell.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushey AM, Ramos E, Corces VG. Three subclasses of a Drosophila insulator show distinct and cell type-specific genomic distributions. Genes & development. 2009;23:1338–50. doi: 10.1101/gad.1798209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Molecular cell. 2005;20:845–54. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Carriere L, Graziani S, Alibert O, Ghavi-Helm Y, Boussouar F, et al. Genomic binding of Pol III transcription machinery and relationship with TFIIS transcription factor distribution in mouse embryonic stem cells. Nucleic acids research. 2012;40:270–83. doi: 10.1093/nar/gkr737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker SE, Dillon LA, Gerstein MB, Gunsalus KC, Henikoff S, et al. Unlocking the secrets of the genome. Nature. 2009;459:927–30. doi: 10.1038/459927a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakalova L, Fraser P. Organization of transcription. Cold Spring Harbor perspectives in biology. 2010;2:a000729. doi: 10.1101/cshperspect.a000729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambeyron S, Bickmore WA. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes & development. 2004;18:1119–30. doi: 10.1101/gad.292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien R, Zeng W, Kawauchi S, Bender MA, Santos R, et al. Cohesin mediates chromatin interactions that regulate mammalian beta-globin expression. The Journal of biological chemistry. 2011;286:17870–8. doi: 10.1074/jbc.M110.207365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Molecular cell. 2011;44:667–78. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comet I, Schuettengruber B, Sexton T, Cavalli G. A chromatin insulator driving three-dimensional Polycomb response element (PRE) contacts and Polycomb association with the chromatin fiber. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:2294–9. doi: 10.1073/pnas.1002059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T, Cremer M. Chromosome territories. Cold Spring Harbor perspectives in biology. 2010;2:a003889. doi: 10.1101/cshperspect.a003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome research. 2009;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Ambrosio C, Schmidt CK, Katou Y, Kelly G, Itoh T, et al. Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes & development. 2008;22:2215–27. doi: 10.1101/gad.1675708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–11. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- Donohoe ME, Zhang LF, Xu N, Shi Y, Lee JT. Identification of a Ctcf cofactor, Yy1, for the X chromosome binary switch. Molecular cell. 2007;25:43–56. doi: 10.1016/j.molcel.2006.11.017. [DOI] [PubMed] [Google Scholar]
- Donze D, Adams CR, Rine J, Kamakaka RT. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes & development. 1999;13:698–708. doi: 10.1101/gad.13.6.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donze D, Kamakaka RT. RNA polymerase III and RNA polymerase II promoter complexes are heterochromatin barriers in Saccharomyces cerevisiae. The EMBO journal. 2001;20:520–31. doi: 10.1093/emboj/20.3.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drissen R, Palstra RJ, Gillemans N, Splinter E, Grosveld F, et al. The active spatial organization of the beta-globin locus requires the transcription factor EKLF. Genes & development. 2004;18:2485–90. doi: 10.1101/gad.317004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Z, Andronescu M, Schutz K, McIlwain S, Kim YJ, et al. A three-dimensional model of the yeast genome. Nature. 2010;465:363–7. doi: 10.1038/nature08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersole T, Kim JH, Samoshkin A, Kouprina N, Pavlicek A, et al. tRNA genes protect a reporter gene from epigenetic silencing in mouse cells. Cell Cycle. 2011;10:2779–91. doi: 10.4161/cc.10.16.17092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essafi A, Webb A, Berry RL, Slight J, Burn SF, et al. A wt1-controlled chromatin switching mechanism underpins tissue-specific wnt4 activation and repression. Developmental cell. 2011;21:559–74. doi: 10.1016/j.devcel.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficz G, Heintzmann R, Arndt-Jovin DJ. Polycomb group protein complexes exchange rapidly in living Drosophila. Development. 2005;132:3963–76. doi: 10.1242/dev.01950. [DOI] [PubMed] [Google Scholar]
- Filippova GN, Fagerlie S, Klenova EM, Myers C, Dehner Y, et al. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Molecular and cellular biology. 1996;16:2802–13. doi: 10.1128/mcb.16.6.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser P, Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007;447:413–7. doi: 10.1038/nature05916. [DOI] [PubMed] [Google Scholar]
