Abstract
The degree of applicability of chemogenomic approaches to protein families depends on the accuracy and completeness of pharmacological data and the corresponding level of pharmacological similarity observed among their protein members. The recent public domain availability of pharmacological data for thousands of small molecules on 204 G protein-coupled receptors (GPCRs) provides a firm basis for an in-depth cross-pharmacology analysis of this superfamily. The number of protein targets included in the cross-pharmacology profile of the different GPCRs changes significantly upon varying the ligand similarity and binding affinity criteria. However, with the exception of muscarinic receptors, aminergic GPCRs distinguish themselves from the rest of the members in the family by their remarkably high levels of pharmacological similarity among them. Clusters of non-GPCR targets related by cross-pharmacology with particular GPCRs are identified and the implications for unwanted side-effects, as well as for repurposing opportunities, discussed.
Keywords: GPCR network, ligand similarity, target profile, adverse effects, drug repositioning
Introduction
G protein-coupled receptors (GPCRs) are extremely versatile signaling proteins involved in many physiological processes [1]. Because of that, they are highly relevant in a wide range of therapeutic indications and thus they constitute a target superfamily of utmost importance in drug discovery [2]. Sequence analyses have recognised over 800 GPCRs in the human genome, of which approximately 50% of them are expected to exert their biological function in response to endogenous small molecules [3]. At present, ligands have been identified for the majority of these GPCRs but there remain more than 100 orphan GPCRs for which endogenous ligands have yet to be assigned [4]. Complementing phylogenetic relationships with a deeper understanding of the patterns observed in the interaction profile of small molecules across GPCRs may become a useful deorphanisation strategy [5].
In recent years, the capacity to screen large chemical libraries on multiple GPCRs has increased dramatically and, most importantly, in many cases the bioactivity data generated has become available in the public domain [6]. In addition, several informatics efforts aiming at extracting and storing chemical structures for which pharmacological data have been reported in bibliographic sources have contributed significantly to expand our knowledge base on ligand-GPCR interactions [7]. Collectively, these data have enabled the characterisation of the property space relevant for GPCR activity [8,9] and for identifying the presence of privileged substructures in GPCR ligands [10-12]. This information can be exploited in the design of GPCR-directed chemical libraries with optimal coverage across its members [13]. Furthermore, these data have revealed that active GPCR ligands tend to have exceptionally high levels of target promiscuity, in particular for class A aminergic GPCRs [14-17], making chemogenomic strategies especially adequate to GPCR drug discovery [18-23].
The broad target promiscuity observed for GPCR ligands is a reflection of the inherent similarity among the transmembrane binding cavities of GPCRs [18,22]. The idea that closely related targets will bind similar ligands has led to the concept of pharmacological similarity, commonly referred to as cross-pharmacology [24]. In this respect, GPCRs are recognised to have levels of cross-pharmacology significantly above those observed within other protein families [17]. For example, based on a large set of screening data available from proprietary and commercial sources, Paolini et al. [25] identified among the most intense cross-pharmacology relationships those within and between class A aminergic and peptidic GPCRs. Using a much smaller set of pharmacological data from both public and commercial sources, Gregori-Puigjané and Mestres [15] revealed that aminergic GPCRs had particularly intense pharmacological similarities with opioid receptors but also with two non-GPCR proteins, namely, sigma-1 and NMDA. The identification of cross-pharmacology signals among proteins can be naturally exploited to predict putative novel targets for drugs but also to anticipate potential adverse drug reactions. Under this premise, Keiser et al. [26,27] identified several novel drug-GPCR interactions that were then successfully confirmed experimentally and Garcia-Serna and Mestres [28] used recently comparative pharmacology to predict the likely side-effect profile of GPCR drugs.
The current main limitation of cross-pharmacology analyses of proteins is that coverage of both chemical space and pharmacological data along target space are largely incomplete [29]. As new data are collected, they may modulate the levels of cross-pharmacology perceived previously but may reveal also new cross-pharmacology relationships for GPCRs. The recent explosion of publicly available pharmacological data calls for an up-to-date in-depth cross-pharmacology analysis of GPCRs.
Reference Framework
When performing pharmacological similarity analyses of target families, one should keep in mind that the final perception of the cross-pharmacology between two targets depends on the reference framework used [30]. This comprises three aspects: the number and diversity of ligands for which pharmacological data is known, the molecular descriptors used to represent ligands mathematically and the index to assess their similarity, and the chemical similarity and biological activity criteria set.
Chemogenomic Databases
The construction of chemical libraries annotated with pharmacological data opened an avenue to performing family-wide cross-pharmacology analyses of protein targets. Private initiatives, such as BioPrint [31], Wombat [32], and MDL Drug Data Report (MDDR) [33], were pioneering in this respect. Therefore, most of the cross-pharmacology analyses reported to date were based either on internal proprietary data [25] or on those commercial databases [15, 23-26]. However, recent efforts on delivering publicly available well-crated chemogenomic databases have finally opened cross-pharmacology analyses to the entire scientific community. The list of available public databases is in constant growth and it currently includes GLIDA [34], PDSP [35], BindingDB [36], lUPHARdb [37], PubChem [38], ChEMBLdb [39], hGPCR-lig [40], and DrugBank [41]. The cross-pharmacology analysis of GPCRs performed by Gregori-Puigjané and Mestres [15] made use of BindingDB and the recent work by van der Horst et al. [23] was based on GLIDA, PDSP, and ChEMBLdb. As databases are continuously updated, Table 1 contains a summary of the number of ligands, GPCRs, and ligand-GPCR interaction data collected from the latest releases of seven public sources. A total of 196,074 unique interactions between 93,068 ligands and 204 GPCRs were compiled, of which 180 are human GPCRs sharing at least one bioactive ligand with another GPCR (list provided as Supplementary Material). Among the pharmacological data available, affinity data (pKi) were extracted from five public sources, namely, ChEMBLdb, PDSP, IUPHARdb, BindingDB, and PubChem, and represent 48.3%, 81.9%, and 54.9% of the total number of unique ligands, GPCRs, and interactions, respectively. With the exception of PDSP, the other four sources contain also an important number of additional functional data (pIC50 and pEC50) that together represent an additional 41.7%, 12.6%, and 36.9% of the total number of unique ligands, GPCRs, and interactions, respectively. Finally, two public sources, namely, hGPCR-lig and DrugBank, do not contain quantitative pharmacological data but contribute to expand GPCR space with additional non-numeric bioactivity annotations. It ought to be clarified that if a ligand had different values of the same interaction type for exactly the same target interaction (either within the same database or across databases), an average interaction value was assigned. A systematic analysis of the variations found in compounds with multiple interaction data of the same type for the same target revealed an average standard deviation of ca. 0.5 log units, irrespective of the value range.