- Gerasimova TI, Byrd K, Corces VG. A chromatin insulator determines the nuclear localization of DNA. Molecular cell. 2000;6:1025–35. doi: 10.1016/s1097-2765(00)00101-5. [DOI] [PubMed] [Google Scholar]
- Gerasimova TI, Corces VG. Polycomb and trithorax group proteins mediate the function of a chromatin insulator. Cell. 1998;92:511–21. doi: 10.1016/s0092-8674(00)80944-7. [DOI] [PubMed] [Google Scholar]
- Gerasimova TI, Lei EP, Bushey AM, Corces VG. Coordinated control of dCTCF and gypsy chromatin insulators in Drosophila. Molecular cell. 2007;28:761–72. doi: 10.1016/j.molcel.2007.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh D, Gerasimova TI, Corces VG. Interactions between the Su(Hw) and Mod(mdg4) proteins required for gypsy insulator function. The EMBO journal. 2001;20:2518–27. doi: 10.1093/emboj/20.10.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillen AE, Harris A. The role of CTCF in coordinating the expression of single gene loci. Biochemistry and cell biology = Biochimie et biologie cellulaire. 2011;89:489–94. doi: 10.1139/o11-040. [DOI] [PubMed] [Google Scholar]
- Grau DJ, Chapman BA, Garlick JD, Borowsky M, Francis NJ, Kingston RE. Compaction of chromatin by diverse Polycomb group proteins requires localized regions of high charge. Genes & development. 2011;25:2210–21. doi: 10.1101/gad.17288211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaud C, Bantignies F, Pal-Bhadra M, Ghana P, Bhadra U, Cavalli G. RNAi components are required for nuclear clustering of Polycomb group response elements. Cell. 2006;124:957–71. doi: 10.1016/j.cell.2006.01.036. [DOI] [PubMed] [Google Scholar]
- Grosberg A, Nechaev SK, Shakhnovich EI. The role of topological limitations in the kinetics of homopolymer collapse and self-assembly of biopolymers. Biofizika. 1988;33:247–53. [PubMed] [Google Scholar]
- Gruzdeva N, Kyrchanova O, Parshikov A, Kullyev A, Georgiev P. The Mcp element from the bithorax complex contains an insulator that is capable of pairwise interactions and can facilitate enhancer-promoter communication. Molecular and cellular biology. 2005;25:3682–9. doi: 10.1128/MCB.25.9.3682-3689.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–51. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- Gurudatta BV, Corces VG. Chromatin insulators: lessons from the fly. Briefings in functional genomics & proteomics. 2009;8:276–82. doi: 10.1093/bfgp/elp032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, et al. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460:410–3. doi: 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldar D, Kamakaka RT. tRNA genes as chromatin barriers. Nature structural & molecular biology. 2006;13:192–3. doi: 10.1038/nsmb0306-192. [DOI] [PubMed] [Google Scholar]
- Handoko L, Xu H, Li G, Ngan CY, Chew E, et al. CTCF-mediated functional chromatin interactome in pluripotent cells. Nature genetics. 2011;43:630–8. doi: 10.1038/ng.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nature genetics. 2007;39:311–8. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Holohan EE, Kwong C, Adryan B, Bartkuhn M, Herold M, et al. CTCF genomic binding sites in Drosophila and the organisation of the bithorax complex. PLoS genetics. 2007;3:e112. doi: 10.1371/journal.pgen.0030112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iborra FJ, Pombo A, Jackson DA, Cook PR. Active RNA polymerases are localized within discrete transcription ‘factories’ in human nuclei. Journal of cell science. 1996;109(Pt 6):1427–36. doi: 10.1242/jcs.109.6.1427. [DOI] [PubMed] [Google Scholar]
- Ishihara K, Oshimura M, Nakao M. CTCF-dependent chromatin insulator is linked to epigenetic remodeling. Molecular cell. 2006;23:733–42. doi: 10.1016/j.molcel.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–5. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Kang K, Kim J. AEBP2 as a potential targeting protein for Polycomb Repression Complex PRC2. Nucleic acids research. 2009;37:2940–50. doi: 10.1093/nar/gkp149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–45. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–7. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Cecchini KR, Kim TH. Conserved, developmentally regulated mechanism couples chromosomal looping and heterochromatin barrier activity at the homeobox gene A locus. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:7391–6. doi: 10.1073/pnas.1018279108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenova EM, Nicolas RH, Paterson HF, Carne AF, Heath CM, et al. CTCF, a conserved nuclear factor required for optimal transcriptional activity of the chicken c-myc gene, is an 11-Zn-finger protein differentially expressed in multiple forms. Molecular and cellular biology. 