Table 1. Number of Ligands Interacting with GPCR Targets According to Bioactivity Data and Annotations Available from Public Sources.
| Database | No. Ligands | No. GPCRs | No. Interactions |
|---|---|---|---|
| Affinity Data (pKi) | |||
|
| |||
| ChEMBLdb | 43,440 | 151 | 98,820 |
| PDSP | 1,667 | 127 | 9,309 |
| IUPHARdb | 514 | 74 | 1,625 |
| BindingDB | 464 | 30 | 855 |
| PubChem | 16 | 2 | 16 |
| Total unique | 44,960 | 167 | 107,638 |
|
| |||
| Additional Functional Data (pIC50, pECSO) | |||
|
| |||
| ChEMBLdb | 38,261 | 166 | 71,405 |
| IUPHARdb | 144 | 46 | 203 |
| BindingDB | 201 | 7 | 259 |
| PubChem | 140 | 7 | 213 |
| Total unique | 38,531 | 172 | 71,800 |
| Cumulative unique | 77,123 | 191 | 170,511 |
|
| |||
| Additional Bioactivity Annotations | |||
|
| |||
| hGPCR-lig | 18,581 | 152 | 29,450 |
| DrugBank | 380 | 97 | 827 |
| Total unique | 18,821 | 167 | 30,053 |
| Cumulative unique | 93,068 | 204 | 196,074 |
Molecular Descriptors
Similarity assessment between ligands requires that chemical structures are encoded using some sort of mathematical descriptors. The choice of a particular type of molecular descriptor may ultimately have a subtle influence on our perception of the cross-pharmacology between targets. Interestingly, recent cross-pharmacology analyses of GPCRs have been performed on essentially different types of molecular descriptors. Hert et al. [24] and Keiser et al. [26] used up to six types of topological fingerprints, including 2048-bit Daylight [42], 988-bit Unity [43], 166-bit MDL keys [44], 1024-bit ECFP4 [45], 1024-bit FCFP4 [45], 1200-bit CATS [46], and one type of three-dimensional structural fingerprint, FEPOPS [47]. In contrast, van der Horst et al. [23] utilised frequencies of substructures present in bioactive ligands [48], whereas Gregori-Puigjané and Mestres [15] employed a reduced set of 10 feature-based topological Shannon entropy descriptors (SHED) [49]. The latter will be applied here in the cross-pharmacology analysis of GPCRs based on the most updated publicly available chemogenomic databases (vide infra). In addition, the (dis)similarity index used on the particular selection of molecular descriptors may have also an effect on the final perception of the cross-pharmacology between targets. In this respect, while Hert et al. [24] and Keiser et al. [26] relied on the use of Tanimoto coefficients to assess ligand similarity, van der Horst et al. [23] applied Pearson correlation coefficients and Gregori-Puigjané and Mestres [15] used a Euclidean distances. The cross-pharmacology analysis of GPCRs presented here (vide infra) will describe ligands with SHED and will use Euclidean distances to assess their (dis)similarity.
Cross-Pharmacology
The number of similar bioactive ligands between two targets determines its level of cross-pharmacology. However, the exact definition of both similar and bioactive needs to be specified, as those criteria will have a direct impact on the strength of the cross-pharmacology signals detected. In this respect, Paolini et al. [25] took the similarity criteria to the limit of identity and used an activity window instead of a threshold to decide whether a compound is shared between two targets only if itself has less than an n log difference in potency. In contrast, Keiser et al. [26] considered that two compounds contribute to the cross-pharmacology of a pair of targets if the similarity between their Daylight fingerprints is above a Tanimoto coefficient of 0.57 and both have at least 10 μM affinity for their respective targets, the same bioactivity cut-off applied also by Gregori-Puigjané and Mestres [15] and van der Horst et al. [23]. However, a systematic study of the effect that similarity and bioactivity criteria have on the cross-pharmacology between targets is still missing.