1993;13:7612–24. doi: 10.1128/mcb.13.12.7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruhlak MJ, Celeste A, Dellaire G, Fernandez-Capetillo O, Muller WG, et al. Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. The Journal of cell biology. 2006;172:823–34. doi: 10.1083/jcb.200510015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurukuti S, Tiwari VK, Tavoosidana G, Pugacheva E, Murrell A, et al. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:10684–9. doi: 10.1073/pnas.0600326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong C, Adryan B, Bell I, Meadows L, Russell S, et al. Stability and dynamics of polycomb target sites in Drosophila development. PLoS genetics. 2008;4:e1000178. doi: 10.1371/journal.pgen.1000178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzuolo C, Roure V, Dekker J, Bantignies F, Orlando V. Polycomb response elements mediate the formation of chromosome higher-order structures in the bithorax complex. Nature cell biology. 2007;9:1167–74. doi: 10.1038/ncb1637. [DOI] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–13. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei EP, Corces VG. RNA interference machinery influences the nuclear organization of a chromatin insulator. Nature genetics. 2006;38:936–41. doi: 10.1038/ng1850. [DOI] [PubMed] [Google Scholar]
- Li G, Margueron R, Ku M, Chambon P, Bernstein BE, Reinberg D. Jarid2 and PRC2, partners in regulating gene expression. Genes & development. 2010;24:368–80. doi: 10.1101/gad.1886410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HB, Muller M, Bahechar IA, Kyrchanova O, Ohno K, et al. Insulators, not Polycomb response elements, are required for long-range interactions between Polycomb targets in Drosophila melanogaster. Molecular and cellular biology. 2011;31:616–25. doi: 10.1128/MCB.00849-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–93. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Mortensen UH, Rothstein R. Colocalization of multiple DNA double-strand breaks at a single Rad52 repair centre. Nature cell biology. 2003;5:572–7. doi: 10.1038/ncb997. [DOI] [PubMed] [Google Scholar]
- MacPherson MJ, Beatty LG, Zhou W, Du M, Sadowski PD. The CTCF insulator protein is posttranslationally modified by SUMO. Molecular and cellular biology. 2009;29:714–25. doi: 10.1128/MCB.00825-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder P, Boss JM. Cohesin regulates MHC class II genes through interactions with MHC class II insulators. J Immunol. 2011;187:4236–44. doi: 10.4049/jimmunol.1100688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao YS, Zhang B, Spector DL. Biogenesis and function of nuclear bodies. Trends in genetics : TIG. 2011;27:295–306. doi: 10.1016/j.tig.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–9. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maston GA, Evans SK, Green MR. Transcriptional regulatory elements in the human genome. Annual review of genomics and human genetics. 2006;7:29–59. doi: 10.1146/annurev.genom.7.080505.115623. [DOI] [PubMed] [Google Scholar]
- Meister P, Mango SE, Gasser SM. Locking the genome: nuclear organization and cell fate. Current opinion in genetics & development. 2011;21:167–74. doi: 10.1016/j.gde.2011.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekhail K, Moazed D. The nuclear envelope in genome organization, expression and stability. Nature reviews. Molecular cell biology. 2010;11:317–28. doi: 10.1038/nrm2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnik S, Deng B, Papantonis A, Baboo S, Carr IM, Cook PR. The proteomes of transcription factories containing RNA polymerases I, II or III. Nature methods. 2011;8:963–8. doi: 10.1038/nmeth.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–60. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirny LA. The fractal globule as a model of chromatin architecture in the cell. Chromosome research : an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology. 2011;19:37–51. doi: 10.1007/s10577-010-9177-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misulovin Z, Schwartz YB, Li XY, Kahn TG, Gause M, et al. Association of cohesin and Nipped-B with transcriptionally active regions of the Drosophila melanogaster genome. Chromosoma. 2008;117:89–102. doi: 10.1007/s00412-007-0129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon H, Filippova G, Loukinov D, Pugacheva E, Chen Q, et al. CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab-8 insulator. EMBO reports. 2005;6:165–70. doi: 10.1038/sj.embor.7400334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moqtaderi Z, Struhl K. Genome-wide occupancy profile of the RNA polymerase III machinery in Saccharomyces cerevisiae reveals loci with incomplete transcription complexes. Molecular and cellular biology. 2004;24:4118–27. doi: 10.1128/MCB.24.10.4118-4127.