Cross-Pharmacology Profiles and Scores
The cross-pharmacology profile of a given target is defined here as the list of targets having at least one similar bioactive ligand. For each one of the 180 human GPCRs (provided as Supplementary Material), two cross-pharmacology profiles were derived: one within GPCRs (internal) and another one within non-GPCRs (external). The concept of Shannon entropy [50] is then applied to determine the variability in the number of similar bioactive ligands in a cross-pharmacology profile. Within this approach, the entropy, S, of a total number of similar bioactive ligands, L, shared with a certain number of targets, T, is given by
where pi and li are, respectively, the probability and the number of similar bioactive ligands at each target i of the cross-pharmacology profile. The values of S range between 0, reflecting the situation of all similar bioactive ligands being concentrated in a single target, and a maximum number, Smax = lnT, reflecting the situation of a uniformly distributed population of similar bioactive ligands among multiple targets. In order to have a more intuitive measure that can be linearly related to the situation of full uniform occupancy, entropy values are transformed into projected entropy values, E = es . Correspondingly, E values provide a measure of the expected maximum uniform occupancy from the corresponding S value. Now, for any given ligand population L > 0, the values of E can vary from 1, reflecting the situation of zero entropy in which the population is totally concentrated in a single target, to T, reflecting the situation of maximum entropy in which the population is uniformly distributed among all targets. In the limit case of L = 0, then E will be assigned to E = 0. This E value will be used here as a cross-pharmacology score. The bias, B, in the distribution of the ligand population is given by 1 − E/T.
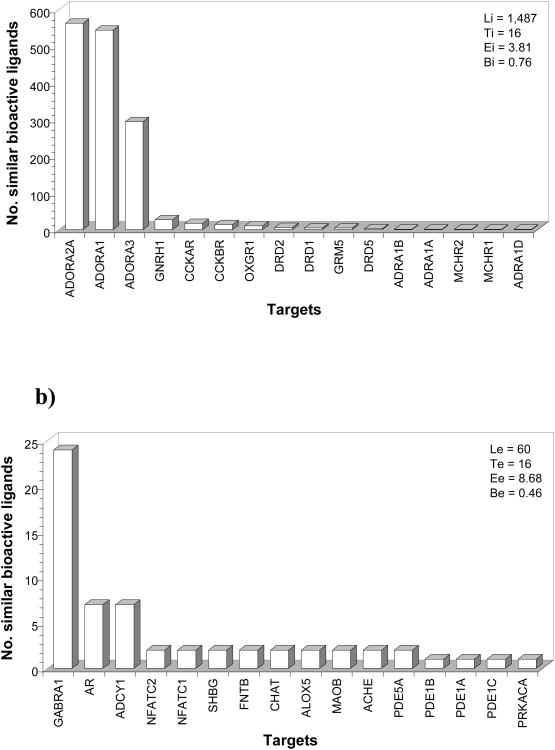
To illustrate the different concepts, Fig. (1) depicts the internal and external cross-pharmacology profiles for the adenosine 2B receptor (ADORA2B) when taking a distance threshold of 0.2 (ligands between two targets are considered similar if their SHED Euclidean distance is less than or equal to 0.2) and a bioactivity threshold of 7.0 (ligands between two targets are considered bioactive if their similar ligands have pKi, pIC50, or pEC50 values larger than or equal to 7.0 for both targets). The internal cross-pharmacology profile Fig. (1a) is composed of 1,487 similar bioactive ligands distributed across 16 GPCR targets. As can be observed, the distribution is highly biased (Bi = 0.76) towards the three other adenosine receptor subtypes, as reflected by a cross-pharmacology score close to 3 (Ei = 3.81). In comparison, the external cross-pharmacology profile Fig. (1b) contains only 60 similar bioactive ligands distributed across 16 non-GPCR targets. Whereas the maximum number of similar bioactive ligands with a GPCR target (adenosine 2A receptor) was 563, the corresponding number with a non-GPCR target (the ion channel Gamma-aminobutyric acid receptor subunit alpha-1) is 24, which gives a good impression of the significantly different degree of internal and external cross-pharmacology. In comparison with the internal profile, the external cross-pharmacology profile is less biased (Be = 0.46) and the population of similar bioactive ligands is more evenly distributed across the targets (Ee = 8.68). This type of analysis was performed for the 180 human GPCR targets covered in this work.
Figure 1.
Cross-pharmacology profiles among (a) GPCR and (b) non-GPCR targets for the adenosine 2B receptor (ADORA2B). All numerical bioactivity data available (pKi, pIC50, and pEC50) were considered and the criteria for shared ligands were set to d ≤ 0.2 and pAct ≥ 7.0. See text for the definition of the different parameters (Li, Ti, Ei, Bi and Le, Te, Ee, Be). Description of gene names is available as Supplementary Material.
Cross-Pharmacology Analysis
In an attempt to address the issue of how different similarity and bioactivity criteria affect the cross-pharmacology of targets, variations in the cross-pharmacology profiles of all GPCR targets were studied through systematic scanning of distance (d) and bioactivity (pAct) criteria. Seven distance criteria were applied. The tightest one (d=0.0) considered cross-pharmacology only when a “descriptor collision” between ligands bioactive to any two targets occurred. Note here that descriptor collision does not strictly correspond to structural identity since some atomic mutations may ultimately lead to exactly the same feature-pair distribution [49]. A distance window of d ≤ t allows for contributing to the cross-pharmacology of targets all bioactive ligands within a Euclidean distance smaller than or equal to a given threshold t. Based on previous validation studies [15], threshold t values ranged from 0.1 to a maximum of 0.6, in intervals of 0.1. Five bioactivity criteria were applied, with bioactivity windows of pAct≥ t, threshold t values ranging from 5 (10 μM) to 9 (1 nM), in intervals of 1 log unit (using the negative log scale). In total, all cross-pharmacology profiles obtained from the thirty-five combinations of distance and bioactivity thresholds were explored.
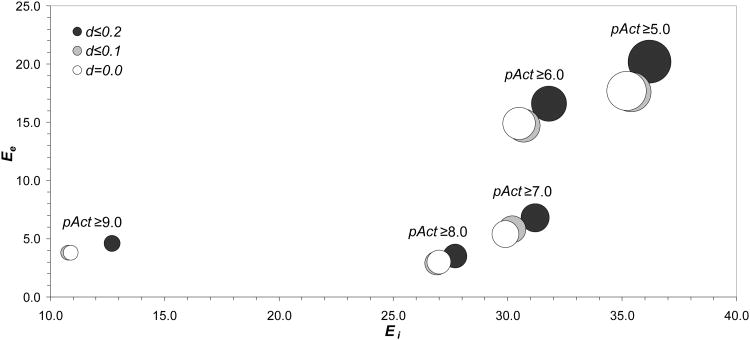
As an illustrative example, changes in the cross-pharmacology perception of the histamine HI receptor (H1R) upon varying distance and bioactivity thresholds are shown in Fig. (2). Two main effects, reproduced in the cross-pharmacology profiles of the other GPCRs, are worth discussing. First, as the bioactivity threshold is set towards higher potency (pAct from 5 to 9), the cross-pharmacology scores (Ei and Ee) decrease significantly, meaning that fewer GPCRs, but also fewer non-GPCR targets, are related to H1R by cross-pharmacology. Notably, the level of cross-pharmacology of H1R with other GPCRs is, under all pAct thresholds, clearly higher than with non-GPCR targets. In this particular case, when the bioactivity threshold changes from pAct ≥ 6.0 to pAct ≥ 7.0, the cross-pharmacology relative to non-GPCR targets is severely reduced, whereas the cross-pharmacology relative to GPCR remains largely unaffected. This is most indicative of the fact that as one moves towards more stringent bioactivity criteria, the probability of hitting any unwanted non-GPCR target decreases significantly. Second, as the distance threshold is enlarged to allow for similar compounds to be considered, both internal and external cross-pharmacology scores increase steadily. As can be observed, the H1R cross-pharmacology profile shrinks from 150 targets for the most relaxed criteria (d ≤ 0.2 and pAct ≥ 5.0) to just 19 targets for the most stringent criteria (d = 0.0 and pAct ≥ 9.0). In this respect, more stringent criteria are likely to highlight potent promiscuous GPCR antagonists for which, more often than not, H1R antagonism is an unwanted side effect (drowsiness), for centrally acting H1R antagonists, rather than the therapeutic effect in itself (antiallergic), for peripherally acting H1R blockers [51].
Figure 2.
Changes in the internal, Ei, and external, Ee, cross-pharmacology scores for the histamine H1 receptor (H1R) upon varying distance and bioactivity thresholds. Size of circles reflect the relative number of targets involved in the cross-pharmacology profile of H1R.
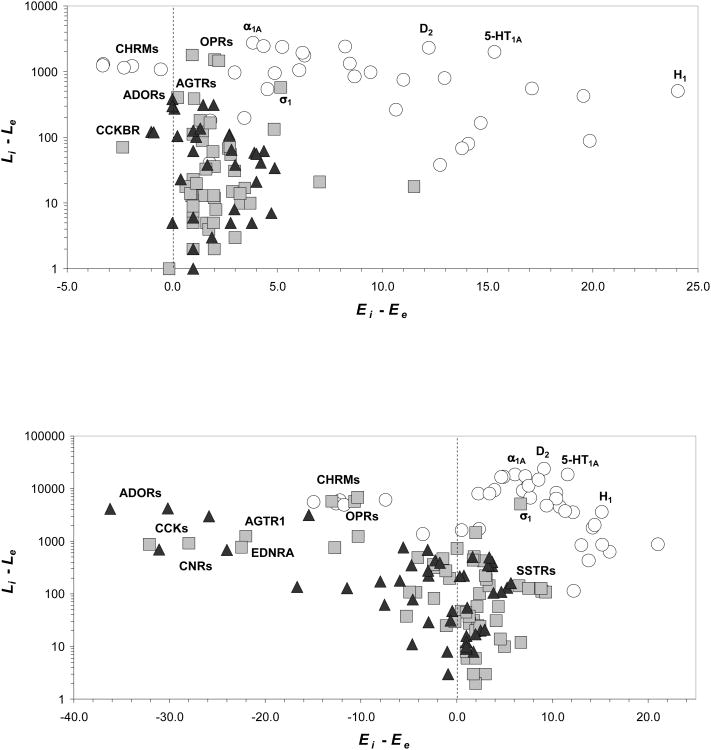
Having explored the effects of the various distance and bioactivity criteria on the final perception of the cross-pharmacology of targets, the relationship between the degree of internal and external cross-pharmacology (as measured by Ei − Ee) and the number of ligands considered to evaluate the respective cross-pharmacology scores (as measured by Li − Le) was revised. The results under two essentially different sets of criteria are illustrated in Fig. (3). Using a set of rather stringent criteria (d = 0.0 and pAct ≥ 8.0), it is observed that class A aminergic GPCRs clearly differentiate from the rest of GPCRs Fig. (3a). On one hand, they show a comparably large bias towards wider cross-pharmacology among GPCRs than among non-GPCR targets, with 57% of class A aminergic GPCRs having Ei − Ee ≥ 5.0 compared to the 5% and 0% of peptidic and other GPCRs, respectively. On the other hand, they also are amongst the ones showing the largest differences in the amount of ligands shared with GPCRs relative to non-GPCR targets, with 65% of class A aminergic GPCRs having Li−Le≥500 compared to the 6% and 0% of peptidic and other GPCRs, respectively. In particular, the histamine H1 receptor is the aminergic GPCR with the largest difference between internal and external cross-pharmacology (Ei − Ee= 24.0) and the adrenoceptor α1A is the one with the largest difference between the amount of ligands shared with GPCRs and non-GPCRs (Li − Le = 2,729). Of mention is the case of muscarinic receptors (CHRMs), the only aminergic GPCRs showing negative values for the difference in cross-pharmacology scores. Close inspection of their cross-pharmacology profiles reveals that, unlike the rest of aminergic GPCRs, this is caused by the relatively low strength of the cross-pharmacology outside their own subfamily, very much in agreement with the results from recent clustering analyses of GPCR binding site sequences [22,23] and the difficulties for obtaining subtype selective compounds via an orthosteric mechanism [52].