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moqtaderi Z, Wang J, Raha D, White RJ, Snyder M, et al. Genomic binding profiles of functionally distinct RNA polymerase III transcription complexes in human cells. Nature structural & molecular biology. 2010;17:635–40. doi: 10.1038/nsmb.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshkovich N, Nisha P, Boyle PJ, Thompson BA, Dale RK, Lei EP. RNAi-independent role for Argonaute2 in CTCF/CP190 chromatin insulator function. Genes & development. 2011;25:1686–701. doi: 10.1101/gad.16651211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai S, Davoodi N, Gasser SM. Nuclear organization in genome stability: SUMO connections. Cell research. 2011;21:474–85. doi: 10.1038/cr.2011.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nativio R, Wendt KS, Ito Y, Huddleston JE, Uribe-Lewis S, et al. Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS genetics. 2009;5:e1000739. doi: 10.1371/journal.pgen.1000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negre N, Brown CD, Shah PK, Kheradpour P, Morrison CA, et al. A comprehensive map of insulator elements for the Drosophila genome. PLoS genetics. 2010;6:e1000814. doi: 10.1371/journal.pgen.1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negre N, Hennetin J, Sun LV, Lavrov S, Bellis M, et al. Chromosomal distribution of PcG proteins during Drosophila development. PLoS biology. 2006;4:e170. doi: 10.1371/journal.pbio.0040170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma K, Cam HP, Maraia RJ, Grewal SI. A role for TFIIIC transcription factor complex in genome organization. Cell. 2006;125:859–72. doi: 10.1016/j.cell.2006.04.028. [DOI] [PubMed] [Google Scholar]
- Noordermeer D, de Wit E, Klous P, van de Werken H, Simonis M, et al. Variegated gene expression caused by cell-specific long-range DNA interactions. Nature cell biology. 2011a;13:944–51. doi: 10.1038/ncb2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordermeer D, Leleu M, Splinter E, Rougemont J, De Laat W, Duboule D. The dynamic architecture of Hox gene clusters. Science. 2011b;334:222–5. doi: 10.1126/science.1207194. [DOI] [PubMed] [Google Scholar]
- Oktaba K, Gutierrez L, Gagneur J, Girardot C, Sengupta AK, et al. Dynamic regulation by polycomb group protein complexes controls pattern formation and the cell cycle in Drosophila. Developmental cell. 2008;15:877–89. doi: 10.1016/j.devcel.2008.10.005. [DOI] [PubMed] [Google Scholar]
- Ong CT, Corces VG. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nature reviews. Genetics. 2011;12:283–93. doi: 10.1038/nrg2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nature genetics. 2004;36:1065–71. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–33. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Paro R. Imprinting a determined state into the chromatin of Drosophila. Trends in genetics : TIG. 1990;6:416–21. doi: 10.1016/0168-9525(90)90303-n. [DOI] [PubMed] [Google Scholar]
- Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SW, Solovei I, et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Molecular cell. 2010;38:603–13. doi: 10.1016/j.molcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickersgill H, Kalverda B, de Wit E, Talhout W, Fornerod M, van Steensel B. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nature genetics. 2006;38:1005–14. doi: 10.1038/ng1852. [DOI] [PubMed] [Google Scholar]
- Pirrotta V, Li HB. A view of nuclear Polycomb bodies. Current opinion in genetics & development. 2011 doi: 10.1016/j.gde.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab JR, Chiu J, Zhu J, Katzman S, Kurukuti S, et al. Human tRNA genes function as chromatin insulators. The EMBO journal. 2011 doi: 10.1038/emboj.2011.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter C, Oktaba K, Steinmann J, Muller J, Knoblich JA. The tumour suppressor L(3)mbt inhibits neuroepithelial proliferation and acts on insulator elements. Nature cell biology. 2011;13:1029–39. doi: 10.1038/ncb2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruben GJ, Kirkland JG, MacDonough T, Chen M, Dubey RN, et al. Nucleoporin mediated nuclear positioning and silencing of HMR. PloS one. 2011;6:e21923. doi: 10.1371/journal.pone.0021923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, et al. CTCF physically links cohesin to chromatin. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:8309–14. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma K, Margueron R, Ivanov A, Pirrotta V, Reinberg D. Ezh2 requires PHF1 to efficiently catalyze H3 lysine 27 trimethylation in vivo. Molecular and cellular biology. 2008;28:2718–31. doi: 10.1128/MCB.02017-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawarkar R, Paro R. Interpretation of developmental signaling at chromatin: the Polycomb perspective. Developmental cell. 2010;19:651–61. doi: 10.1016/j.devcel.2010.10.012. [DOI] [PubMed] [Google Scholar]
- Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, et al. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nature genetics. 2010;42:53–61. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B, Ganapathi M, Leblanc B, Portoso M, Jaschek R, et al. Functional anatomy of polycomb and trithorax chromatin landscapes in Drosophila embryos. PLoS biology. 2009;7:e13. doi: 10.1371/journal.pbio.1000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, et al. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nature genetics. 2006;38:700–5. doi: 10.1038/ng1817. [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Kahn TG, Stenberg P, Ohno K, Bourgon R, Pirrotta V. Alternative epigenetic chromatin states of polycomb target genes. PLoS genetics. 2010;6:e1000805. doi: 10.1371/journal.pgen.1000805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott KC, Merrett SL, Willard HF. A heterochromatin barrier partitions the fission yeast centromere into discrete chromatin domains. Current biology : CB. 2006;16:119–29. doi: 10.1016/j.cub.2005.11.065. [DOI] [PubMed] [Google Scholar]
- Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, et al. Three-Dimensional Folding and Functional Organization Principles of the Drosophila Genome. Cell. 2012 doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- Sigrist CJ, Pirrotta V. Chromatin insulator elements block the silencing of a target gene by the Drosophila polycomb response element (PRE) but allow trans interactions between PREs on different chromosomes. Genetics. 1997;147:209–21. doi: 10.1093/genetics/147.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J, Chiang A, Bender W, Shimell MJ, O’Connor M. Elements of the Drosophila bithorax complex that mediate repression by Polycomb group products. Developmental biology. 1993;158:131–44. doi: 10.1006/dbio.1993.1174. [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nature reviews. Molecular cell biology. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nature genetics. 2006;38:1348–54. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- Song L, Zhang Z, Grasfeder LL, Boyle AP, Giresi PG, et al. Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome research. 2011;21:1757–67. doi: 10.1101/gr.121541.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter E, Heath H, Kooren J, Palstra RJ, Klous P, et al. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes & development. 2006;20:2349–54. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanizawa H, Iwasaki O, Tanaka A, Capizzi JR, Wickramasinghe P, et al. Mapping of long-range associations throughout the fission yeast genome reveals global genome organization linked to transcriptional regulation. Nucleic acids research. 2010;38:8164–77. doi: 10.1093/nar/gkq955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiry M, Lafontaine DL. Birth of a nucleolus: the evolution of nucleolar compartments. Trends in cell biology. 2005;15:194–9. doi: 10.1016/j.tcb.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Thompson M, Haeusler RA, Good PD, Engelke DR. Nucleolar clustering of dispersed tRNA genes. Science. 2003;302:1399–401. doi: 10.1126/science.1089814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari VK, Cope L, McGarvey KM, Ohm JE, Baylin SB. A novel 6C assay uncovers Polycomb-mediated higher order chromatin conformations. Genome research. 2008a;18:1171–9. doi: 10.1101/gr.073452.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari VK, McGarvey KM, Licchesi JD, Ohm JE, Herman JG, et al. PcG proteins, DNA methylation, and gene repression by chromatin looping. PLoS biology. 2008b;6:2911–27. doi: 10.1371/journal.pbio.0060306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B, Blom M, Kerkhoven RM, Pagie L, Teunissen H, et al. Interactions among Polycomb domains are guided by chromosome architecture. PLoS genetics. 2011;7:e1001343. doi: 10.1371/journal.pgen.1001343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B, de Wit E, Muijrers I, Teunissen H, Talhout W, et al. Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nature genetics. 2006;38:694–9. doi: 10.1038/ng1792. [DOI] [PubMed] [Google Scholar]
- Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Molecular cell. 2002;10:1453–65. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- Trojer P, Cao AR, Gao Z, Li Y, Zhang J, et al. L3MBTL2 protein acts in concert with PcG protein-mediated monoubiquitination of H2A to establish a repressive chromatin structure. Molecular cell. 2011;42:438–50. doi: 10.1016/j.molcel.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–93. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakoc CR, Letting DL, Gheldof N, Sawado T, Bender MA, et al. Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Molecular cell. 2005;17:453–62. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- Valenzuela L, Dhillon N, Kamakaka RT. Transcription independent insulation at TFIIIC-dependent insulators. Genetics. 2009;183:131–48. doi: 10.1534/genetics.109.106203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bemmel JG, Pagie L, Braunschweig U, Brugman W, Meuleman W, et al. The insulator protein SU(HW) fine-tunes nuclear lamina interactions of the Drosophila genome. PloS one. 2010;5:e15013. doi: 10.1371/journal.pone.0015013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bortle K, Ramos E, Takenaka N, Yang J, Wahi J, Corces V. Drosophila CTCF synergizes with Su(Hw) and BEAF-32 insulators at the borders of H3K27me3 domains. Submitted. 2012 doi: 10.1101/gr.136788.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastenhouw NL, Zhang Y, Woods IG, Imam F, Regev A, et al. Chromatin signature of embryonic pluripotency is established during genome activation. Nature. 2010;464:922–6. doi: 10.1038/nature08866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernimmen D, Lynch MD, De Gobbi M, Garrick D, Sharpe JA, et al. Polycomb eviction as a new distant enhancer function. Genes & development. 2011;25:1583–8. doi: 10.1101/gad.16985411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Garcia-Bassets I, Benner C, Li W, Su X, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474:390–4. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–8. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nature genetics. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- Werner F, Grohmann D. Evolution of multisubunit RNA polymerases in the three domains of life. Nature reviews. Microbiology. 2011;9:85–98. doi: 10.1038/nrmicro2507. [DOI] [PubMed] [Google Scholar]
- Weth O, Renkawitz R. CTCF function is modulated by neighboring DNA binding factors. Biochemistry and cell biology = Biochimie et biologie cellulaire. 2011;89:459–68. doi: 10.1139/o11-033. [DOI] [PubMed] [Google Scholar]
- Wiblin AE, Cui W, Clark AJ, Bickmore WA. Distinctive nuclear organisation of centromeres and regions involved in pluripotency in human embryonic stem cells. Journal of cell science. 2005;118:3861–8. doi: 10.1242/jcs.02500. [DOI] [PubMed] [Google Scholar]
- Wilkinson FH, Park K, Atchison ML. Polycomb recruitment to DNA in vivo by the YY1 REPO domain. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:19296–301. doi: 10.1073/pnas.0603564103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo CJ, Kharchenko PV, Daheron L, Park PJ, Kingston RE. A region of the human HOXD cluster that confers polycomb-group responsiveness. Cell. 2010;140:99–110. doi: 10.1016/j.cell.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AM, Van Bortle K, Ramos E, Takenaka N, Rohrbaugh M, et al. Regulation of chromatin organization and inducible gene expression by a Drosophila insulator. Molecular cell. 2011;44:29–38. doi: 10.1016/j.molcel.2011.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe E, Tanay A. Probabilistic modeling of Hi-C contact maps eliminates systematic biases to characterize global chromosomal architecture. Nature genetics. 2011;43:1059–65. doi: 10.1038/ng.947. [DOI] [PubMed] [Google Scholar]
- Yang L, Lin C, Liu W, Zhang J, Ohgi KA, et al. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147:773–88. doi: 10.1016/j.cell.2011.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H, Brick K, Evrard Y, Xiao T, Camerini-Otero RD, Felsenfeld G. Mediation of CTCF transcriptional insulation by DEAD-box RNA-binding protein p68 and steroid receptor RNA activator SRA. Genes & development. 2010;24:2543–55. doi: 10.1101/gad.1967810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusufzai TM, Tagami H, Nakatani Y, Felsenfeld G. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Molecular cell. 2004;13:291–8. doi: 10.1016/s1097-2765(04)00029-2. [DOI] [PubMed] [Google Scholar]
- Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, et al. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Molecular cell. 2010;40:939–53. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Barolo S, Szymanski P, Levine M. The Fab-7 element of the bithorax complex attenuates enhancer-promoter interactions in the Drosophila embryo. Genes & development. 1996;10:3195–201. doi: 10.1101/gad.10.24.3195. [DOI] [PubMed] [Google Scholar]
- Zippo A, Serafini R, Rocchigiani M, Pennacchini S, Krepelova A, Oliviero S. Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell. 2009;138:1122–36. doi: 10.1016/j.cell.2009.07.031. [DOI] [PubMed] [Google Scholar]
- Zlatanova J, Caiafa P. CTCF and its protein partners: divide and rule? Journal of cell science. 2009;122:1275–84. doi: 10.1242/jcs.039990. [DOI] [PubMed] [Google Scholar]