Figure 3.
Difference between GPCR and non-GPCR cross-pharmacology, Ei − Ee, versus difference in the number of ligands considered to evaluate the respective cross-pharmacology scores, Li − Le, under two sets of criteria: (a) d = 0.0 and pAct ≥ 8.0 and (b) d ≤ 0.2 and pAct ≥ 6.0. Description of gene names and subfamily abbreviations is available as Supplementary Material, ○: aminergic GPCRs, □: peptidic GPCRs, Δ: other GPCRs.
The use of more relaxed criteria (d ≤ 0.2 and pAct ≥ 6.0) offers a different view of the cross-pharmacology of GPCRs discussed above Fig. (3b). The most significant change is the clear spread by some non-aminergic GPCRs towards larger negative values of the difference between internal and external cross-pharmacology scores, reflecting wider cross-pharmacology profiles for non-GPCR targets. Among them, cholecystokinin (CCKs), adenosine (ADORs) and cannabinoid (CNRs) receptors are the ones being most affected. In all cases, this is due to the fact that, while their external cross-pharmacology has expanded when similar low-potent ligands have been included in the analysis, their internal cross-pharmacology has remained mainly concentrated within the members of each subfamily. Remarkably, inclusion of similar low-potent ligands in the analysis had little effect on the cross-pharmacology of aminergic GPCRs perceived under more stringent criteria Fig. (3a) and, apart from the already noted odd muscarinic receptors, they all keep on appearing well discriminated from the rest of GPCRs in a similar region of the picture.
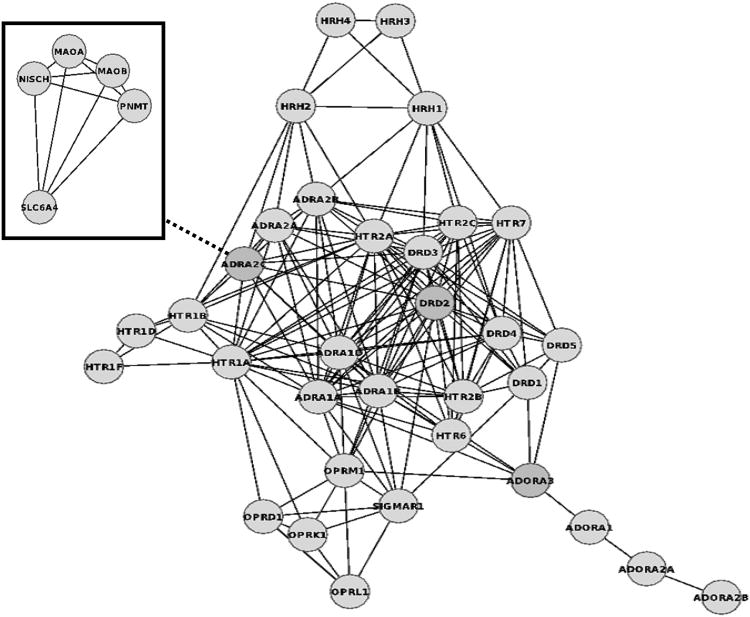
To summarise in a more illustrative manner some of the results obtained from the cross-pharmacology analysis of GPCRs, a cross-pharmacology network was constructed Fig. (4). The use of stringent criteria (d ≤ 0.2 and pAct ≥ 9.0) allowed for focussing on some of the strongest cross-pharmacology relationships identified. The central network in Fig. (4) contains GPCR targets that are linked if they share highly potent similar ligands. As can be observed, this network is composed mainly of class A aminergic GPCRs. Muscarinic receptors are however notably absent in this network, due to their previously noted inbreeding cross-pharmacology. The only non-aminergic GPCRs present in the central network are the sigma-1 receptor and all opioid and adenosine receptors.
Figure 4.
Cross-pharmacology network between GPCR targets and a cluster of non-GPCR targets connected by cross-pharmacology to GPCRs. GPCR targets linked share ligands under the criteria of d ≤ 0.2 and pAct ≥ 9.0; non-GPCR targets linked share ligands under the same criteria with the adrenoceptor α2C (marked with a dashed line). Description of gene names is available as Supplementary Material.
Also added in Fig. (4) is a related network composed solely of non-GPCR targets, namely, sodium-dependent serotonin transporter (SLC6A4, also referred to as SERT or 5HTT), nischarin (NISCH), monoamine oxidases (MAOA and MAOB) and phenylethanolamine N-methyltransferase (PNMT). Targets in this network are linked because they share highly potent similar ligands with GPCR targets and, in particular, they all share ligands with the adrenoceptor α2C. SERT is likely to be linked to α2C due to the cross-link with the sodium-dependent noradrenaline transporter (SLC6A2, also referred to as NET); nischarin appears to be a functional imidazoline I1 receptor [53] and related to α2C antagonists that have an antihypertensive effect (e.g., moxonidine and tolazoline); MAO enzymes are known to have a catabolic role in adrenergic pathways; and PNMT is also acknowledged to have an anabolic role in adrenergic pathways (epinephrine biosynthesis). Therefore, this cluster appears to be organised around ligands with antihypertensive and antidepressive indications, which substantiates the need for stronger scrutiny with respect to unwanted side-effects but also to repurposing opportunities. This provides a good representative example of the potential implications that cross-pharmacology analyses can have for GPCR drug discovery.
Conclusions
Perhaps the main lesson learned from all the pharmacological similarity studies reported thus far is that cross-pharmacology analyses are context-sensitive, as they depend on the amount, quality, and type of pharmacological data available, the molecular descriptors used, and the similarity and bioactivity criteria applied. With respect to data, one ought to consider that in many instances the data available may reflect in part that pharmacological testing of GPCR ligands outside the realm of its own family is more the exception rather than the norm and thus, any conclusions drawn should be taken with caution and balanced with regards to data completeness [29]. With respect to descriptors, one should be warned by the different artifacts that may arise from the use of any type of mathematical representation of ligands and that can partly distort the perception of cross-pharmacology for certain (or all) targets. Finally, with respect to criteria, stringent similarity and bioactivity thresholds offer a highly focused view of cross-pharmacology, often centered in a sub-family of receptors (e.g., class A amine GPCRs). This type of analysis could be used in prophetic patents, in particular when one lacks the resources to perform extended coverage for multiple receptor types. On the other hand, more relaxed criteria (lower similarity and bioactivity thresholds) highlight out-of-target-class potential interactions, which may indeed prove relevant in the context of adverse events or drug repurposing, as well as a potential platform for lead hopping.
Major technical advancements have allowed recently the determination of the first high-resolution X-ray crystal structures of GPCRs and in the not so distant future representative structures of key subfamily members are expected to become available [54]. With this structural information in hand, the different cross-pharmacology relationships between GPCRs observed at present indirectly from pharmacological similarity analysis of bioactive ligands are likely to be rationally explained through comparative analyses of binding site structures [55], opening an avenue for the design of safer, more efficacious, GPCR ligands with customised pharmacological profiles [56].
Supplementary Material
Acknowledgments
This research was funded by the Instituto de Salud Carlos III, the Spanish Ministerio de Ciencia e Innovación (project reference BIO200 8-023 29) and the NIH grant 5U54MH084690-02.
Footnotes
Supplementary Material: Supplementary material is available on the publishers Web site along with the published article
References and Notes
- 1.Kristiansen K. Molecular mechanisms of ligand binding, signaling, and regulation within the superfamily of G protein-coupled receptors: molecular modeling and mutagenesis approaches to receptor structure and function. Pharmacol Ther. 2004;103:21–80. doi: 10.1016/j.pharmthera.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 2.Jacoby E, Bouhelal R, Gerspaher M, Seuwen K. The 7TM G protein-coupled receptor target family. ChemMedChem. 2006;1:760–782. doi: 10.1002/cmdc.200600134. [DOI] [PubMed] [Google Scholar]
- 3.Vassilatis DK, Hohmann JG, Zeng H, Li F, Ranchalis JE, Mortrud MT, Brown A, Rodriguez SS, Weller JR, Wright AC, Bergmann JE, Gaitanaris GA. The G protein-coupled receptor repertoires of human and mouse. Proc Nail Acad Sci USA. 2003;100:4903–4908. doi: 10.1073/pnas.0230374100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chung S, Funakoshi T, Civelli O. Orphan GPCR research. Br J Pharmacol. 2008;153:S339–S346. doi: 10.1038/sj.bjp.0707606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levoye A, Jockers R. Alternative drug discovery approaches for orphan GPCRs. Drug Discov Today. 2008;13:52–58. doi: 10.1016/j.drudis.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 6.Strachan RT, Ferrara G, Roth BL. Screening the receptorome: an efficient approach for drug discovery and target validation. Drug Discov Today. 2006;11:708–716. doi: 10.1016/j.drudis.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 7.Savchuk NP, Balakin KV, Tkachenko SE. Exploring the chemogenomic knowledge space with annotated chemical libraries. Curr Opin Chem Biol. 2004;8:412–417. doi: 10.1016/j.cbpa.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 8.Balakin KV, Tkachenko SE, Lang SA, Okun I, Ivashchenko AA, Savchuk NP. Property-based design of GPCR-targeted library. J Chem Inf Compui Sci. 2002;42:1332–1342. doi: 10.1021/ci025538y. [DOI] [PubMed] [Google Scholar]
- 9.Lowrie JF, Delisle RK, Hobbs DW, Diller DJ. The different strategies for designing GPCR and kinase targeted libraries. Comb Chem High Throughput Screen. 2004;7:495–510. doi: 10.2174/1386207043328625. [DOI] [PubMed] [Google Scholar]
- 10.Klabunde T, Hessler G. Drug design strategies for targeting G protein-coupled receptors. ChemBioChem. 2002;3:928–944. doi: 10.1002/1439-7633(20021004)3:10<928::AID-CBIC928>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 11.Bondensgaard K, Ankersen M, Thogersen H, Hansen BS, Wulff BS, Bywater RP. Recognition of privileged structures by G protein-coupled receptors. J Med Chem. 2004;47:888–899. doi: 10.1021/jm0309452. [DOI] [PubMed] [Google Scholar]
- 12.van der Horst E, Okuno Y, Bender A, Ijzerman AP. Substructure mining of GPCR ligands reveals activity-class specific functional groups in an unbiased manner. J Chem Inf Model. 2009;49:348–360. doi: 10.1021/ci8003896. [DOI] [PubMed] [Google Scholar]
- 13.Gregori-Puigjané E, Mestres J. Coverage and bias in chemical library design. Curr Opin Chem Biol. 2008;12:359–365. doi: 10.1016/j.cbpa.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 14.Roth BL, Sheffler DJ, Kroeze WK. Magic shotguns versus magic bullets: selectively non-selective drugs for mood disorders and schizophrenia. Nat Rev Drug Discov. 2004;3:353–359. doi: 10.1038/nrd1346. [DOI] [PubMed] [Google Scholar]
- 15.Gregori-Puigjané E, Mestres J. A ligand-based approach to mining the chemogenomic space of drugs. Comb Chem High Throughput Screen. 2008;11:669–676. doi: 10.2174/138620708785739952. [DOI] [PubMed] [Google Scholar]
- 16.Peters JU, Schnider P, Mattei P, Kansy M. Pharmacological promiscuity: dependence on compound properties and target specificity in a set of recent Roche compounds. ChemMedChem. 2009;4:680–686. doi: 10.1002/cmdc.200800411. [DOI] [PubMed] [Google Scholar]
- 17.Mestres J, Gregori-Puigjané E, Valverde S, Solé RV. The topology of drug-target interaction networks: implicit dependence on drug properties and target families. Mol BioSys. 2009;5:1051–1057. doi: 10.1039/b905821b. [DOI] [PubMed] [Google Scholar]
- 18.Surgand JS, Rodrigo J, Kellenberger E, Rognan D. A chemogenomic analysis of the transmembrane binding cavity of human G protein-coupled receptors. Proteins. 2006;62:509–538. doi: 10.1002/prot.20768. [DOI] [PubMed] [Google Scholar]
- 19.Jacob L, Hoffmann B, Stoven V, Vert JP. Virtual screening of GPCRs: an in silico chemogenomics approach. BMC Bioinformatics. 2008;9:363. doi: 10.1186/1471-2105-9-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weill N, Rognan D. Development and validation of a novel protein-Iigand fingerprint to mine chemogenomic space: application to G protein-coupled receptors and their ligands. J Chem Inf Model. 2009;49:1049–1062. doi: 10.1021/ci800447g. [DOI] [PubMed] [Google Scholar]
- 21.Heilker R, Wolff M, Tautermann CS, Bieler M. G protein-coupled receptor-focused drug discovery using a target class platform approach. Drug Discov Today. 2009;14:231–240. doi: 10.1016/j.drudis.2008.11.011. [DOI] [PubMed] [Google Scholar]
- 22.Gloriam DE, Foord SM, Blaney FE, Garland SL. Definition of the G protein-coupled receptor transmembrane bundle binding pocket and calculation of receptor similarities for drug design. J Med Chem. 2009;52:4429–4442. doi: 10.1021/jm900319e. [DOI] [PubMed] [Google Scholar]
- 23.van der Horst E, Peironcely JE, Ijzerman AP, Beukers MW, Lane JR, van Vlijmen HWT, Emmerich MTM, Okuno Y, Bender A. A novel chemogenomics analysis of G protein-coupled receptors (GPCRs) and their ligands: a potential strategy for receptor de-orphanization. BMC Bioinformatics. 2010;11:316. doi: 10.1186/1471-2105-11-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hert J, Keiser MJ, Irwin JJ, Oprea TI, Shoichet BK. Quantifying the relationships among drug classes. J Chem Inf Model. 2008;48:755–765. doi: 10.1021/ci8000259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paolini GV, Shapland RHB, van Hoorn WP, Mason JS, Hopkins AL. Global mapping of pharmacological space. Nat Biotechnol. 2006;24:805–815. doi: 10.1038/nbt1228. [DOI] [PubMed] [Google Scholar]
- 26.Keiser MJ, Roth BL, Armbruster BN, Ernsberger P, Irwin JJ, Shoichet BK. Relating protein pharmacology by ligand chemistry. Nat Biotechnol. 2007;25:197–206. doi: 10.1038/nbt1284. [DOI] [PubMed] [Google Scholar]
- 27.Keiser MJ, Setola V, Irwin JJ, Laggner C, Abbas AI, Hufeisen SJ, Jensen NH, Kuijer MB, Matos RC, Tran TB, Wlialey R, Glennon RA, Hert J, Thomas KLH, Edwards DD, Shoichet BK, Roth BL. Predicting new molecular targets for known drugs. Nature. 2009;462:175–182. doi: 10.1038/nature08506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcia-Serna R, Mestres J. Anticipating drug side effects by comparative pharmacology. Expert Opin Drug Metab Toxicol. 2010;6:1253–1263. doi: 10.1517/17425255.2010.509343. [DOI] [PubMed] [Google Scholar]
- 29.Mestres J, Gregori-Puigjané E, Valverde S, Solé RV. Data completeness: the Achilles heel of drug-target networks. Nat Biotechnol. 2008;26:983–984. doi: 10.1038/nbt0908-983. [DOI] [PubMed] [Google Scholar]
- 30.Oprea TI. Chemical space navigation in lead discovery. Curr Opin Chem Biol. 2002;6:384–389. doi: 10.1016/s1367-5931(02)00329-0. [DOI] [PubMed] [Google Scholar]
- 31.Krejsa CM, Horvath D, Rogalski SL, Penzotti JE, Mao B, Barbosa F, Migeon JC. Predicting ADME properties and side effects: the BioPrint approach. Curr Opin Drug Discov Devel. 2003;6:470–480. [PubMed] [Google Scholar]
- 32.Olah M, Mracec M, Ostopovici L, Rad R, Bora A, Hadaruga N, Olah I, Banda M, Simon Z, Oprea TI. WOMBAT: World of Molecular Bioactivity. In: Oprea TI, editor. Chemoinformatics in Drug Discovery. Wiley-VCH; New York: 2004. pp. 223–239. [Google Scholar]
- 33.MDL Information Systems, Inc. MDL Drug Data Report database. http://mdl.com.
- 34.Okuno Y, Tamon A, Yabuuchi H, Niijima S, Minowa Y, Tonomura K, Kunimoto R, Feng C. GLIDA: GPCR ligand database for chemical genomics drug discovery – database and tools update. Nucl Acids Res. 2008;36:D907–D912. doi: 10.1093/nar/gkm948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jensen NH, Roth BL. Massively parallel screening of the receptorome. Comb Chem High Throughput Screen. 2008;11:420–427. doi: 10.2174/138620708784911483. Ki determinations were generously provided by the National Institute of Mental Health's Psychoactive Drug Screening Program, Contract HHSN-271-2008-00025-C. http://pdsp.med.unc.edu. [DOI] [PubMed] [Google Scholar]
- 36.Liu T, Lin Y, Wen X, Jorrisen RN, Gilson MK. BindingDB: a web-accessible database of experimentally determined protein-ligand binding affinities. Nucl Acids Res. 2007;35:D198–D201. doi: 10.1093/nar/gkl999. http://www.bindingdb.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harmar AJ, Hills RA, Rosser EM, Jones M, Buneman OP, Dunbar DR, Greenhill SD, Hale VA, Sharman JL, Bonner TI, Catterall WA, Davenport AP, Delagrange P, Dollery CT, Foord SM, Gutman GA, Laudet V, Neubig RR, Ohlstein EH, Olsen RW, Peters J, Pin JP, Ruffolo RR, Searls DB, Wright MW, Spedding M. IUPHAR-DB: the IUPHAR database of G protein-coupled receptors and ion channels. Nucl Acids Res. 2009;37:D680–D685. doi: 10.1093/nar/gkn728. http://www.iuphar-db.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y, Bolton E, Dracheva S, Karapetyan K, Shoemaker BA, Suzek TO, Wang J, Xiao J, Zhang J, Bryant SH. An overview of the PubChem bioassay resource. Nucl Acids Res. 2010;38:D255–D266. doi: 10.1093/nar/gkp965. http://pubchem.ncbi.nlm.nih.gov/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.ChEMBLdb database. http://www.ebi.ac.uk/chembldb.
- 40.hGPCR-lig database. http://bioinfo-pharma.u-strasbg.fr/hGPCR-Iig.
- 41.Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, Gautam B, Hassanali M. DrugBank: a knowledge base for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36:D901, D906. doi: 10.1093/nar/gkm958. http://www.drugbank.ca/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Daylight, Inc. Daylight Toolkit. http://www.daylight.com.
- 43.Tripos, Inc. Unity. http://www.tripos.com.
- 44.Durant JL, Leland BA, Henry DR, Nourse JG. Reoptimization of MDL keys for use in drug discovery. J Chem Inf Model. 2002;42:1273–1280. doi: 10.1021/ci010132r. [DOI] [PubMed] [Google Scholar]
- 45.Accelrys, Inc. Pipeline Pilot. http://www.accelrys.com.
- 46.Schneider G, Neidhart W, Giller T, Schmid G. Scaffold hopping by topological pharmacophore search: a contribution to virtual screening. Angew Chem Int Ed. 1999;38:2894–2896. [PubMed] [Google Scholar]
- 47.Jenkins JL, Glick M, Davies JW. A 3D similarity method for scaffold hopping from known drugs or natural ligands to new chemotypes. J Med Chem. 2004;47:6144–6159. doi: 10.1021/jm049654z. [DOI] [PubMed] [Google Scholar]
- 48.Wörlein M, Meinl T, Fischer I, Philippsen M. A quantitative comparison of the subgraph miners MoFa, gSpan, FFSM, and Gaston. Knowledge Discov Dalab. 2005:392–403. [Google Scholar]
- 49.Gregori-Puigjané E, Mestres J. SHED: Shannon entropy descriptors from topological feature distributions. J Chem Inf Model. 2006;46:1615–1622. doi: 10.1021/ci0600509. [DOI] [PubMed] [Google Scholar]
- 50.Shannon CE, Weaver W. The Mathematical Theory of Communication. University of Illinois; Urbana: 1949. [Google Scholar]
- 51.Broccatelli F, Carosati E, Cruciani G, Oprea TI. Transporter-mediated efflux influences CNS side effects: ABCB1, from antitarget to target. Mol Inf. 2010;29:16–26. doi: 10.1002/minf.200900075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van Zwieten PA, Doods HN. Muscarinic receptors and drugs in cardiovascular medicine. Cardiovascular Drugs Ther. 1995;9:159–167. doi: 10.1007/BF00877757. [DOI] [PubMed] [Google Scholar]
- 53.Zhang J, Abdel-Rahman AA. Nischarin as a functional imidazoline I1 receptor. FEBS Lett. 2006;580:3070–3074. doi: 10.1016/j.febslet.2006.04.058. [DOI] [PubMed] [Google Scholar]
- 54.Mustafi D, Palczewski K. Topology of class A G protein-couple receptors: insights gained from crystal structures of rhodopsins, adrenergic and adenosine receptors. Mol Pharmacol. 2009;75:1–12. doi: 10.1124/mol.108.051938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Congreve M, Marshall F. The impact of GPCR structures on pharmacology and structure-based drug design. Br J Pharmacol. 2010;159:986–996. doi: 10.1111/j.1476-5381.2009.00476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morphy R, Rankovic Z. Designed multiple ligands: an emerging drug discovery paradigm. J Med Chem. 2005;48:6524–6543. doi: 10.1021/jm058225d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.